Abstract
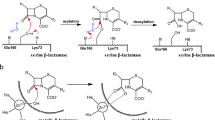
The treatment of bacterial infections is currently threatened by the emergence of pathogenic bacteria producing β-lactamase, which catalyzes the hydrolysis of β-lactams. Although the hydrolysis of the substrate nitrocefin by a metallo-β-lactamase, namely β-lactamase N1 from USA300 (a typical methicillin-resistant Staphylococcus aureus), has previously been reported in the literature, its mechanism remains elusive. Here, we show that molecular modeling and quantum-mechanical/molecular mechanics (QM/MM) calculations describing the complex of β-lactamase N1 with nitrocefin (the substrate of β-lactamase N1) can predict the catalytic mechanism of nitrocefin hydrolysis by β-lactamase N1. Molecular dynamics simulation shows that the catalytic reaction begins with hydrogen bond formation between Gln171 and a water molecule, which is thereby captured for nitrocefin hydrolysis by β-lactamase N1. In addition, the carboxyl group coordinates Zn2 in a chelating fashion. The binding energy decompositions suggest that Phe169 anchors nitrocefin by π-stacking interactions between the benzene rings. Specifically, Phe169 and Zn2 position the nitrocefin in specific orientations. The active site of β-lactamase N1 contains two residues (Gln171 and Phe169) that we expected to be crucial for guiding the nitrocefin hydrolysis reaction. Compelling evidence is provided that the mutants F169A and Q171A show lower enzymatic activity than the wild-type protein. On the basis of the QM/MM calculations, we propose that nitrocefin hydrolysis is initiated by the interaction between the oxygen atom of water and the C18 atom of nitrocefin, leading to the opening of the four-membered ring of nitrocefin and the formation of a substrate intermediate. In the next step, a hydrogen atom transfers from the nitrogen atom to the C11 atom of nitrocefin, resulting in the stable product.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Antibiotic-resistant bacterial infection has become a concerning global threat to human health, according to the World Health Organization (WHO) reports [1, 2]. For decades, the treatment of bacterial infections has relied mainly on β-lactam antibiotics, such as penicillins, cephalosporins, and carbapenems. However, the emergence and spread of pathogenic bacteria producing β-lactamase have seriously threatened the treatment effectiveness of β-lactam antibiotics [3, 4]. The β-lactamases that can hydrolyze the signature β-lactam ring are divided into four classes based on sequence and mechanism: A, B, C, and D [5, 6]. Owing to the involvement of a serine residue in the catalytic active site, classes A, C, and D are termed serine β-lactamases (SBLs). In hydrolysis catalyzed by SBL, the hydroxyl group of the serine residue attacks the carbonyl carbon atom of β-lactam. In contrast, class B enzymes, which are referred to as metallo-β-lactamases (MBLs), utilize one or two active site Zn(II) ions to catalyze the hydrolysis of β-lactam antibiotics [7]. The MBL superfamily is a large and diverse group [8, 9]; however, the level of sequence homology of MBLs is very low, and they have broad and diverse activities [10, 11]. After the identification of the first β-lactamase N1 obtained from Bacillus cereus, MBLs were considered somewhat obscure and posed little clinical threat. The increasing discovery of new MBLs, however, has revealed that they can hydrolyze a wide range of β-lactam antibiotics, including inhibitors of SBLs, cephamycins and imipenem [12,13,14,15]. Moreover, the traditional β-lactamase inhibitors, such as clavulanic acid and sulbactam, show little or no inhibitory activity against MBLs.
To aid in the development of new and more effective anti-β-lactamase inhibitors, researchers have explored the three-dimensional structures of MBLs. As early as 1996, the crystal structure of metallo-β-lactamase from Bacteroides fragilis was determined [16]. Knowledge of the 3D structure can identify residues in the active site of the enzyme that can bind with zinc. Subsequently, the interaction between metallo-β-lactamase from Bacteroides fragilis and an inhibitor, 4-morpholinoethanesulfonic acid (MES), was studied by the determination of a complex X-ray crystal structure of metallo-β-lactamase with MES [17]. The information from the crystal structure revealed that the binding of MES with the protein led to a significant change in the conformation of the β-strand, which can be an important consideration in the design of inhibitors targeting MBLs. In most cases, MBLs can work in their zinc-bound form, but activity is also observed in the forms bound to other ions [12, 18, 19]. An entire new family of MBL-like metalloenzymes, termed the UlaG family, has been described on the basis of the crystal structures of UlaG [20]. In these new MBL-like enzymes, the metal binding site of the protein contains a single metal ion, Mn(II). Furthermore, the same researchers found that zinc(II) can bind to UlaG without activating the hydrolysis of β-lactam antibiotics. In 2016, the new subclass B3 of MBLs from environmental species, referred to as Rm3, was identified [21]. On the basis of the analysis of the Rm3 crystal structure, the αβ/βα fold in the MBL superfamily was found to show closer resemblance than other chromosomal enzymes. In 2015, Ahmad Ghavami reported that a metallo-β-lactamase from USA300 (a typical methicillin-resistant Staphylococcus aureus), possesses a strong ability to catalyze the hydrolysis of nitrocefin [22]. However, the hydrolysis mechanism at the atomic level still remains elusive.
In this study, the 3D structure of the metallo-β-lactamase (named β-lactamase N1 in this paper) from USA300 was constructed by a homology approach, and the interactions among the substrate, nitrocefin, and β-lactamase N1 was explored through molecular modeling. Two amino acids were identified as essential for the enzymatic activity. Furthermore, the hydrolysis mechanism of β-lactamase N1 was investigated on the basis of QM/MM calculations. Because the structures of most metallo-β-lactamases are conserved, our study may provide a basis for future research on metallo-β-lactamase inhibitors.
Methods
Protein expression and purification
The DNA fragment encoding WT-β-lactamase N1 was amplified using the genome of BAA1717 as a template, and the primers are listed in Table 1. Next, the amplified fragment was cloned into a pGEX-6p-1 plasmid, and the protocol was performed as described by Qiu et al. [23]. The plasmids expressing F169A-β-lactamase N1 and Q171A-β-lactamase N1 were mutated using the QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA, USA), and the mutagenic primers are listed in Table 1.
The plasmids encoding WT-β-lactamase N1, F169A-β-lactamase N1, and Q171A-β-lactamase N1 were transformed into E. coli BL21 (DE3). The protein expression was induced by IPTG, and the protein was purified by affinity chromatography. The protocol was performed as described by Qiu et al. [23].
β-Lactamase N1 activity assay
To investigate the catalytic hydrolysis ability of WT-β-lactamase N1 and its mutants, purified WT-β-lactamase N1, F169A-β-lactamase N1, and Q171A-β-lactamase N1 proteins were used in this assay. First, 100 ng/mL of protein was added to PBS buffer (pH 7.2) in the presence of 25 μg/mL nitrocefin and incubated at 37 °C for 10 min. Then, the reaction mixtures were monitored at 492 nm.
Protein model
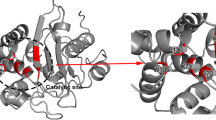
The β-lactamase N1 from Staphylococcal aureus USA300 shares 44.63% homology with ribonuclease J (PDB code: 3ZQ4) [24], which is the highest homology of any protein with a crystal structure in the Swiss-Model Template Library. Therefore, we chose ribonuclease J (PDB code: 3ZQ4) as the template for homology structure modeling by the SWISS-MODEL Server (http://www.swissmodel.expasy.org/). The 3D structural and sequence alignment between the model and the template ribonuclease J are shown in Fig. 1. The model structure is highly consistent with that of the template, implying that the homology model structure based on ribonuclease J is reliable.
Subsequently, the homology model was used as the initial structure for classical molecular dynamics simulations. The model structure was immersed in a periodic cubic box containing water molecules and neutralized with sodium ions and chloride ions. A minimum distance of 0.9 nm between periodically replicated images of the protein was used to build the box. The Amber99sb force field and tip3p water model were used to perform the MD simulation with Gromacs 5.0.7 software [25, 26]. The electrostatic term was described with the particle mesh Ewald algorithm. The LINCS algorithm was used to constrain all of the bond lengths. The SETTLE algorithm was used for the water molecules. The dielectric permittivity ε was set to 1, and a time step of 2 fs was used. All atoms were given an initial velocity as determined from the Maxwell distribution at the desired initial temperature of 300 K. The density of the system was adjusted during the first equilibration runs under NPT conditions by weak coupling to a bath of constant pressure (P0 = 1 bar, coupling time τP = 0.5 ps). The protein simulation was run for 1 μs.
Molecular docking
The substrate of β-lactamase N1, nitrocefin, was docked into the active site of the protein using the Autodock 4.0 [27]. Further details are provided in the Supporting Information.
Classical molecular dynamics
The parameters of nitrocefin were estimated with the antechamber program [28] and the RESP partial atomic charges from the Amber suite [29]. The molecular dynamics simulation was carried out for the complex system of the protein with nitrocefin, and the parameter settings of the MD simulation were consistent with those of the free protein MD simulation.
QM/MM calculation
In this work, the Gaussian 09 and Gromacs 5.0.7 packages were used to carry out QM/MM calculation for the β-lactamase N1-nitrocefin complex. The nitrocefin molecule and one water molecule were treated using QM methods by Gaussian. The remaining water and the protein were treated using a Gromacs force field and an Amber99sb force field. The QM region was treated with the DFT method at the BLYP level.
Calculation of the decomposition binding free energy
In this work, the binding free energies were calculated using the MM-GBSA approach [30, 31] supplied with the Amber 10 package. We chose a total number of 100 snapshots evenly from the last 70 ns on the MD trajectory with an interval of 10 ps.
The interactions between nitrocefin and each residue in the binding site of β-lactamase N1 were analyzed using the decomposition process in Amber 10. The binding interaction of each ligand-residue pair includes three terms, namely, the van der Waals contribution (ΔEvdw), electrostatic contribution (ΔEele), and solvation contribution (ΔEsol). All of the energy components were calculated using the same snapshots used for the free energy calculation.
Results and discussion
Homology model of β-lactamase N1
The homology model of β-lactamase N1 was used as the initial structure for the classical MD simulation. After a 1000 ns simulation, the stable 3D structure of β-lactamase N1 was determined. Figure 2a shows that after 600 ns of the simulation, the average RMSD of the protein was 0.5 nm, implying that the protein reached equilibrium at 600 ns. The protein equilibrium structure is shown in Fig. 2b. Interestingly, an active pocket containing two zinc ions was found in β-lactamase N1. This active pocket may become the hydrolytically active pocket for the substrate. These results showed that Zn1 atoms can be anchored via the hydrogen bonds with Ile239 and Gln242. Moreover, Zn2 atoms can also form strong hydrogen bonds with Ala367 and Ser368.
The stable 3D structure of β-lactamase N1 based on the homology model and molecular dynamics simulation. (a) RMSD of the protein backbone; (b) the whole 3D structure of β-lactamase N1 and the binding sites of zinc atoms with β-lactamase N1 in the free protein. The shaded region represents the site of catalytic activity
Molecule docking of nitrocefin with β-lactamase N1
First, to explore the interaction of the protein with the substrate, the molecular docking calculation was performed for nitrocefin and β-lactamase N1 using Autodock 4.0 software. On the basis of the docking results, 150 binding poses were obtained. The total binding poses were ranked according to the lowest-energy representative from each cluster. Then, three ligand docking poses were named pose 1, pose 2, and pose 3. Pose 1 had the lowest-energy conformation (−9.3 kJ mol−1) and the most populated cluster (53) compared with pose 2 (−8.2 kJ mol−1, 38) and pose 3 (−6.8 kJ mol−1, 19). Furthermore, pose 1 places the nitrocefin exactly in the β-lactamase N1 active pocket containing the two zinc ions, as expected. Therefore, pose 1 was chosen for further study. The lowest-energy conformation in the most populated cluster was chosen for further study. The best-ranked docking pose places the substrate, nitrocefin, in the binding pocket.
MD simulation of nitrocefin with β-lactamase N1
To explore the information on the interaction between protein residues and nitrocefin and to relax the simulation system for QM/MM calculation, molecular modeling of the complex system was performed. The initial structure was obtained from pose 1 described above.
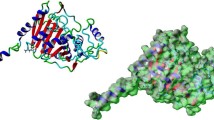
During the microsecond-scale simulation, a preferential binding model of β-lactamase N1 and nitrocefin was determined. The substrate, nitrocefin, remains in the active pocket of lactamase, as mentioned in the “Homology model of β-lactamase N1” section. On the basis of the classical molecular dynamics simulation, it was found that two zinc atoms remain in the active pocket of the protein, as shown in Fig. 3c. However, the detailed binding sites of zinc atoms in the complex are different compared with those of zinc atoms in the free protein. Figure 3d shows that except for nitrocefin, only the Zn2 atom can form the hydrogen bond with Arg183, due to the binding of the substrate, nitrocefin. The RMSD analysis showed that the complex reached equilibrium at 300 ns, and the average RMSD was 0.52 nm, indicating that the conformation of the complex structure achieved stability, as shown in Fig. 3a. The chemical structure of nitrocefin is shown in Fig. 3b. As shown in Fig. 3c, nitrocefin is bound to the active pocket of β-lactamase N1 via H-bonding and hydrophobic interactions.
General view of the complex of β-lactamase N1 with nitrocefin based on MD simulation. (a) RMSD of the protein backbone with nitrocefin from their initial coordinates as a function of time; (b) the chemical structure of nitrocefin; (c) electrostatic potentials (−60.78 to +60.78 kT/e) for the final binding cavity (blue = positive regions, red = negative regions, and white = neutral regions). The structure of the binding mode of nitrocefin with β-lactamase N1 shows the interactions of nitrocefin with Pro147, Lys168, Phe169, Gln171, and Met184; (d) the binding sites of zinc atoms with β-lactamase N1 in the complex system
Figure 3c shows in detail that Phe169 was close to the benzene ring groups on the right side of nitrocefin, thus forming strong π-π interactions between this residue and nitrocefin. In addition, two zinc ions formed interactions with the carboxyl group in the central section and the nitro group in the benzene ring, respectively. These results showed that the substrate (nitrocefin) can be anchored in the active pocket of β-lactamase N1 by interactions with Phe169 and two zinc ions. Moreover, we found that a water molecule was fixed above the β-lactam ring of nitrocefin by the formation of a hydrogen bond with Gln171 (Fig. 3c). The result of this hydrogen bond between Gln171 and water is shown in Table 2, indicating that this hydrogen bond can exist stably. To achieve a more comprehensive understanding, spatial distribution function (SDF) and radial distribution function (RDF) calculations were performed for Gln171 and the water molecule to give the probability of finding a molecule in a 3D space around a center molecule. The SDF and RDF of the water molecule around the side chain amino group of Gln171 are depicted in Fig. 4. As expected, Fig. 4a shows the RDF of the center of mass (COM) of the amino group with the main peak at 3.8 Å. This water–amino distance indicates that the amino group is embedded within the cavity formed by the water molecules. The SDF centered on the amino group of Gln171 provides a clear picture of the local structure of the water molecules. As shown in Fig. 4b, the distribution of water molecules mainly centers around the central amino group and the amino-water interactions at contour level 3, confirming the RDF results. This result revealed that the hydrogen atoms in the NH2 group show the strongest tendencies to form hydrogen bonds with water molecules, which is consistent with the above results.
To confirm the residues in the active pocket of β-lactamase N1 and determine the contributions of different residues to substrate binding, the total binding energy contributed by the residues to protein-substrate binding was calculated by the molecular mechanics generalized Born surface area (MM-GBSA) (Fig. 5a) and the molecular mechanics/Poisson-Boltzmann surface area (MM-PBSA) methods (Fig. 5b). Figure 5 shows that Phe169 had the strongest binding energy with nitrocefin, with ΔEtotal values of −3.561 kcal mol−1, indicating that Phe169 interacted strongly with the benzene ring groups in the right side of nitrocefin. Moreover, Gln171 also contributed to the binding free energy with a ΔEtotal value of −1.1305 kcal mol−1, indicating strong interactions between Gln171 and nitrocefin. The distances between all residues and nitrocefin were calculated during the last 20 ns of the simulation. As shown in Fig. 6, the shortest distances to nitrocefin were from Phe169 and Gln171 (less than 0.2 nm), which was consistent with the above results. The residues Phe169 and Gln171 are likely critical for the binding and hydrolysis of the substrate nitrocefin based on the MD simulation results.
To confirm this conclusion, the hydrolysis activity of wild-type β-lactamase N1 and two mutants (F169A, Q171A) was compared (Fig. 7). Figure 7 shows that the mutants’ hydrolysis activity is clearly lower than that of wild-type β-lactamase N1, supporting the above conclusion.
QM/MM calculations for the hydrolysis mechanism of β-lactamase N1
As shown above, on the basis of the MD simulations, the positions of the water molecule and residues in the active site of β-lactamase N1 were predicted. For further insight into the hydrolysis mechanism of β-lactamase N1, we performed ab initio QM/MM calculations for the substrate (nitrocefin) coupled with the water molecule located in the active site of the protein. Starting from the reactant structures (point I in Fig. 8), the reaction route to the hydrolysis product was modeled. According to the computed minimum energy profile, three elementary steps were identified in this reaction route. The respective local intermediate and transition states were connected based on optimizations obtained by using QM (BLYP/6-31G*) calculation. The complex systems’ free energies of β-lactamase N1 with ligands were also calculated, as shown in Fig. 9. The complex systems’ free energies were −622,060, −622,012, −622,087, −622,008, and −622,155 kcal mol−1 for reactor, TS1, IM1, TS2, and rroduct, respectively. As illustrated in Fig. 10, the distance from the oxygen atom of the water molecule to the carbon atom (C18) of nitrocefin is 2.50 Å in the original conformation, so the oxygen atom can be expected to attack the carbon atom. Analysis of the β-lactam ring hydrolysis step from the reactants to IM1 reveals that the cleavage of the C18-N15 bond is promoted by the formation of the transition state TS1. A four-membered ring chain, including C18, H, O, and N15 atoms, provides a route for the bond cleavage between C18 and N15 coupled with the formation of an H–N bond. The energy of TS1 is 40 kcal mol−1 greater than that of the intermediate IM1. Subsequently, the transfer of a proton from N15 of IM1 to C11 of the product occurred via the transition state TS2. Presumably, this step is required to generate the transition state including H–N bond cleavage and H–C bond formation. In TS2, the distances between the H atom and the N15 atom and C11 atom are 1.54 and 1.18 Å, respectively. The energy of TS2 is 50 kcal mol−1 higher than that of the intermediate IM1. On the basis of the QM/MM calculation, we finally successfully demonstrate that all chemical transformations involved in the hydrolysis of nitrocefin are well localized.
Conclusions
In this work, the interaction between residues in the active pocket of β-lactamase N1 and substrate (nitrocefin) was explored by classical molecular modeling. Molecular modeling revealed that the substrate (nitrocefin) can be anchored into the active pocket by interactions with Phe169 and two zinc ions, while Gln171 supplies the water molecule for hydrolysis. Two mutants, F169A and Q171A, were expressed for hydrolysis activity experiments. The results showed that the mutants had clearly lower hydrolysis activity than wild-type β-lactamase N1, supporting the above conclusion. Then, the chemical reaction sequences of β-lactamase N1-catalyzed hydrolysis at the atomic level were clarified by molecular modeling and quantum-mechanical/molecular mechanics (QM/MM) calculations. Here, the reaction begins with the attack of the O atom on the C18 atom of nitrocefin. Subsequently, H atom transfer from an N15 atom to a C11 atom was completed, leading to the formation of the product. Understanding this hydrolysis mechanism at the atomic level can aid in the design of new and more effective antibacterial agents.
References
The antibiotic alarm (2013). Nature 495(7440):141
Berendonk TU, Manaia CM, Merlin C, Fatta-Kassinos D, Cytryn E, Walsh F, Burgmann H, Sorum H, Norstrom M, Pons MN, Kreuzinger N, Huovinen P, Stefani S, Schwartz T, Kisand V, Baquero F, Martinez JL (2015) Tackling antibiotic resistance: the environmental framework. Nat Rev Microbiol 13(5):310–317. https://doi.org/10.1038/nrmicro3439
Bush K (2013) Proliferation and significance of clinically relevant beta-lactamases. Ann N Y Acad Sci 1277:84–90. https://doi.org/10.1111/nyas.12023
King DT, Strynadka NC (2013) Targeting metallo-beta-lactamase enzymes in antibiotic resistance. Future Med Chem 5(11):1243–1263. https://doi.org/10.4155/fmc.13.55
Hall BG, Barlow M (2005) Revised ambler classification of {beta}-lactamases. J Antimicrob Chemother 55(6):1050–1051. https://doi.org/10.1093/jac/dki130
Bush K, Jacoby GA (2010) Updated functional classification of beta-lactamases. Antimicrob Agents Chemother 54(3):969–976. https://doi.org/10.1128/AAC.01009-09
Drawz SM, Bonomo RA (2010) Three decades of beta-lactamase inhibitors. Clin Microbiol Rev 23(1):160–201. https://doi.org/10.1128/CMR.00037-09
Neuwald AF, Liu JS, Lipman DJ, Lawrence CE (1997) Extracting protein alignment models from the sequence database. Nucleic Acids Res 25(9):1665–1677
Carfi A, Pares S, Duee E, Galleni M, Duez C, Frere JM, Dideberg O (1995) The 3-D structure of a zinc metallo-beta-lactamase from Bacillus cereus reveals a new type of protein fold. EMBO J 14(20):4914–4921
Aravind L (1999) An evolutionary classification of the metallo-ß-lactamase fold proteins. In Silico Biol 1(2):69–91
Daiyasu H, Osaka K, Ishino Y, Toh H (2001) Expansion of the zinc metallo-hydrolase family of the beta-lactamase fold. FEBS Lett 503(1):1–6
Yang Y, Rasmussen BA, Bush K (1992) Biochemical characterization of the metallo-beta-lactamase CcrA from Bacteroides fragilis TAL3636. Antimicrob Agents Chemother 36(5):1155–1157
Felici A, Amicosante G, Oratore A, Strom R, Ledent P, Joris B, Fanuel L, Frere JM (1993) An overview of the kinetic parameters of class B beta-lactamases. Biochem J 291(Pt 1):151–155
Felici A, Amicosante G (1995) Kinetic analysis of extension of substrate specificity with Xanthomonas maltophilia, Aeromonas hydrophila, and Bacillus cereus metallo-beta-lactamases. Antimicrob Agents Chemother 39(1):192–199
Rasmussen BA, Yang Y, Jacobus N, Bush K (1994) Contribution of enzymatic properties, cell permeability, and enzyme expression to microbiological activities of beta-lactams in three Bacteroides fragilis isolates that harbor a metallo-beta-lactamase gene. Antimicrob Agents Chemother 38(9):2116–2120
Concha NO, Rasmussen BA, Bush K, Herzberg O (1996) Crystal structure of the wide-spectrum binuclear zinc beta-lactamase from Bacteroides fragilis. Structure 4(7):823–836
Fitzgerald PM, Wu JK, Toney JH (1998) Unanticipated inhibition of the metallo-beta-lactamase from Bacteroides fragilis by 4-morpholineethanesulfonic acid (MES): a crystallographic study at 1.85-a resolution. Biochemistry 37(19):6791–6800. https://doi.org/10.1021/bi9730339
Davies RB, Abraham EP (1974) Metal cofactor requirements of beta-lactamase II. Biochem J 143(1):129–135
Bandoh K, Muto Y, Watanabe K, Katoh N, Ueno K (1991) Biochemical properties and purification of metallo-beta-lactamase from Bacteroides fragilis. Antimicrob Agents Chemother 35(2):371–372
Garces F, Fernandez FJ, Montella C, Penya-Soler E, Prohens R, Aguilar J, Baldoma L, Coll M, Badia J, Vega MC (2010) Molecular architecture of the Mn2+−dependent lactonase UlaG reveals an RNase-like metallo-beta-lactamase fold and a novel quaternary structure. J Mol Biol 398(5):715–729. https://doi.org/10.1016/j.jmb.2010.03.041
Salimraj R, Zhang L, Hinchliffe P, Wellington EM, Brem J, Schofield CJ, Gaze WH, Spencer J (2016) Structural and biochemical characterization of Rm3, a subclass B3 metallo-beta-lactamase identified from a functional metagenomic study. Antimicrob Agents Chemother 60(10):5828–5840. https://doi.org/10.1128/AAC.00750-16
Ghavami A, Labbe G, Brem J, Goodfellow VJ, Marrone L, Tanner CA, King DT, Lam M, Strynadka NC, Pillai DR, Siemann S, Spencer J, Schofield CJ, Dmitrienko GI (2015) Assay for drug discovery: synthesis and testing of nitrocefin analogues for use as beta-lactamase substrates. Anal Biochem 486:75–77. https://doi.org/10.1016/j.ab.2015.06.032
Qiu J, Niu X, Dong J, Wang D, Wang J, Li H, Luo M, Li S, Feng H, Deng X (2012) Baicalin protects mice from Staphylococcus aureus pneumonia via inhibition of the cytolytic activity of alpha-hemolysin. J Infect Dis 206(2):292–301. https://doi.org/10.1093/infdis/jis336
Newman JA, Hewitt L, Rodrigues C, Solovyova A, Harwood CR, Lewis RJ (2011) Unusual, dual endo- and exonuclease activity in the degradosome explained by crystal structure analysis of RNase J1. Structure 19(9):1241–1251. https://doi.org/10.1016/j.str.2011.06.017
Hess B, Kutzner C, van der Spoel D, Lindahl E (2008) GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput 4(3):435–447. https://doi.org/10.1021/ct700301q
Pronk S, Pall S, Schulz R, Larsson P, Bjelkmar P, Apostolov R, Shirts MR, Smith JC, Kasson PM, van der Spoel D, Hess B, Lindahl E (2013) GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 29(7):845–854. https://doi.org/10.1093/bioinformatics/btt055
Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30(16):2785–2791
Wang J, Wang W, Kollman PA, Case DA (2006) Automatic atom type and bond type perception in molecular mechanical calculations. J Mol Graph Model 25(2):247–260. https://doi.org/10.1016/j.jmgm.2005.12.005
Jakalian A, Jack DB, Bayly CI (2002) Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II. Parameterization and validation. J Comput Chem 23(16):1623–1641. https://doi.org/10.1002/jcc.10128
Punkvang A, Saparpakorn P, Hannongbua S, Wolschann P, Beyer A, Pungpo P (2010) Investigating the structural basis of arylamides to improve potency against M. Tuberculosis strain through molecular dynamics simulations. Eur J Med Chem 45(12):5585–5593. https://doi.org/10.1016/j.ejmech.2010.09.008
Schaffner-Barbero C, Gil-Redondo R, Ruiz-Avila LB, Huecas S, Lappchen T, den Blaauwen T, Diaz JF, Morreale A, Andreu JM (2010) Insights into nucleotide recognition by cell division protein FtsZ from a mant-GTP competition assay and molecular dynamics. Biochemistry 49(49):10458–10472. https://doi.org/10.1021/bi101577p
Acknowledgments
The authors acknowledge the financial support by the National Nature Science Foundation of China [Grant no. 31572566 to X. D. N].
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 36 kb)
Rights and permissions
About this article
Cite this article
Yu, Y., Wang, X., Gao, Y. et al. Molecular modeling and QM/MM calculation clarify the catalytic mechanism of β-lactamase N1. J Mol Model 25, 118 (2019). https://doi.org/10.1007/s00894-019-4001-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-019-4001-z