Abstract
Class B1 metallo-β-lactamases (MBLs) are metalloenzymes found in drug resistant bacteria. The enzyme requires zinc ions, along with conserved amino acid coordination for nucleophilic attack of the lactam ring to induce hydrolysis and inactivation of β-lactam and some carbapenem antibiotics. To this date there are no clinically relevant class B1 MBL inhibitors, however L-captopril has shown significant results against NDM-1, the most difficult MBL to inhibit. Herein, we report the synthesis and evaluation of novel nucleoside analogues modified with polyethylene glycolamino (PEGA) as potential inhibitors for class B1 MBLs. Molecular dynamics simulations, using internal coordinate mechanics (ICM) algorithm, were performed on subclass B1 enzyme complex models screened with twenty-one possible PEGA-nucleosides. Analogue A, 3′-deoxy-3′-(2-(2-hydroxyethoxy)ethanamino)-β-D-xylofuranosyluracil showed superior binding, with high specificity to the conserved zinc ions in the class B1 MBL active site by utilizing key β-lactam mimic points in the uridine nucleobase. The PEGA moiety showed chelating activity with zinc and disrupted the metal-binding amino acid geometry. In all subclass B1 proteins tested, analogue A had the most effective inhibition when compared to penicillin or L-captopril. Chemical synthesis was performed by condensation of the corresponding keto ribonucleoside with PEGA, followed by enantioselective reduction of the formed imine to produce the amino derivative with desired configuration. Pharmacokinetic and pharmacodynamic screenings revealed that PEGA-pyrimidine nucleosides are not toxic, nor violate Lipinski’s rules. These results suggested that analogue A can be proposed as a potential metalloenzyme inhibitor against the widespread antibiotic resistant bacteria and is worth further in vitro and in vivo investigations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Recent warnings from the World Health Organization state that the rapid spread of drug resistant bacteria across the world will cause a projected 10 million deaths per year by 2050 (Dixit et al. 2019). Bacteria regularly mutate to develop resistance, which indicates the crucial need for new antimicrobial agents (Zhang et al. 2017). Antibiotics such as carbapenems, a class of β-lactams, are often used as a last resort in treating infections, however with the emergence of multi-drug resistant bacteria and the repeated exploitation of antibiotics this last line of defense is now under attack (Chandar et al. 2018; Zhang et al. 2018; Snyder et al. 2019). β-Lactam antibiotics have been rendered inactive due to the rapid spread of class B metallo-β-lactamases, di-zinc enzymes that catalyze hydrolysis of the β-lactam ring (Docquier and Mangani 2018). Subclass B1 MBLs consist of New Delhi Metallo-β-lactamase-1(NDM-1), Verona Integrin-encoded Metallo-β-lactamase 2 (VIM-2), and Imipenemase Metallo-β-lactamase 1 (IMP-1) (Ju et al. 2018). These enzymes are characterized by a unique αβ/βα sandwich fold and have become the most relevant “superbugs” due to the lack of clinically relevant inhibitors (Mojica et al. 2016; Brem et al. 2016). MBL’s unique broad substrate profiles are characterized by flexible loops towering over two zinc ions, with the first zinc ion coordinated in tetrahedral geometry to three histidine residues and the second zinc ion configured in trigonal bipyramidal geometry with histidine, cystine and aspartic acid (Antony et al. 2002; Shen et al. 2013; González et al. 2016; Tehrani et al. 2020). The unique active site structure, water bridge hydrolysis mechanism, and di-zinc coordination have allowed subclass B1 MBLs to accommodate for inactivation of a variety of substrates (Shen et al. 2013). The B1 MBL’s ability to execute specific interactions targeting β-lactam core ring has suggested that inhibitors with capability to mimic the β-lactam core would increase their binding affinity (Thomas et al. 2019).
Nucleosides are fundamental molecules found in metabolic processes of all living organisms, where they display antibacterial, antiviral, immunostimulative, immunosuppressive and anticancer properties (Niu and Tan 2015). Nucleoside analogues such as 3′-azidothymidine (AZT) and idoxuridine (Fig. 1) have been reported as effective therapeutics against K. pneumonae, the same strain responsible for the rapid spread of MBLs (Thomson and Lamont 2019; Langley et al. 2019). Tunicamycins and polyoxins are also uracil based nucleosides that have been used as antibacterial agents due to their ability to mimic essential substrates in bacterial enzyme survival, with no observed human toxicity (Serpi et al. 2016).
To exploit their structure similarity to β-lactam antibiotics, herein we report the evaluation and synthesis of novel nucleoside analogues as B1 MBLs inhibitors. Uridine and adenosine nucleosides were both examined for their binding affinity towards MBL enzymes, as models of either pyrimidine or purine nitrogenous base, respectively. Polyethylene glycolamino (PEGA) modified nucleosides with the modification introduced at the 2′, 3′, or 5′ positions were assessed through molecular simulations, aiming to identify analogues with effective active site binding along with low toxicity, and optimal solubility without violation of Lipinski’s rules (Benet et al. 2016). The proposed PEGA moiety was hypothesized to enhance binding affinity and interfere with the function of B1 MBLs by chelating and disrupting conserved zinc ion interactions. In-silico molecular modeling, using internal coordinate mechanics (ICM) algorithm (Neves et al. 2012), was performed and lead-like inhibitors with stronger B1 MBL binding affinity were predicted. Virtual ligand screening (VLS) was applied based on its ability to account for active site structure flexibility with comparably high accuracy (Bursulaya et al. 2003; Wang et al. 2015; Daina et al. 2017). ProToxII and SwissADME were used to predict pharmacological properties of selected inhibitors (Perola et al. 2004; Daina et al. 2017). Nucleoside analogues displayed strong MBL substrate binding affinity and zinc ion coordination with favorable pharmacokinetic and pharmacodynamic properties were identified, and the top PEGA analogue was synthesized to establish a protocol for the chemical synthesis of the proposed structures and for further in vitro and vivo investigations as a potential metalloenzyme inhibitor.
Materials and methods
Molecular docking study and nucleosides tested
Internal coordinate mechanics molecular modeling package was used to investigate the electrostatic sphere which includes: bond lengths, bond angles, torsion angles, and phase dihedral angles between PEGA-nucleosides and the B1 MBL’s active site (Abagyan et al. 1994; Fernández-Recio et al. 2003). Each enzyme structure was retrieved using the Protein Data Bank for efficient local minimization of polar amino acids to produce more accurate long range electrostatic interactions (Abagyan et al. 1994; Lateef et al. 2017). Monte Carlo global energy minimization procedure was assured by running three independent runs per compound. Nucleosides screened were both adenosine and uridine to compare the effect of the nitrogenous base size and lactam mimicking core interactions with the MBL active site (Katritch et al. 2010). The analogues included PEGA modifications at the 2′, 3′ or 5′ positions; where 2′ and 3′ were explored in both xylo and arabinofuranoside forms. Two modifications were examined to produce a total of twenty nucleosides resulting from condensation of the keto nucleoside with either ethanolamine or 2-(2-aminoethoxy)ethanol. The effect of the side chain length on the substrate binding was further investigated by testing an additional analogue modified with 2-[2-(2-aminoethoxy)ethoxy]ethanol at its 3′ position. Analogues displaying the most favorable binding energy for each of the three B1 MBLs, A among the pyrimidine derivatives, B among the purine derivatives, and the pyrimidine derivative with longer side chain C (Fig. 2) were analyzed for key active site interactions and for disruption of the di-zinc water bridge. Their interactions were evaluated to determine those that maintain a high level of binding affinity to the active site of all three B1 MBL proteins, when compared to the references penicillin and L-captopril (Li et al. 2014; Mbaye et al. 2019).
Conformational methods and generation of the protein model
Each crystal structure was retrieved and prepared through the PDB using the molecular conversion procedure executed by ICM docking. To adequately prepare the structures for simulation, to the PDB receptors (NDM-1, PDB ID: 4EXS; IMP-1, PDB ID: 5EV6; VIM-2 PDB ID: 4BZ3) were added hydrogen atoms to ensure that all Zn2+ water bridge interactions were accounted for during molecular simulation. His, Pro, Asn, Gln, and Cys were added, disulfide bonds were integrated, and the best hydrogen energy conformation of three possible rotameric states were included to preserve the covalent geometry in the ICM object derived from the PDB crystal structure (Abagyan et al. 1994). The models were then monomerized to chain A where chain B was deleted to reduce redundancy in the crystal structure.
Selection of ligand binding pockets
The NDM-1 active site was established independent of L-captopril through direct removal from the structure’s pocket. ICM PocketFinder allowed for calculation of electrostatic, hydrophobicity, hydrogen binding, and Van der Waals maps of the enzyme binding site without limitations created from ligands previously bound to the crystal structure (Sheridan et al. 2010; Lam et al. 2018). Charge depths in electrostatic energy calculations produced a grid of points, the electrostatic sphere, which filled the concavity beginning at the pocket surface of the binding site with a tolerance of 4.6 (Abagyan et al. 1994). Tolerance was used to identify the threshold for the search in pockets, the lower the tolerance the shallower the enzyme substrate binding pocket. Volume, area, buridness and hydrophobicity of the pockets were accounted for and included with drug-like density (DLID) in the selection of pocket 1. A selection grid of 18 to 20 mols, a water radius of 1.4 Å, and a hydrogen cut off distance of 3.0 Å from the electrostatic sphere was implemented (Abagyan et al. 1994). VIM-2 and IMP-1, having no bound inhibitor at the time of crystallization, were prepared using ICM PocketFinder following extraction from PDB and conversion to ICM objects. The 3D structure of the pocket receptor was retrieved for all proteins. The selection of pockets for VIM-2 and IMP-1, were generated upon the same standards as in NDM-1. Optimal binding pockets for all B1 MBL proteins docked were also selected based upon proximity to the di-zinc active site.
Virtual ligand screening
Virtual ligand screening through MolSoft ICM 3.8 was used to test the ligand binding energy. A 3D grid of energy potential maps were calculated using Van der Waals, H-bonding, hydrophobic and electrostatic interactions between B1 MBLs and the nucleoside analogues. The ligands and receptors were accounted for flexibility in the calculated energy potential grid, where the derivatives were independently re-docked three times each (Katritch et al. 2010). VLS was conducted on all proteins following identification of the binding pocket. All ligands were each loaded onto the ligand batch after being imported from Chem Draw. Following VLS, refinement was run on both the ligand and the enzyme to allow for full calculations of translational and rotational degrees of freedom during simulation (Arnautova et al. 2018). Subsequently, the most favorable interaction for each nucleoside analogue per protein were recorded in kJ/mol for comparison and selection. Pyrimidine and purine analogues with binding energy closest to − 30 kJ/mol were considered to display enhanced active site binding affinity to B1 MBL enzymes. To further validate the ICM docking methods and scoring functions, the pose selection was implemented where ICM was used to re-dock L-captopril to NMD-1. The docking result was then superimposed to the L-captopril/NDM-1 structure co-crystallized in-vitro and the root mean square deviation (RMSD) value was calculated. It is known that a program able to give RMSD value below 2 Å are considered to perform successfully (Hevener et al. 2009; Ramírez and Caballero 2018). The returned RMSD value was 1.18 Å indicating good docking solution, thus validate the ability of ICM to accurately predict scoring function that reproduced the crystallographic binding orientation.
Pharmacokinetic and pharmacodynamic screening
Drug likeness of analogues A, B and C were executed by SwissADME, where the analogue with superior oral bioavailability was chosen based on Lipinski’s rule of five. The LD50, acute toxicity, hepatotoxicity, carcinogenicity, cytotoxicity, mutagenicicty, and toxicity were assessed using ProTox-II (Banerjee et al. 2018). Each platform returned a suite of information that provided a profile for each compound to determine its potential as a viable and non-toxic drug (Daina et al. 2017). The analogue with the most favorable profile was chosen to establish a protocol for the chemical synthesis as well as for future in-vivo and in-vitro investigations.
Chemical synthesis
To establish a synthetic protocol for the proposed PEGA nucleosides, here we described a method towards the synthesis of analogue A (Scheme 1). The 2′ and 5′ hydroxyl groups of uridine was protected utilizing TBDMS protective group, and the 3′ alcohol was oxidized to produce the corresponding keto nucleoside (I) according to the previously reported method (Utley et al. 2018). The synthesis of the 2′,5′-TBDMS protected PEGA nucleoside (III) involved a stereoselective reduction of the 3′-imine moiety. Initially, the 3′-keto uridine was converted into the 3′-imine derivative (II) by treatment with 2-(2-aminoethoxy)ethanol in glacial acetic acid. The subsequent conversion of keto nucleosides into oxime derivatives by treatment with hydroxylamine hydrochloride was previously reported (Kojima et al. 2002), a method which was adapted for the imine formation in this study. The stereoselective reduction of the 3′-diethylene glycolimino group to 3′-diethylene glycolamino by using Corey-Bakshi-Shibata Oxazaborolidine (CBS) catalyst produced the desired 3′-2-(2-hydroxyethoxy)ethanamino-β-D-xylofuranosyl configuration III in 87% yield. CBS oxazaborolidine has been reported as a chiral catalyst that mediates the borane reduction of ketones to secondary alcohols with excellent enantioselestivity (Corey et al. 1987). The TBDMS protective groups can be easily removed with tetrabutylammonium fluoride in THF to afford the unprotected analogue A according to the standard procedure (Utley et al. 2018).
1H and 13C NMR spectra were recorded on Varian-500 instrument (500 MHz 1H and 125 MHz 13C) using CDCl3 or DMSO-d6 as a solvent. Chemical shifts are reported in parts per million (δ), and signals are expressed as s (singlet), d (doublet), t (triplet), q (quartet), m (multiplet), and coupling constants (J) are given in Hz. Thin layer chromatography (TLC) was carried out on Silica Gel 60 F254 pre-coated plates and visualization of the products was performed under UV light. Silica gel used for column chromatography was Scientific Silica Gel (particle size 0.035–0.070 mm). ESI mass spectra were recorded on Micromass QTOF2 Quadrupole/Time of Flight Tandem mass spectrometer with Turbo Ion Spray ionization source. All reactions were performed under nitrogen atmosphere using anhydrous solvents.
1-[2′,5′-bis-O-(tert-butyldimethylsilyl)-β-D-erythro-pentofuran-3′-2-(2-hydroxyethoxy)ethanimin-3′-ulosyl]uracil (II): Glacial acetic acid (2.85 mL, 49.84 mmol) was added dropwise over 30 min to 2-(2-aminoethoxy)ethanol (5 mL, 49.84 mmol). The solution was stirred at room temperature until viscosity increased and turned to gel-like mixture. Ethanol (1 mL) was added, mixed, and the resulting solution was concentrated under reduced pressure. To the mixture was added a solution of I (2.75 g, 5.84 mmol) in anhydrous pyridine (5 mL), and the reaction was allowed to stir under N2 at room temperature for 20 h. The solvent was removed under reduced pressure, and the residue was dissolved in EtoAc, washed with H2O and brine, dried over anhydrous Na2SO4 and concentrated in vacuum. The product was stirred in anhydrous ether and filtered to afford pure compound II (1.92 g, 59% yield). HRMS (ESI) m/z Calcd for C25H47N3O7NaSi2 (M + Na)+ 580.2850, found 580.2744. 1H NMR (500 MHz, CDCl3): 7.39 d, 1H, J = 7.8 (H-6); 5.82 br s, 1H (H-5); 5.21 s, 1H (H-1′); 4.35 d, J = 3.9 1H (OH); 3.65–3.51 m, 10H (H-2′, H-4′, H-5′, 3CH2O); 3.31 br s, 2H (CH2N); 0.81 s, 9H, (SiC(CH3)3); 0.78 s, 9H (SiC(CH3)3); 0.10 s, 3H (SiCH3); 0.06 s, 3H (SiCH3); − 0.004 s, 3H (SiCH3); − 0.039 s, 3H (SiCH3); 13C NMR (125 MHz, CDCl3) 187.7, 174.8, 163.2, 137.1, 130.3, 128.6, 123.9, 78.4, 72.8, 70.9, 67.6, 61.5, 48.2, 25.9, 25.8, 18.3, 18.2, − 4.9, − 5.0, − 5.4, − 5.5.
1-[2′,5′-bis-O-(tert-butyldimethylsilyl)-3′-deoxy-3′-2-(2-hydroxyethoxy)ethanamino-β-D-xylofuranosyl]uracil (III): To a solution of (R)-( +)-2-methyl-CBS-oxazaborolidine catalyst (0.03 g, 0.1 mmol) in THF (1 mL) was added BH3, 1 M in THF (1.0 mL, 1.0 mmol). The mixture was stirred under N2 at room temperature for 30 min. A solution of II (0.56 g, 1.0 mmol) in anhydrous THF (3.0 mL) was added dropwise over 30 min, and the resulting solution was stirred at room temperature for 4 h. The reaction was cooled to 0 °C and quenched with 1 mL of cold methanol. The solution was concentrated under reduced pressure, and the residue was purified by silica gel column chromatography (70% ethyl acetate in hexanes) to afford pure compound III (0.49 g, 87% yield). HRMS (ESI) m/z Calcd for C25H49N3O7KSi2 (M + Na)+ 582.2746, found 582.2822. 1H NMR (500 MHz, CDCl3): 7.41 d, 1H, J = 12.2 (H-6); 5.75 br s, 1H (H-5); 5.25 s, 1H (H-1′); 4.40 t, J = 4.4 1H (OH); 3.70–3.55 m, 11H (H-2′, H-3′, H-4′, H-5′, 3CH2O); 3.35 br s, 2H (CH2N); 0.84 s, 9H, [SiC(CH3)3]; 0.81 s, 9H [SiC(CH3)3]; 0.10 s, 3H (SiCH3); 0.03 s, 3H (SiCH3); − 0.03 s, 3H (SiCH3); − 0.04 s, 3H (SiCH3); 13C NMR (125 MHz, DMSO-d6) 162.4, 151.5, 142.2, 140.1, 103.1, 100.8, 85.1, 83.7, 76.1, 75.8, 66.8, 61.5, 45.2, 40.4, 26.2, 25.8, 18.6, 18.3, − 4.6, − 4.9, − 5.0, − 5.3.
Results and discussion
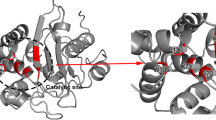
Molecular docking is an important step in the development of enzyme-inhibiting drugs because of the reduction in costs it entails, minimizing waste, and enhancing the discovery process by identifying compounds that are likely to have appropriate binding conformations within their targets (Yadav et al. 2017). An imperative method used to estimate the druggability of a compound requires calculation of binding energy of the proposed molecule to the target protein, where a more negative kJ/mol is favored (Zhu et al. 2013). ICM runs were conducted independently, for each of the PEGA modified nucleosides, to compare for binding efficacy and zinc ion destabilization with each of the three MBLs tested. To test the binding affinity of purine versus pyrimidine nucleosides, adenosine and uridine were both examined. Two PEGA moieties were examined employing either ethanolamine or 2-(2-aminoethoxy)ethanol. To produce molecules with mimicking lactam ring hybridization and strong binding energy, PEGA moieties were applied to the 2′, 3′ or 5′ position of the nucleosides; where 2′ and 3′ positions were explored in both xylo- and arabinofuranoside forms. For both uridine and adenosine analogues, the 3′ position in the xylofuranoside form (Fig. 2a, b, respectively) exhibited the best binding affinity across all B1 MBLs tested. In addition, the uridine analogue (A) has shown the favorable binding energy to all three MBL enzymes (compound 10 in Table 1). Analogue B, although showing weaker binding affinity than uridine analogues, was the superior binding analogue among the purines tested (compound 22 in Table 1). Analogue C, was constructed by adding another ethylene glycol unit to the 3′ PEGA side chain, to test whether an extended PEGA modification would allow for enhanced binding energy and better binding affinity (compound 12 in Table 1). Molecular docking simulations showed that the binding energies were not improved when 2-[2-(2-aminoethoxy)ethoxy]ethanol was attached to the uridine 3′ position thus 2-(2-aminoethoxy)ethanol has been chosen as the favorable length for the PEGA-nucleoside/substrate binding (compound 10 vs. compound 12). Notably, uridine modified at the 5′-position with ethanolamine has also shown remarkable binding affinity with all three MBL enzymes, represented by binding energies of − 29.6, − 29.2, and − 32.5 kJ/mol (compound 6 in Table 1). In addition to the less favorable binding energies compared to analogue A, one important reason that this structure was omitted is the fact that nucleoside analogues are prodrugs that require activation by phosphorylation at the 5′-hydroxyl group for cytotoxicity. The sugar moiety may remain intact by leaving the 5′-hydroxyl group free to be phosphorylated by the corresponding nucleoside kinase (Youssef and Lien 2015). The docking interactions of analogues, A and B in Fig. 2 to the conserved zinc binding active site of all three enzymes NDM-1, VIM-2 and IMP-1 were then analyzed.
NDM-1 active site interactions
Carbonyl Oxygen-2 of A formed a coordination bond to Zn2, with a length of 2.04 Å (Fig. 3a). Three other Zn2 hydrogen bonds were formed with the sulfide group on residue C208 (2.36 Å), the nitrogen group on residue H250 (2.2 Å), and with the oxygen of the carboxyl group on residue D124 (2.01 Å). A hydrogen bond was formed between hydrogen-3 and D124 oxygen (1.78 Å) while a polar bond was formed between oxygen-4 and the amino hydrogen on Q123 (2.24 Å). Disruptions of the zinc coordination to the water bridge and it’s stabilizing amino acids allow for impeding of the B1 MBL hydrolysis in NDM-1. It is known that in β-lactam hydrolysis the oxygen atom of the lactam carbonyl orients towards Zn1 as the carboxylate is oriented towards Zn2, this orientation is disrupted by the three mimic points on the uracil which orient the oxygen-2 carbonyl towards Zn2 while the oxygen-4 carbonyl is bound to Q123, away from Zn1 (Tooke et al. 2019). In addition, analogue A also bonds to D124, a conserved zinc binding active site residue across all B1 MBLs that involve in β-lactam hydrolysis. This residue forms part of the bipyramidal geometry required for effective interactions of the Zn2 with the water bridge, which leads to hydrolysis (Tooke et al. 2019).
Nitrogen-7 of the purine analogue B formed a coordination bond to Zn2 with a length of 2.33 Å (Fig. 3b). Three other Zn2 coordination bonds were formed with the sulfide group on residue C208 (2.2 Å), the nitrogen on residue H250 (2.19 Å), and with the oxygen of the carboxyl group on residue D124 (1.92 Å). Other hydrogen bonds were formed between the 2′ oxygen on B and the nitrogen atom on Q123 (1.95 Å). Oxygen-6 on B and the amino on residue Q222 (1.88 Å), and hydrogen-6 with the oxygen on residue H189 (2.17 Å) also formed bonds during VLS simulations. Analogue B, similarly to analogue A, occupied a Zn2 coordination bond, a bond which is crucial in the hydrolysis mechanism of lactam antibiotics. Nitrogen-7 added a bond to conserved Zn2 binding active site residues, assisting in the destabilization of the trigonal bipyramidal geometry required for the metal ions to perform nucleophilic β-lactam hydrolysis. Consequently, analogue B changed the Zn2 geometry in the active site from trigonal bipyramidal to tetrahedral.
VIM-2 active site interactions
Oxygen-6 on analogue A bound simultaneously to the amino group on residue W87 with a polar bond length of 1.98 Å (Fig. 3c) and to the carbonyl on the residue D117 (2.41 Å). Oxygen-4 formed a H-bonding (1.86 Å) with the amino hydrogen on residue R205. Other coordination bonds were observed between Zn1, Zn2 and the oxygen on the water bridge. A H-bonding was also observed between the carbonyl oxygen on residue D118 and one of the hydrogens on the water bridge (1.85 Å). It is known that VIM-2 inhibitor fragments utilize carbonyl groups to create tighter binding with active site metal ions while the presence of nitrogen groups aid in tighter binding to NDM-1 (Christopeit and Leiros 2016). This is evident in the VLS active site interactions noted across all uridine B1 MBL complex docking simulations, where the carbonyl groups are actively involved in coordination to the metal ions creating tighter binding across the entire class. This pattern was not emulated within adenosine B1 MBL complex simulations.
Oxygen-6 on analogue B was H-bonded to the amino hydrogen on residue N148 with a bond length of 1.78 Å (Fig. 3d). Coordination bonds were observed between Zn1, Zn2 and the water bridge. The nitrogen on residue H240 and the sulfide C198 bound to Zn2 with a distance of 2.09 Å and 2.25 Å, respectively. One oxygen on residue D118 formed a coordination bond with Zn2 while the other oxygen formed H-bonding with a hydrogen on the water bridge with distance of 2.18 and 1.85 Å, respectively. When docked to VIM-2, analogue B displayed activity through binding site interactions rather than metal-ion binding, which was also observed in the simulations for analogue A. Due to the weaker binding affinity to VIM-2, compared to analogue A, and the lack of metal ion binding, a characteristic which have shown to inactivate NDM-1 lactam hydrolysis where NDM-1 is the most difficult B1 MBL to inhibit (Yarlagadda et al. 2018), analogue B was not selected for further investigations.
IMP-1 active site interactions
Oxygen-2 on analogue A formed a coordination bond to Zn1 with a bond length of 2.29 Å (Fig. 3e). Three other Zn1 interactions were formed between the nitrogen on residues H77, H79, and H139 with bond lengths of 2.13 Å, 2.05 Å, and 2.05 Å, respectively. Three Zn2 interactions were formed with the sulfide on residue C158 (2.26 Å), the nitrogen on residue H197 (2.14 Å), and the carbonyl oxygen on residue D81 (1.98 Å). A H-bonding was also formed between hydrogen-6 of analogue A and the oxygen on residue G164. The interactions noted for analogue A describe a similar trend to that with this same analogue when docked to NDM-1. The analogue coordinates with Zn1 to disrupt the tetrahedral geometry held during hydrolysis. It has been proven that binding to the metal ions in the IMP-1 active site have disrupted the stability of the active site geometry, thus inactivating hydrolysis of lactams (Gardonio and Siemann 2009).
Oxygen-6 on analogue B coordinated to Zn1 and Zn2 forming coordination bonds with lengths of 2.22 Å and 2.20 Å, respectively (Fig. 3f). Three other Zn1 bonds were formed with the nitrogen on residues H77, H79, and H139 with bond lengths of 2.12 Å, 2.05 Å, and 2.05 Å, respectively. Zn2 coordinated to the sulfide on residue C158 (2.26 Å), the nitrogen on residue H197 (2.14 Å), and the carbonyl on residue D81 (1.98 Å). Analogue B’s PEGA hydroxyl group bound to both zinc ions in the active site, displacing the water bridge and thus provokes bonding, therefore, disrupting the hydrolysis water bridge mechanism within the enzyme active site.
Binding energy
Adenosine and its PEGA analogue underperformed compared to their corresponding uridine counterparts (Table 1). Results showed that introducing 2-(2-aminoethoxy)ethanol to uridine in the xylo configuration, compound 10 in Table 1, provided the most effective analogue, A, with the strongest binding energy of − 29.8, − 31.8, − 36.6 kJ/mol towards all of the three enzymes NDM-1, VIM-2 and IMP-1, respectively. Separate runs were performed to examine the binding affinity of uridine modified with a longer PEGA moiety, 2-[2-(2- aminoethoxy)ethoxy]ethanol in the xylo configuration, compound 12, where the binding energies were − 26.6, − 28.4, − 27.5 kJ/mol indicating weaker binding affinity compared to analogue A. Uridine composed of three β-lactam mimic points on its base, found between atoms 2, 3 and 4 (Fig. 2). Due to analogue A’s ability to bind and engage with the conserved active site components (Fig. 3) in a similar fashion as β-lactams, the molecule interrupted the di-zinc geometry leading to destabilization of the B1 MBL hydrolysis mechanism. It is known that β-lactams are susceptible to hydrolysis by antibiotic resistant bacteria because of torsional and angle strain caused by the 109.5° angle of the sp3 hybridized carbons (Lisa et al. 2017). The trigonal pyramidal geometry prevents resonance stabilization and allows for facile hydrolysis. In turn, uridine holds three mimic points that draw the nucleoside into the B1 MBL active site while the resonance stabilized base deactivates the hydrolysis mechanism. Analogue A, with the strongest binding affinity across all class B1 MBL enzymes, was then selected for chemical synthesis.
Chemical synthesis
A synthetic protocol for the proposed PEGA nucleosides was described (Scheme 1). The 3′-keto nucleoside (I) was converted into the corresponding 3′-imine derivative (II) by treatment with 2-(2-aminoethoxy)ethanol in glacial acetic acid. The conversion of keto nucleosides into oxime derivatives was previously reported, and was adapted for the imine formation in this study (Kojima et al. 2002). The stereoselective reduction of the 3′-imino to 3′-amino derivative using Corey-Bakshi-Shibata oxazaborolidine (CBS) as a chiral catalyst produced the desired configuration 3′-2-(2-hydroxyethoxy)ethanamino-β-D-xylofuranosyl III in 87% yield. CBS oxazaborolidine has been reported as a chiral catalyst that mediates the borane reduction of ketones to secondary alcohols with excellent enantioselestivity (Corey et al. 1987). The diastereoselective reduction of C-2′ and C-3′-keto ribofuranoside derivatives, using the (R)-isomer of the CBS, to the corresponding arabino and xylofuranosides in greater than 95% diastereomeric excess has been also previously reported (Utley et al. 2018). Removal of the TBDMS protective groups from III can be easily achieved to afford the unprotected analogue A according to the standard procedure (Utley et al. 2018).
Comparison of binding energy to market drugs
To compare analogue A to known antibiotics on the market, penicillin and L-captopril were independently docked to all three of the B1 MBL proteins. L-Captopril is a proposed NDM-1 inhibitor which acts through metal ion chelation (King et al. 2012), parallel to analogue A that has also shown to bind to the NDM-1 conserved di-zinc active site. Penicillin and L-captopril should binding energy of − 2.6 kJ/mol and − 21.9 kJ/mol, respectively, towards NDM-1. VIM-2 had a binding energy measured at − 1.9 kJ/mol and − 20.5 kJ/mol, and IMP-1 had a binding energy measured at − 15.7 kJ/mol and − 33.3 kJ/mol for penicillin and L-captopril, respectively (Table 1). Analogue A showed superior binding energy compared to penicillin and L-captopril, − 29.8, − 31.8, − 36.6 kJ/mol for NDM-1, VIM-2, and IMP-1, respectively. Penicillin is known to be completely hydrolyzed by MBLs and the VLS binding energy correlated with this fact. However, L-captopril was previously submitted as an NDM-1 inhibitor but did not surpass analogue A in binding to the active site of class B1 MBLs.
Toxicity evaluation and ADME analysis
To further investigate the analogues with strongest B1 MBL binding energy as a potential inhibitors, toxicity screenings were conducted, using ProTox-II web tool, for selection of the analogue with the greatest likelihood of passing in-vivo toxicity tests. SwissADME predictor was used to evaluate the ADMET properties, like aqueous solubility, synthetic accessibility score, percentage absorption, pharmacokinetics, and drug-likeness properties of small molecules (Table 2). All analogues did not display hepatotoxicity, carcinogenicity, cytotoxicity, mutagenicity, blood brain barrier permeation or hepatotoxicity. The lack of blood brain barrier (BBB) permeation is a desired quality based on location of the target, where the unnecessary permeation should be avoided (Daina et al. 2017). Analogue B was predicted to have a toxicity class lower than the other analogues, an unfavorable characteristic in drug development, due to the toxicity caused with a low amount of drug quantity which can also be seen in its LD50 value (Toropova et al. 2014). The toxicity profiles suggested that analogues A and C are worth further investigations as potential small molecule drugs.
CYP450 is one of the main enzymes involved in the metabolism of cellular chemicals (Ogu and Maxa 2017). CYP substrates would be ruled out as potential drugs due to the possibility of creating toxic environment when prescribed alongside other medications. The pyrimidine analogues A and C are predicted as non CYP substrate inhibitors, however the purine analogue B showed to be a CYP3A subfamily substrate. These data suggested that analogues A and C remained the most favorable choices, and their solubility profiles were then evaluated. Small molecule drugs must be highly soluble to be taken orally (Savjani et al. 2012). All proposed analogues are water-soluble and have predicted high intestinal absorption, giving high probability of these analogues being efficient as an oral drug in cocktail with antibiotics. Organic synthesis PAINS yielded zero alerts for all analogues (Table 2), demonstrating no problematic pharmacokinetic or pharmacodynamic issues arising from the use of their functional groups during developmental stages (Vidler et al. 2018). Analogues B and C, however, possess ten nitrogen and oxygen groups, violating Lipinski’s rule of five. The clear toxicity profile of analogue A and the superiority in binding affinity with the B1 MBL active site support this molecule as a potential B1 MBL inhibitor that worth further in-vitro and in-vivo studies.
Conclusion
Metallo-β-lactamases have acquired the ability to inactivate β-lactam antibiotics through zinc mediated hydrolysis of the sp3 hybridized amide bond. In this study we described evaluation and synthesis of novel nucleoside analogues modified with polyethylene glycolamine as class B1 MBL inhibitors. Twenty-one analogues were constructed for in-silico molecular modeling simulations and pharmacological property screening. Three analogues that exhibited strongest binding affinities were identified, where analogue A and C were pyrimidine derivatives and analogue B was a purine. Analogue B held the weakest binding energy of the three candidates and was also found to be a CYP450 substrate and violated Lipinski’s rule of five, excluding it as lead-like inhibitor. The longer PEGA moiety in analogue C did not enhance binding affinity and displayed a violation of Lipinski’s rule. Due to its ability to mimic β-lactams at three locations with resonance stability, its superior binding with high specificity to the conserved active site, and destabilization of the di-zinc geometry coordination hindering the ability of B1 MBLs to hydrolyze the lactam ring, analogue A has been chosen as a potential B1 MBLs inhibitor, and was synthesized to establish a protocol for the chemical synthesis of the proposed PEGA nucleosides and for further in vitro and in-vivo studies on targeting zinc or other metal-containing enzymes.
References
Abagyan R, Totrov M, Kuznetsov D (1994) ICM—a new method for protein modeling and design: applications to docking and structure prediction from the distorted native conformation. J Comput Chem. https://doi.org/10.1002/jcc.540150503
Antony J, Gresh N, Olsen L, Hemmingsen L, Schofield CJ, Bauer R (2002) Binding of D and L-captopril inhibitors to metallo-β-lactamase studied by polarizable molecular mechanics and quantum mechanics. J Comput Chem. https://doi.org/10.1002/jcc.10111
Arnautova YA, Abagyan R, Totrov M (2018) Protein-RNA docking using ICM. J Chem Theory Comput 14:4971–4984. https://doi.org/10.1021/acs.jctc.8b00293
Banerjee P, Eckert AO, Schrey AK, Preissner R (2018) ProTox-II: a webserver for the prediction of toxicity of chemicals. Nucleic Acids Res 46:W257–W263. https://doi.org/10.1093/nar/gky318
Benet LZ, Hosey CM, Ursu O, Oprea TI (2016) BDDCS, the rule of 5 and drugability. Adv Drug Deliv Rev 101:89–98. https://doi.org/10.1016/j.addr.2016.05.007
Brem J, Van Berkel SS, Zollman D, Lee SY, Gileadi O, McHugh PJ, Walsh TR, McDonough MA, Schofield CJ (2016) Structural basis of metallo-β-lactamase inhibition by captopril stereoisomers. Antimicrob Agents Chemother. https://doi.org/10.1128/AAC.01335-15
Bursulaya BD, Totrov M, Abagyan R, Brooks CL (2003) Comparative study of several algorithms for flexible ligand docking. J Comput Aided Mol Des 17:755–763. https://doi.org/10.1023/B:JCAM.0000017496.76572.6f
Chandar B, Pati S, Bhattacharya D (2018) An overview of New Delhi metallo-beta lactamase-1 and extended spectrum beta lactamase producing bacteria: need for an alternate. J Antimicrob Agents. https://doi.org/10.4172/2472-1212.1000155
Christopeit T, Leiros HKS (2016) Fragment-based discovery of inhibitor scaffolds targeting the metallo-β-lactamases NDM-1 and VIM-2. Bioorganic Med Chem Lett. https://doi.org/10.1016/j.bmcl.2016.03.004
Corey EJ, Bakshi RK, Shibata S (1987) Highly enantioselective borane reduction of ketones catalyzed by chiral oxazaborolidines Mechanism and synthetic implications. J Am Chem Soc 109:5551–5553. https://doi.org/10.1021/ja00252a056
Daina A, Michielin O, Zoete V (2017) SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep. https://doi.org/10.1038/srep42717
Dixit A, Kumar N, Kumar S, Trigun V (2019) Antimicrobial resistance: progress in the decade since emergence of New Delhi metallo-β-lactamase in India. Indian J Community Med. https://doi.org/10.4103/ijcm.IJCM_217_18
Docquier JD, Mangani S (2018) An update on β-lactamase inhibitor discovery and development. Drug Resist Updat. https://doi.org/10.1016/j.drup.2017.11.002
Fernández-Recio J, Totrov M, Abagyan R (2003) ICM-DISCO docking by global energy optimization with fully flexible side-chains. Proteins Struct Funct Genet. https://doi.org/10.1002/prot.10383
Gardonio D, Siemann S (2009) Chelator-facilitated chemical modification of IMP-1 metallo-β-lactamase and its consequences on metal binding. Biochem Biophys Res Commun. https://doi.org/10.1016/j.bbrc.2009.02.021
González LJ, Bahr G, Nakashige TG, Nolan EM, Bonomo RA, Vila AJ (2016) Membrane anchoring stabilizes and favors secretion of New Delhi metallo-β-lactamase. Nat Chem Biol. https://doi.org/10.1038/nchembio.2083
Hevener KE, Zhao W, Ball DM, Babaoglu K, Qi J, White SW, Lee RE (2009) Validation of molecular docking programs for virtual screening against dihydropteroate synthase. J Chem Inf Model. https://doi.org/10.1021/ci800293n
Ju LC, Cheng Z, Fast W, Bonomo RA, Crowder MW (2018) The continuing challenge of metallo-β-lactamase inhibition: mechanism matters. Trends Pharmacol Sci 39:635–647. https://doi.org/10.1016/j.tips.2018.03.007
Katritch V, Jaakola VP, Lane JR, Lin J, Ijzerman AP, Yeager M, Kufareva I, Stevens RC, Abagyan R (2010) Structure-based discovery of novel chemotypes for adenosine A2A receptor antagonists. J Med Chem. https://doi.org/10.1021/jm901647p
King DT, Worrall LJ, Gruninger R, Strynadka NCJ (2012) New delhi metallo-β-lactamase: structural insights into β-lactam recognition and inhibition. J Am Chem Soc. https://doi.org/10.1021/ja303579d
Kojima N, Szabo IE, Bruice TC (2002) Synthesis of ribonucleic guanidine: replacement of the negative phosphodiester linkages of RNA with positive guanidinium linkages. Tetrahedron 58:867–879. https://doi.org/10.1016/S0040-4020(01)01185-1
Lam PCH, Abagyan R, Totrov M (2018) Ligand-biased ensemble receptor docking (LigBEnD): a hybrid ligand/receptor structure-based approach. J Comput Aided Mol Des 32:187–198. https://doi.org/10.1007/s10822-017-0058-x
Langley GW, Cain R, Tyrrell JM, Hinchliffe P, Calvopiña K, Tooke CL, Widlake E, Dowson CG, Spencer J, Walsh TR, Schofield CJ, Brem J (2019) Profiling interactions of vaborbactam with metallo-β-lactamases. Bioorganic Med Chem Lett. https://doi.org/10.1016/j.bmcl.2019.05.031
Lateef M, Azhar A, Siddiqui BS, Zarina S, Uddin N, Anwar MF, Siddiqui K, Azhar KF, Iqbal L, Mehmood R, Perveen S (2017) New anthrarobin acyl derivatives as butyrylcholinesterase inhibitors: synthesis, in vitro and in silico studies. Heliyon. https://doi.org/10.1016/j.heliyon.2017.e00350
Li N, Xu Y, Xia Q, Bai C, Wang T, Wang L, He D, Xie N, Li L, Wang J, Zhou H, Xu F et al (2014) Simplified captopril analogues as NDM-1 inhibitors. Bioorg Med Chem Lett. https://doi.org/10.1016/j.bmcl.2013.10.068
Lisa MN, Palacios AR, Aitha M, González MM, Moreno DM, Crowder MW, Bonomo RA, Spencer J, Tierney DL, Llarrull LI, Vila AJ (2017) A general reaction mechanism for carbapenem hydrolysis by mononuclear and binuclear metallo-β-lactamases. Nat Commun. https://doi.org/10.1038/s41467-017-00601-9
Mbaye MN, Gilis D, Rooman M (2019) Rational antibiotic design: in silico structural comparison of the functional cavities of penicillin-binding proteins and ß-lactamases. J Biomol Struct Dyn. https://doi.org/10.1080/07391102.2017.1418678
Mojica MF, Bonomo RA, Fast W (2016) B1-Metallo-beta-Lactamases : where do we stand ? Curr Drug Targets. https://doi.org/10.2174/1389450116666151001105622
Neves MAC, Totrov M, Abagyan R (2012) Docking and scoring with ICM: the benchmarking results and strategies for improvement. J Comput Aided Mol Des. https://doi.org/10.1007/s10822-012-9547-0
Niu G, Tan H (2015) Nucleoside antibiotics: biosynthesis, regulation, and biotechnology. Trends Microbiol. https://doi.org/10.1016/j.tim.2014.10.007
Ogu CC, Maxa JL (2017) Drug interactions due to cytochrome P450. Pharmacol Notes. https://doi.org/10.1080/08998280.2000.11927719
Perola E, Walters WP, Charifson PS (2004) A detailed comparison of current docking and scoring methods on systems of pharmaceutical relevance. Proteins Struct Funct Genet. https://doi.org/10.1002/prot.20088
Ramírez D, Caballero J (2018) Is it reliable to take the molecular docking top scoring position as the best solution without considering available structural data? Molecules. https://doi.org/10.3390/molecules23051038
Savjani KT, Gajjar AK, Savjani JK (2012) Drug solubility: importance and enhancement techniques. ISRN Pharmaceutics. https://doi.org/10.5402/2012/195727
Serpi M, Ferrari V, Pertusati F (2016) Nucleoside derived antibiotics to fight microbial drug resistance: New utilities for an established class of drugs? J Med Chem. https://doi.org/10.1021/acs.jmedchem.6b00325
Shen B, Yu Y, Chen H, Cao X, Lao X, Fang Y, Shi Y, Chen J, Zheng H (2013) Inhibitor discovery of full-length New Delhi Metallo-β-Lactamase-1 (NDM-1). PLoS ONE. https://doi.org/10.1371/journal.pone.0062955
Sheridan RP, Maiorov VN, Holloway MK, Cornell WD, Gao YD (2010) Drug-like density: a method of quantifying the “bindability” of a protein target based on a very large set of pockets and drug-like ligands from the protein data bank. J Chem Inf Model. https://doi.org/10.1021/ci100312t
Snyder BM, Montague BT, Anandan S, Madabhushi AG, Pragasam AK, Verghese VP, Balaji V, Simões EAF (2019) Risk factors and epidemiologic predictors of blood stream infections with New Delhi Metallo-b-lactamase (NDM-1) producing Enterobacteriaceae. Epidemiol Infect. https://doi.org/10.1017/S0950268819000256
Tehrani KHME, Fu H, Brüchle NC, Mashayekhi V, Prats Luján A, Van Haren MJ, Poelarends GJ, Martin NI (2020) Aminocarboxylic acids related to aspergillomarasmine A (AMA) and ethylenediamine-: N, N′-disuccinic acid (EDDS) are strong zinc-binders and inhibitors of the metallo-beta-lactamase NDM-1. Chem Commun. https://doi.org/10.1039/d0cc00356e
Thomas PW, Cammarata M, Brodbelt JS, Monzingo AF, Pratt RF, Fast W (2019) A lysine-targeted affinity label for Serine-β-Lactamase also covalently modifies New Delhi Metallo-β-lactamase-1 (NDM-1). Biochemistry. https://doi.org/10.1021/acs.biochem.9b00393
Thomson JM, Lamont IL (2019) Nucleoside analogues as antibacterial agents. Front Microbiol. https://doi.org/10.3389/fmicb.2019.00952
Tooke CL, Hinchliffe P, Bragginton EC, Colenso CK, Hirvonen VHA, Takebayashi Y, Spencer J (2019) β-Lactamases and β-lactamase inhibitors in the 21st century. J Mol Biol. https://doi.org/10.1016/j.jmb.2019.04.002
Toropova AA, Toropova AP, Raska I Jr, Leszczynska D, Leszczynski J (2014) Comprehension of drug toxicity: software and databases. Comput Biol Med. https://doi.org/10.1016/j.compbiomed.2013.11.013
Utley LM, Maldonado J, Awad AM (2018) A practical synthesis of xylo- and arabinofuranoside precursors by diastereoselective reduction using Corey-Bakshi-Shibata catalyst, Nucleosides. Nucleotides Nucleic Acids. https://doi.org/10.1080/15257770.2017.1414240
Vidler LR, Watson IA, Margolis BJ, Cummins DJ, Brunavs M (2018) Investigating the behavior of published PAINS alerts using a pharmaceutical company data set. ACS Med Chem Lett. https://doi.org/10.1021/acsmedchemlett.8b00097
Wang X, Lu M, Shi Y, Ou Y, Cheng X (2015) Discovery of novel new delhi metallo-β-lactamases-1 inhibitors by multistep virtual screening. PLoS ONE. https://doi.org/10.1371/journal.pone.0118290
Yadav S, Pandey SK, Singh VK, Goel Y, Kumar A, Singh SM (2017) Molecular docking studies of 3-bromopyruvate and its derivatives to metabolic regulatory enzymes: Implication in designing of novel anticancer therapeutic strategies. PLoS ONE. https://doi.org/10.1371/journal.pone.0176403
Yarlagadda V, Sarkar P, Samaddar S, Manjunath GB, Das Mitra S, Paramanandham K, Shome BR, Haldar J (2018) Vancomycin analogue restores meropenem activity against NDM-1 gram-negative pathogens. ACS Infect Dis. https://doi.org/10.1021/acsinfecdis.8b00011
Youssef KM, Lien EJ (2015) Design and synthesis of potential ribonucleotide reductase enzyme(RNR) inhibitors as antileukemic and/or antiviral 2’-deoxymethylenenucleosides. Future J Pharm Sci. https://doi.org/10.1016/j.fjps.2015.10.001
Zhang J, Wang S, Wei Q, Guo Q, Bai Y, Yang S, Song F, Zhang L, Lei X (2017) Synthesis and biological evaluation of Aspergillomarasmine A derivatives as novel NDM-1 inhibitor to overcome antibiotics resistance. Bioorganic Med Chem. https://doi.org/10.1016/j.bmc.2017.07.025
Zhang E, Wang MM, Huang SC, Xu SM, Cui DY, Bo YL, Bai PY, Hua YG, Xiao CL, Qin S (2018) NOTA analogue: a first dithiocarbamate inhibitor of metallo-β-lactamases. Bioorganic Med Chem Lett. https://doi.org/10.1016/j.bmcl.2017.10.074
Zhu K, Lu J, Ye F, Jin L, Kong X, Liang Z, Chen Y, Yu K, Jiang H, Li JQ, Luo C (2013) Structure-based computational study of the hydrolysis of New Delhi metallo-β-lactmase-1. Biochem Biophys Res Commun. https://doi.org/10.1016/j.bbrc.2012.12.141
Acknowledgements
The authors acknowledge the California State University Chancellor's office RSCA awards, and the SURF program at California State University Channel Islands for their generous financial support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Mendoza, J.A., Pineda, R.Y., Nguyen, M. et al. Molecular docking studies, in-silico ADMET predictions and synthesis of novel PEGA-nucleosides as antimicrobial agents targeting class B1 metallo-β-lactamases. In Silico Pharmacol. 9, 33 (2021). https://doi.org/10.1007/s40203-021-00092-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s40203-021-00092-z