Abstract
Metallo-β-lactamases (MβLs) are Zn(II)-based bacterial enzymes that hydrolyze β-lactam antibiotics, hampering their beneficial effects. In the most relevant subclass (B1), X-ray crystallography studies on the enzyme from Bacillus Cereus point to either two zinc ions in two metal sites (the so-called ‘3H’ and ‘DCH’ sites) or a single Zn(II) ion in the 3H site, where the ion is coordinated by Asp120, Cys221 and His263 residues. However, spectroscopic studies on the B1 enzyme from B. Cereus in the mono-zinc form suggested the presence of the Zn(II) ion also in the DCH site, where it is bound to an aspartate, a cysteine, a histidine and a water molecule. A structural model of this enzyme in its DCH mononuclear form, so far lacking, is therefore required for inhibitor design and mechanistic studies. By using force field based and mixed quantum–classical (QM/MM) molecular dynamics (MD) simulations of the protein in aqueous solution we constructed such structural model. The geometry and the H-bond network at the catalytic site of this model, in the free form and in complex with two common β-lactam drugs, is compared with experimental and theoretical findings of CphA and the recently solved crystal structure of new B2 MβL from Serratia fonticola (Sfh-I). These are MβLs from the B2 subclass, which features an experimentally well established mono-zinc form, in which the Zn(II) is located in the DCH site. From our simulations the εεδ and δεδ protomers emerge as possible DCH mono-zinc reactive species, giving a novel contribution to the discussion on the MβL reactivity and to the drug design process.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Metallo-β-lactamases (MβLs) are zinc enzymes capable of hydrolyzing the most widely used class of antibiotics, the β-lactams. The hydrolysis of these drugs renders them ineffective [1]. MβLs are able to inactivate even the latest generation of β-lactam drugs, the carbapenems. This situation is worsened by the absence of clinically useful inhibitors [2, 3]. The large structural diversity among MβLs, classified into subclasses B1, B2 and B3, makes it difficult to identify common structural features [4], which could be exploited for drug design. The B1 subclass is the most relevant. In fact, large part of the enzymes belonging to this subclass are encoded in mobile genetic elements and, therefore, are spread across geographical and species boundaries. The recent rapid worldwide dissemination of the B1 lactamase NDM-1 is the most representative case [5].
The structural diversity within B1 MβLs includes the number of Zn(II) ions that can be bound to the active sites. This issue has given rise to an intense controversy about the essentiality of the different metal binding sites. The first reported X-ray structure of an MβL was that of the B1 enzyme from Bacillus Cereus (BcII) [6]. It showed a single Zn(II) ion coordinated to three histidine residues, and a metal-bound water. Subsequent structural and spectroscopic studies on BcII and other B1 MβLs disclosed that this enzyme can coordinate two Zn(II) ions, one in the 3H site (Fig. 1, pink carbon atoms) and a second one bound to an aspartate, a cysteine and a histidine (DCH site, Fig. 1, green carbon atoms), with a bridging solvent molecule (Wat-Z in Fig. 1) between the two metal ions [7, 8]. This observation, along with the lack of X-ray structures of the mononuclear form with the Zn(II) ion in the DCH site, has led to the suggestion that both the mononuclear 3H form and the binuclear form exist [9] and the metal ion at the 3H site has been suggested to be essential in delivering the attacking nucleophile both in the mononuclear [7, 8] and in the binuclear forms [10].
Metal binding site of the di-zinc BcII MβL X-ray structure (PDB code 3I13[11]). The carbon atoms of the residues are colored in pink and green for the 3H and for the DCH site, respectively
Recently, spectroscopic investigations in the Co(II)-substituted enzyme supported the hypothesis that the mononuclear species with the metal ion only in the DCH site can also be present [12]. The fact that a mononuclear DCH site may be active is further bolstered by the following facts: (1) B2 MβLs are active with only one zinc ion bound to the DCH site [13, 14]; (2) the B3 enzyme GOB is active with one Zn(II) located in a DCH-like site [14, 15]; (3) the only reaction intermediate described so far is an anionic specie, which is stabilized by interaction with the metal ion at the DCH site, as supported by experimental evidences [13, 16–19]; (4) Substrate binding is driven by the metal ions, particularly that at the DCH site [20].
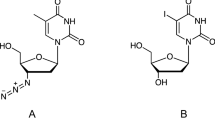
This scenario prompts for structural characterization of the B1 β-lactamase with the metal ion located at the DCH site and for a comparison between the latter and all the proposed metallated forms of the same B1 enzyme (in the free form and in complex with antibiotics), encompassing metal stoichiometry and location as well [21–23]. The absence of experimental structural information can be circumvented by the use of computational methods, as we report here. We have used a variety of computational techniques ranging from QM calculations on small models of the catalytic site to force field (FF) based molecular dynamics (MD) simulations of the entire protein and, finally, to hybrid quantum–classical (QM/MM) MD simulations. These techniques have been employed to explore all possible protonation states of the 3H site and to refine the geometry of the metal coordination site, respectively. The calculations are based on the most recent crystal structure of BcII in complex with two zinc ions, which is at the highest resolution so far (1.74 Å) [11]. Moreover, QM/MM MD calculations allowed us to predict a structural model of the protein in complex with cephalexin and imipenem (Fig. 2), two antibiotics belonging to the two most important classes of β-lactams (cephalosporins and carbapenems) hydrolyzed by BcII. The main goals of this study have been (1) to explore the structural feasibility of the mono-zinc DCH site within the protein framework of BcII; (2) to check its ability to properly bind different β-lactam substrates.
To validate our computational findings, a comparison has been made with the X-ray structures and previous computational findings of the mechanism of CphA as well as with the recently solved crystal structure of the MβL from Serratia fonticola (Sfh-I) [4, 24, 25]. These are B2 MβLs natively active only as mono-zinc DCH form, actually Gln replaces His116 in the 3H sites. To carry out this comparison, among the different mechanisms proposed for CphA, we have selected the one-step hydrolysis mechanism (Figure S1 A of the Supporting Information), recently proposed by some of us [4, 21, 26], which appears as a general path common also to the dinuclear BcII and CcrA [26]. This assumption does not dismiss other proposed enzymatic mechanisms consistent with the available experimental data (see Figure S2) [25, 27].
Methods
All models of the mono-zinc DCH BcII were built by removing the Zn(II) ion from the 3H site present in the X-ray structure of the di-zinc BcII (PDB code: 3I13 [11]).
During this work several models, differing in the protonation state of the 3H site were built and simulated. They are identified with a three letters code where every letter is associated with the protonation state (P = doubly protonated, ε = His protonated on ε position and δ = His protonated on δ position) of His116, His118 and His149, respectively.
QM calculations
DFT calculations were carried out on eight models (εεε, δεδ, δεε, εεδ, Pεε, δPδ, Pεδ, δεP). The models were constructed by removing the zinc ion at the 3H site. They included the water molecule (Wat-Z) bound to the Zn ion, the three His ligands of the 3H site: His116, His118 and His148, and the ligands of the DCH site (Asp120, Cys221, His263) along with Arg121, which is directly involved in the H-bond network at the metal site. All residues were truncated at the β-carbon atom, as in [28–30]. In these models we considered all possible combination of protomers with the exception of: (i) the protomers in which His118 is protonated only on Nδ (His118 Nε H-bonds to Asp213 in all BcII X-ray structures); (ii) the δPε, εPε and the δPε protomers, based on the very short distance between the protons; (iii) the protomers with two or three positively charged histidines at the 3H site, which give rise to a high electrostatic repulsion. This resulted in 8 possible models. These models were geometry optimized at the DFT-B3LYP[31, 32]/6-31G* using the Gaussian03 code and the default setting for the convergence (see SI). The positions of the β-carbon were kept fixed during the calculations to mimic the protein frame [28, 30].
To identify the most stable structure we used the energy criterion as in [28]. This requires the calculation of the energy loss due to dehydration of a proton from the water environment (hydration enthalpy, ΔE(H+)). In [28], the calculations were done using the BLYP [31, 33] exchange–correlation functional, using plane wave (PW) basis set. Here, the same calculation was carried out at the B3LYP [31, 32]/6-31G* level of theory. The calculated ΔE(H+) turns out to be 250 kcal/mol, which is in excellent agreement with the value obtained at the BLYP-PW level of theory [28].
Force field based MD simulations
Twenty-six different protomers (δδδ, δδε, δδP, δεδ, δεP, δPδ, δPε, δPP, εδδ, εδε, εεε, εεP, εPδ, εPε, εPP, Pδε, PδP, Pεε, PPδ, PPε, εεδ, PεP, Pδδ, δεε, εPδ, Pεδ) of the 3H site were considered, resulting from all possible combinations of the three His protonation states.
Loop L3 (from Gly60 to Ala66), which was not detected in the X-ray structure due to its mobility, was modeled as previously reported [22]. The histidines outside the catalytic site were protonated as in previous MD simulations [28]: His55 in ε, His136 in both δ and ε, His285 in ε. All Asp and Glu were assumed to be ionized. All the 210 crystallographic waters present in the PDB file were conserved and included in the structure used as starting point for the MD simulations. All 26 protomers were immersed in a water box whose edges were located 10 Å apart from the closest atom of the protein. The systems contained about 11,000 water molecules. A different number of Cl− ions, depending on the charge of each protomer, was added to system to assure the neutrality.
The PARM99 [34] version of the AMBER force field was used for the protein and the counterions, the general AMBER force field (gaff) [35] was used for the drugs depicted in Fig. 2, whereas the TIP3P [36] model was employed to explicitly represent the water molecules. Atomic charges for the two drugs investigated were obtained using the RESP [37] methodology, fitting an electrostatic potential calculated at the HF/6-31G* level of theory with the software Gaussian03 [38]. As for the metal active site, the force field parameters were obtained following the procedure proposed by Merz et al. [39, 40]. In this approach, the Zn(II) ion was explicitly connected with Asp120, Cys221, His263 trough a covalent bond. The atomic charges for the Zn coordination sphere were taken from previous calculations performed by Simona et al. [24] for an identical Zn binding site. The systems were constructed using the leap module of AMBER11 [41]. Periodic boundary conditions were applied. Long-range electrostatic interactions were assessed by using the particle mesh Ewald method, van der Waals and short-range electrostatic interactions were calculated within a 10 Å cutoff. Bonds involving hydrogen atoms were constrained by using the SHAKE algorithm [42] with a relative geometric tolerance for coordinate resetting of 0.00001 Å. A time-step of 2.0 fs was used. 10000 steps minimization were initially performed for each system. All systems were simulated at constant temperature, Langeving dynamics with a collision frequency of 1 ps−1, and constant pressure using an anisotropic scaling. The water shell and counterions were then equilibrated for 30 ps at 298.15 K, applying weak constraints (20 kcal/mol) on the backbone atoms of the protein. Subsequently, the 26 protomers underwent initially 4.5 ns of MD simulations in isothermal-isobaric ensemble at 298.15 K and 1 atm pressure. After the preliminary set of MD simulations three protomers, δεδ, εPε, εεδ were selected based on structural similarity with the CphA structure (pdb code 1X8G [43]). Then, we employed these structures to study the binding of cephalexin and imipenem. The enzyme/drug complexes were manually built placing the drug inside the catalytic site of the classically equilibrated structure of the enzyme, considering the known binding poses of β-lactam drugs on the active site of BcII [22, 44]. The initial structures of the complexes were minimized for 10000 steps. After this minimization the complex structures were relaxed for 1 ns applying weak harmonic constraints (20 kcal/mol) on the distance between the Zn(II) ion and the nitrogen atom of the β-lactam ring and the distance between the carboxylic group of the drug and the nitrogen atom of the side chain of Lys224. After the equilibration phase, the harmonic restraints were removed, and the complexes between the three selected protomers and the two drugs were simulated for 7 ns under the same isothermal-isobaric conditions. All MD calculations have been carried out with AMBER9 [45] program.
QM/MM MD simulations
The three protomers of the free enzyme selected after the classical MD simulations (δεδ, εεδ, εPε) have been simulated for 3 ps using Born–Oppenheimer QM/MM MD as implemented in the CP2K code [46–48].
CP2K implements a dual Gaussian-type/Plane Waves basis set (GPW) [46], thus a triple z basis set, an auxiliary basis set with a density cut-off of 280 Ry, Goedecker–Teter–Hutter [49, 50] pseudopotentials were used for all the atoms in the QM region. The BLYP exchange–correlation functional was employed [31, 32]. Before starting the production runs all systems were annealed to relax the structures and then the temperature was slowly increased up to 298.5 K. All QM/MM MD simulations have been done in NVT ensemble using a Nosè-Hoover thermostat [51] and a time step of 0.50 fs.
The side chains atoms of His118, His149, His116, Asp120, Cys221, His263 as well as the zinc ion, Wat-H and Wat-Z have been included in the QM region, thus the QM part of the system contained ~60 atoms. The valence of the atoms involved in the bonds crossing the QM and the MM boundaries was saturated by adding hydrogen atoms.
At the end of the QM/MM MD calculations only the δεδ protomer had a geometry and a network of water molecules compatible with the proposed one-step hydrolysis mechanisms [4, 25], therefore imipenem was docked inside the equilibrated structure of this protomer. However, before starting the QM/MM MD simulation the complex was simulated for 1 ns using a FF based MD simulation where considering that, when only FF based potential, without additional position restraints, is used, Wat-H had an high propensity to exchange its position with other molecules from the bulk, then Wat-H and Wat-Z were kept fixed with harmonic restraints (vide supra).Footnote 1 Then, the resulting conformation was relaxed by 4.5 ps of QM/MM MD, with the same computational setup described above. To limit the size of the QM region only the β-lactam ring was included in the QM part. The resulting model comprised ~85 QM atoms.
The εεδ-imipenem complex, which showed a water network compatible with the one-step hydrolysis mechanism at the end of the classical MD run, was also relaxed by 3 ps of QM/MM MD.
Analysis
Representative structures employed to generate the figures were selected by cluster analysis. H-bond and cluster analysis was done with the ptraj module of Amber11 [41, 52]. In the H-bond analysis the distance cut-off was set to 3.5 Å and the angle to 120°. The cluster analysis was done using a Root Mean Square Deviation (RMSD) cutoff of 1.5 Å and only considering the residues forming the catalytic site namely: His116, His118, His149, Asp120, Cys221 and His263 for the free form. For the BcII-drug complexes also the drug was included in the cluster analysis.
Results and discussion
Structural studies of the free protomers
QM calculations on small models of the catalytic site
Small-truncated models of the active site have been considered a fairly good model of the zinc site in MBLs [24, 28, 29, 53]. Thus, to identify the most stable protomers for the mono-Zn(II) DCH form of the BcII enzyme, we initially performed gradient corrected DFT-based energy minimizations on these models (Figure S3). A positive net charge on the 3H site is energetically disadvantaged (Table 1). Thus, only the neutral His protonation forms were subsequently considered.
The neutral protomers differ in energy by 3–6 kcal/mol, which is within the error of the DFT calculations (Table 1). However, this error here may be amplified by the limitation of considering only reduced models of the active site. Thus, the small energetic differences observed do not allow discarding any of the protomers on this ground and we investigated all possible protomers of the mono-zinc DCH form of BcII using also classical MD simulations.
Classical MD simulations
Since the QM calculations on the small active site models did not give unambiguous results and considering the reduced computational costs of classical MD calculations based on empirical force field we performed these simulations for all possible combinations of the 3H site protonation states. The resulting 26 different protomers were relaxed by performing 4.5 ns long MD simulations. The global protein fold of all protomers was preserved over 4.5 ns. The RMSD, calculated over the backbone atoms of all the protein residues during the last 3 ns of simulation, has a mean value of ~1 Å for all protomers. Moreover, the Zn(II)-bound water molecule (Wat-Z) maintained the expected coordination in all protomers.
The structures of the metal binding site were compared with the X-ray structure of CphA (PDB code 1X8G [43]), a B2 MβL that is natively active in a mono-zinc form [43].
The RMSDs of the δεδ, εPε, εεδ protomers result to be significantly lower than all the others (i.e. <0.6 Å). A visual inspection of the structures allowed us to assess that, below an RMSD of 0.6 Å, the protein structure, and in particular the metal site, is not substantially distorted with respect to both the CphA crystal structure and the initial coordinates of our model. On the other hand, in all protomers in which the RMSD is higher the 0.6 Å the geometry of the metal site is significantly distorted. In particular, the Cys221 side chain significantly moves with resect to the plane defined by the three other Zn ligands.
The structures of the three protomers with the lowest RMSD (Fig. 3) were compared with the structure of CphA-biapenem complex as obtained in the theoretical studies of Simona et al. [43] to check if their structure and their network of water molecules in the catalytic site were compatible with the binding of the β-lactam antibiotics.
Catalytic site in the X-Ray structure of CphA in complex with biapenem (A) (PDB code 1X8I [43]). Selected snapshots extracted from the last nanosecond of the MD trajectory of δεδ (B), εεδ (C), εPε (D) protomers. For the sake of clarity, only the side chain atoms and two water molecules are displayed
For all the three protomers (Fig. 3B–D) we identified two water molecules in the catalytic site consistently with previous theoretical findings on CphA [4, 24]. In particular, in the δεδ protomer a water molecule (Wat-H) is located between His116 and Asp120, H-bonding to the oxygen of Asp120 and the Nε of His116. A second water molecule is coordinated to the Zn(II) ion (Wat-Z), replacing the position occupied by the nitrogen of biapenem in the X-ray structure of CphA [43]. A similar water network is present also in εPε and in εεδ, although these protomers are characterized by a different H-bond network around Wat-H. In fact, in εPε His149-Nε-Η H-bonds to O@Wat-H, and in εεδ Wat-H H-bonds with Oδ1@Asp120. However, in this latter case, Wat-H does not stably interact with any His residue and it exchanges its position with a water molecule from the bulk solvent (Table 2).
Spencer and co-workers [54] recently reported the crystal structure of a new B2 MBL from Serratia fonticola (Sfh-I) with an high (60 %) sequence identity with CphA. The catalytic site of Sfh-I is structurally equivalent to that of CphA, but in the crystal structure of Sfh-I (pdb code 3SD9) [54] a water molecule is placed between His118 and Asp120, supporting the one step mechanism of β-lactam hydrolysis proposed by some of us for CphA [4]. Notably, the position of this water is similar to that of Wat-H in the δεδ and εεδ protomers.
The similarity of the hydration state and the geometry of the 3H site of the three selected protomers with experimental and theoretical data of CphA and Sfh-I confirms that these protomers may be a reliable starting point to study the interactions between the mono-zinc DHC BcII and selected β-lactam drugs. Interestingly, εεδ and δεδ were present also among the most likely protomers obtained from the QM calculations, while the εPε protomer was not included in the QM calculations.
QM/MM MD simulations of δεδ, εεδ and εPε protomers
Our FF based MD simulations with the Zn(II) ion bound to its putative coordination residues, may keep the conformation of the metal coordination site frozen to an initial arbitrary structure. QM/MM MD simulations instead may relax the metal binding site, obtaining more accurate and reliable results [55, 56]. Hence, as next step, we performed QM/MM MD simulations on the three δεδ, εεδ and εPε protomers. In these simulations, the Zn(II) coordination sphere is treated at the QM level, whilst the protein frame and the solvent at the MM level.
During the QM/MM MD simulations, the average distances between the coordinating residues and the Zn(II) ion are in the range measured in the X-ray structure of CphA (Table 3 and Fig. 4A–C).
Also in this case we analyzed the water H-bond network of the active site and we compared it with that observed for CphA [4]. In the εPε protomer (Fig. 4C) only a water molecule (Wat-Z) is detected inside the catalytic site, and it remains stably coordinated to the Zn(II) ion for the 3 ps of QM/MM MD. However, Wat-Z does not H-bond with any of the surrounding His. Therefore, this protonation state, lacking a His as generalized base, does not find correspondence with the one-step hydrolysis mechanisms proposed so far [4, 25, 27].
During the QM/MM MD simulation of εεδ (Fig. 4A), Wat-Z H-bonds with Nε@His149 (average distance 2.80 ± 0.10 Å, occupancy 80 %). Moreover, Wat-H, essential to carry out the one-step catalytic reaction as proposed in [4], was not stably present in the active site.
In the QM/MM MD simulation of the δεδ protomer (Fig. 4B), the Zn(II) ion binds to Wat-Z. Wat-H is placed between Oδ1@Asp120 (average distance with H1@Wat-H 2.74 ± 0.12 Å, occupancy 95 %) and Nε@His116 (average distance with H1@Wat-H = 2.76 ± 0.09 Å, occupancy 96 %), similarly to what described by Simona et al. [4] for CphA. In this case, Wat-H does not interact with His118 as in CphA, being this residue quite flexible and solvent exposed, but it interacts with His116, which is buried in the catalytic site.
Concerning this point is worth to note as the spatial proximity of the three His of the 3H site may allow an easy exchange of their roles.
Notably, the superposition of the X-ray structure of Sfh-I (pdb code 3SD9) and a representative structure of the δεδ protomer obtained from the QM/MM MD (Figure S4) showed that the position of the crystallographic water molecule, placed between His118 and Asp120 in Sfh-I, is similar to the position Wat-H in δεδ. In this latter, however, Wat-H interacts with His116, which in Sfh-I is replaced by Asn116. This suggests that the δεδ protomer may be a good candidate to represent the active species.
Structural studies of the β-lactam/protomers complexes
Classical MD simulations
From the above Section, we conclude that the δεδ, εεδ and εPε forms are (i) structurally similar to CphA and Sfh-I and (ii) δεδ, εεδ have also a similar hydration of the catalytic site, (iii) they are consistent with the one-step catalytic hydrolysis proposed for CphA [4, 26].
As further step, we investigated if these species could bind two β-lactam drugs. We cosidered cephalexin and imipenem (Fig. 2), which are representative of two important classes of β-lactams (cephalosporins and carbapenems). To this goal we have initially performed 7 ns classical MD simulations of the three selected protomers in complex with these drugs.
Stable geometries were found for all the three protomers in complex with cephalexin. However, only the εεδ-cephalexin complex showed a water network compatible with the one-step catalytic mechanism proposed for CphA [4, 25] (Fig. 5 and S1). In particular, in εεδ Wat-Z H-bonds with Nε-His149 (average distance 2.91 ± 0.10 Å, occupancy 99 %). Instead, Wat-H does not constantly H-bonds with His118 and Asp120 and it frequently exchanges its position with the bulk solvent. Therefore, we do not favour a one-step mechanism for the hydrolysis of cephalexin in the εεδ protomer.
Instead, considering imipenem as a substrate only the εεδ protomer exhibited a stable enzyme/antibiotic complex (Fig. 6). In this complex, Wat-Z remains stably coordinated to the zinc and Wat-H is placed between Oδ2@Asp120 (average distance 2.72 ± 0.10 Å, occupancy 26.0 %) and Nδ@His118 (average distance 3.09 ± 0.17 Å, occupancy 36.0 %). In this case His118 may activate the Wat-H during the reaction, making this complex compatible with the one-step reaction mechanism proposed on the basis of previous theoretical findings [4, 25, 26] and experimental data [57]. Moreover, the carboxylic group of the drug establishes a salt-bridge with Lys224, with an average distance between the nitrogen atom of Lys224 and the central carbon of the carboxylate of 3.61 ± 0.28 Å.
QM/MM simulations of BcII-imipenem complexes
QM/MM MD calculations are very expensive form the computational point of view. Therefore, considering that BcII displays a better catalytic activity towards imipenem with respect to cephalexin [58], we performed these simulations only considering imipenem as a substrate.
The QM/MM MD simulations were performed only for the εεδ- and δεδ-imipenem adducts, as for these we obtained geometry consistent with the one-step hydrolysis mechanism [4] from classical MD of the enzyme/antibiotic adduct and from the QM/MM MD of the free protomer, respectively.
For the εεδ-imipenem complex, we used the classically equilibrated structure as starting point for the QM/MM MD. Analysing the QM/MM MD trajectory of this adduct (Fig. 7A), we noted that Wat-Z conserves its interaction with the Zn(II) ion (Table 3), while Wat-H forms a stable H-bond with Nε@His149 (average distance with Wat-H 2.75 ± 0.16 Å, occupancy 70 %), and with Oδ1@Asp120 (average distance with Wat-H 2.13 ± 0.19 Å). However, considering the relative orientation and the distance between the O@Wat-H and the C7@imipenem (average distance between O@Wat-H and C7@imipenem 4.82 ± 0.51 Å), also after the QM/MM MD simulations it is still not clear if this adduct may undergo the one-step enzymatic reaction [25]. In fact, this distance is usually shorter in reactive adducts [4, 21].
Conversely, for δεδ-imipenem complex we did not obtain any reasonable binding pose of the substrate from classical MD. However, since the geometry observed at the end of the QM/MM MD simulation of the free protomer was different from the classically equilibrated one and it was compatible with the one-step enzymatic activity, we docked imipenem in this structure and, after a FF based MD simulation (see Methods), we started a QM/MM MD run. During the equilibration of this complex (Fig. 7B), Wat-Z forms a stable H-bond with Nε@His149 (average distance 2.90 ± 0.14 Å, occupancy 100 %) and Wat-H is stably placed between Oδ1@Asp120 (H-bond average distances 2.88 ± 0.16 Å, occupancy 100 %) and Nε@His116 (H-bond average distance 2.88 ± 0.15 Å, occupancy 100 %). Moreover Asp120, initially coordinated with Oδ2, rearranges its coordination and binds with Oδ1, similarly to what observed by Guo and coworkers in their computational studies on CphA [25].
In this case, the average distance between the O@Wat-Z and C7@imipenem is 3.47 ± 0.19 Å. Moreover, the H-bond between Wat-Z and His149 may activate Wat-Z to attack the C7@imipenem carbonyl carbon. Therefore, we can hypothesize a two-steps catalytic mechanism where the nucleophilic attack is performed by Wat-Z activated by His149. Once Wat-Z attacked C7@imipenem, His149 may protonate the N of the β-lactam moiety or Wat-H may replace Wat-Z acting as proton shuttle between His149 and the N of the β-lactam moiety. This mechanism is not consistent with the one-step hydrolysis proposed by some of us [4] and it is also different from the other mechanisms proposed so far for CphA [4, 25, 27, 53] (Figures S1 and S2). However, considering the previous theoretical studies on the MβL reactivity we think that it may be potentially feasible for the DCH-BcII specie.
Conclusions
A combination of FF based and QM/MM MD simulations has been performed to provide a structural model for a putative a mono-zinc form of the BcII enzyme with the metal ion in the DCH site. The feasibility of the predicted models has been investigated (i) by comparing the structural features of our models with that of CphA and of the recently released structure of Sfh-I (ii) by comparing the catalytic site hydration of our models with that of the CphA and (iii) by checking if the monozinc-DCH form of the BcII enzyme may also share the general mechanism of one-step hydrolysis proposed for CphA, BcII and CCrA [4, 21, 26].
Our simulations pointed out that the model of εεδ-imipenem complex (Fig. 7A), although having in the catalytic site the correct number and orientation of the waters to perform the one-step hydrolysis is characterized by a distance between O@Wat-H and the C7@imipenem larger than those typically observed in reactive adducts of other MβL enzymes [4, 26]. Therefore, it is not clear from this structural study if this adduct may be catalytically active.
Instead, the δεδ-imipenem complex has a geometrical arrangement compatible with a catalytic mechanism of β-lactam hydrolysis. This, however, may involve His149, a different histidine with respect to His118. This latter has been demonstrated to play an enzymatic role both in CphA [4] and Sfh-I [54]. Therefore, although the β-lactam hydrolysis may be in principle possible for the δεδ protomer, this is not likely to occur in a single step [25, 53].
It is worth noting that in the analysis of the catalytic reaction feasibility we have considered only those protomers which presented a network of water molecules compatible with the general one-step mechanism of hydrolysis of CphA proposed by some of us [4, 26]. This mechanism, in fact, seems to be a general enzymatic hydrolysis mechanism of β-lactams antibitotics shared also by BcII and CCrA MβLs and it was recently bolstered also by the resolution of the atomic structure of a Sfh-I [54].
However, considering our simulations, we cannot exclude that the mono-Zn DCH BcII species may be active with the multistep hydrolysis mechanism proposed in [25, 53].
The lack of an experimental structure has limited our investigation only to qualitative hypotheses on the reactivity of the DCH-BcII form. However, the results of our calculations represent an interesting starting point to extend the discussion on the MβLs reactivity. Quantitative predictions could be obtained by computational studies only if the monozinc DCH-BcII structure will be experimentally solved.
Notes
A short MD calculation (400 ps) has been run removing the postion restraints on the water molecules after the equilibration, however Wat-H conserves its propensity to exchange its position with other water molecules from the bulk. Then the structure equilibrated with postion restraints was used in the subsequent QM/MM MD.
References
Crowder MW, Spencer J, Vila AJ (2006) Acc Chem Res 39:721–728
Toney JH, Moloughney JG (2004) Curr Opin Investig Drugs 5:823–826
Perez-Llarena FJ, Bou G (2009) Curr Med Chem 16:3740–3765
Simona F, Magistrato A, Dal Peraro M, Cavalli A, Vila AJ, Carloni P (2009) J Biol Chem 284:28164–28171
Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR (2009) Antimicrob Agents Chemother 53:5046–5054
Carfi A, Pares S, Duee E, Galleni M, Duez C, Frere JM, Dideberg O (1995) EMBO J 14:4914–4921
Orellano EG, Girardini JE, Cricco JA, Ceccarelli EA, Vila AJ (1998) Biochemistry 37:10173–10180
Fabiane SM, Sohi MK, Wan T, Payne DJ, Bateson JH, Mitchell T, Sutton BJ (1998) Biochemistry 37:12404–12411
Bebrone C (2007) Biochem Pharmacol 74:1686–1701
Bounaga S, Laws AP, Galleni M, Page MI (1998) Biochem J 331:703–711
Gonzalez JM, Buschiazzo A, Vila AJ (2010) Biochemistry 49:7930–7938
Llarrull LI, Tioni MF, Vila AJ (2008) J Am Chem Soc 130(47):15842–15851
Garau G, Lemaire D, Vernet T, Dideberg O, Di Guilmi AM (2005) J Biol Chem 280:28591–28600
Lisa M-N, Hemmingsen L, Vila AJ (2010) J Biol Chem 285:4570–4577
Moran-Barrio J, Gonzalez JM, Lisa MN, Costello AL, Peraro MD, Carloni P, Bennett B, Tierney DL, Limansky AS, Viale AM, Vila AJ (2007) J Biol Chem 282:18286–18293
Spencer J, Read J, Sessions RB, Howell S, Blackburn GM, Gamblin SJ (2005) J Am Chem Soc 127:14439–14444
Tioni MF, Llarrull LI, Poeylaut-Palena AA, Marti MA, Saggu M, Periyannan GR, Mata EG, Bennett B, Murgida DH, Vila AJ (2008) J Am Chem Soc 130:15852–15863
Gonzalez JM, Medrano Martin FJ, Costello AL, Tierney DL, Vila AJ (2007) J Mol Biol 373:1141–1156
Tomatis PE, Fabiane SM, Simona F, Carloni P, Sutton BJ, Vila AJ (2008) Proc Natl Acad Sci U S A 105:20605–20610
Rasia RM, Vila AJ (2004) J Biol Chem 279:26046–26051
Dal Peraro M, Llarrull LI, Rothlisberger U, Vila AJ, Carloni P (2004) J Am Chem Soc 126:12661–12668
Dal Peraro M, Vila AJ, Carloni P (2004) Proteins 54:412–423
Olsen L, Jost S, Adolph H-W, Pettersson I, Hemmingsen L, Jorgensen FS (2006) Bioorg Med Chem 14:2627–2635
Simona F, Magistrato A, Vera DMA, Garau G, Vila AJ, Carloni P (2007) Proteins 69:595–605
Wu S, Xu D, Guo H (2010) J Am Chem Soc 132:17986–17988
Dal Peraro M, Vila AJ, Carloni V (2010) In: Matta F (ed) Quantum biochemistry. New York, USA, pp 605–622
Xu D, Zhou Y, Xie D, Guo H (2005) J Med Chem 48:6679–6689
Dal Peraro M, Vila AJ, Carloni P (2002) J Biol Inorg Chem 7:704–712
Magistrato A, DeGrado WF, Laio A, Rothlisberger U, VandeVondele J, Klein ML (2003) J Phys Chem B 107:4182–4188
Hong R, Magistrato A, Carloni P (2008) J Chem Theory Comput 4:1745–1756
Becke AD (1988) Phys Rev A 38:3098–3100
Lee C, Yang W, Parr RG (1988) Phys Rev B 37:785–789
Becke AD (1993) J Chem Phys 98:5648–5652
Wang J, Cieplak P, Kollman P (2000) J Comp Chem 21:1049–1074
Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA (2004) J Comput Chem 25:1157–1174
Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein LM (1983) J Chem Phys 79:926–935
Bayly CI, Cieplak P, Cornell WD, Kollman PA (1993) J Phys Chem 97:10269–10280
Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery JA, Vreven T, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Klene M, Li X, Knox JE, Hratchian HP, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, Laham A, Peng CY, Nanayakkara A, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Gonzalez C, Pople JA (2003) Gaussian 03, Revision C.02
Diaz N, Suarez D, Merz KM (2001) J Am Chem Soc 123:9867–9879
Suarez D, Diaz N, Merz KM (2002) J Comput Chem 23(16):1587–1600
Case DA, Darden TA, Cheatham TE III, Simmerling CL, Wang J, Duke RE, Luo R, Walker RC, Zhang W, Merz KM, Roberts BP, Wang B, Hayik S, Roitberg A, Seabra G, Kolossváry I, Wong KF, Paesani F, Vanicek J, Liu J, Wu X, Brozell SR, Steinbrecher T, Gohlke H, Cai Q, Ye X, Wang J, Hsieh M-J, Cui G, Roe DR, Mathews DH, Seetin MG, Sagui C, Babin V, Luchko T, Gusarov S, Kovalenko A, Kollman PA (2010) AMBER 11. University of California, San Francisco
Ryckaert JP, Ciccotti G, Berendsen HJC (1977) J Comp Phys 23:327–341
Garau G, Bebrone C, Anne C, Galleni M, Frere J-M, Dideberg O (2005) J Mol Biol 345:785–795
Suarez D, Merz KM (2001) J Am Chem Soc 123:3759–3770
Case DA, Darden TA, Cheatham TE III, Simmerling CL, Wang J, Duke RE, Luo R, Merz KM, Pearlman DA, Crowley M, Walker RC, Zhang W, Wang B, Hayik S, Roitberg A, Seabra G, Wong KF, Paesani F, Wu X, Brozell S, Tsui V, Gohlke H, Yang L, Tan C, Mongan J, Hornak V, Cui G, Beroza P, Mathews DH, Schafmeister C, Ross WS, Kollman PA (2006) AMBER 9, University of California. University of California, San Francisco, San Francisco
VandeVondele J, Krack M, Mohamed F, Parrinello M, Chassain T, Hutter J (2005) Comput Phys Commun 167:103–128
Laino T, Mohamed F, Laio A, Parrinello M (2005) J Chem Theory Comput 1:1176–1184
Laino T, Mohamed F, Laio A, Parrinello M (2006) J Chem Theory Comput 2:1370–1378
Goedecker S, Teter M, Hutter J (1996) Phys Rev B 54:1703–1710
Hartwigsen C, Goedecker S, Hutter J (1998) Phys Rev B 58:3641–3662
Hoover WG (1985) Phys Rev A 31:1695–1697
Shao J, Tanner SW, Thompson N, Cheatham TE (2007) J Chem Theory Comput 3:2312–2334
Xu D, Xie D, Guo H (2006) J Biol Chem 281:8740–8747
Fonseca F, Bromley EHC, Saavedra MJ, Correia A, Spencer J (2011) J Mol Biol 411:951–959
Spiegel K, Magistrato A (2006) Org Biomol Chem 4:2507–2517
Sgrignani J, Magistrato A (2012) J Phys Chem B 116:2259–2268
Llarrull LI, Fabiane SM, Kowalski JM, Bennett B, Sutton BJ, Vila AJ (2007) J Biol Chem 282:18276–18285
Tomatis PE, Rasia RM, Segovia L, Vila AJ (2005) Proc Natl Acad Sci USA 102:13761–13766
Acknowledgments
This work has been supported in part by the FIRB project Contract RBLA032ZM7, by the EC contract SPINE II n° 031220 and by grants from HHMI (Howard Hughes Medical Instituteand ANPCyT (Agencia Nacional de Promoción Científica y Tecnológica) to AJV. AJV is a fellow of the John Simon Guggenheim Foundation and an HHMI International Research Scholar. Access to the computational resources supplied by CASPUR and by CINECA is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Paolo Carloni: Joint venture of RWTH Aachen University and Forschungszentrum Jülich, Germany.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sgrignani, J., Magistrato, A., Dal Peraro, M. et al. On the active site of mononuclear B1 metallo β-lactamases: a computational study. J Comput Aided Mol Des 26, 425–435 (2012). https://doi.org/10.1007/s10822-012-9571-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-012-9571-0