Abstract
Targeted selection and continuous self-crossing were utilized to breed a new Saccharina variety. After five-generation selection breeding, the new high-yield variety “Ailunwan” with stable genetic traits was obtained. Blade length, width, thickness, and fresh weight of variety “Ailunwan” were increased by 20.2, 11.0, 49.5, and 27.3 %, respectively, and the dry matter content increased by 11.8 % compared to the control (a widely used commercial variety). Frequency distributions and coefficients of variation of blade length, width, and fresh weight are discussed, and results showed that variety “Ailunwan” had more excellent traits and lower genetic variation than the control. Grey relational analysis of fresh weight-related traits showed that each related trait was in the same correlation order both in the fourth and in the fifth generations of “Ailunwan.” On the basis of sequence-related amplified polymorphism (SRAP) analysis, the fifth generation of “Ailunwan” had the lowest gene diversity (H = 0.234) compared to the other five varieties including “Zaohoucheng” (H = 0.324), “Dongfang No. 2” (H = 0.260), “Dongfang No. 3” (H = 0.265), “Pingbancai” (H = 0.249), and the control (H = 0.270), which displayed the advantage of genetic stability. Unweighted Pair Group Method with Arithmetic Mean (UPGMA) cluster analysis based on SRAP marker also revealed that variety “Ailunwan” can be distinguished from other cultivation varieties in China and its genetic structure is relatively stable.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Saccharina (Laminariales, Phaeophyceae) is one of the most important marine algal genera with respect to both its economic importance and its global distribution (Kain 1979). Due to their rich iodine, iron, calcium, protein, vitamins, alginate, mannitol, fucoidin, and other economic compositions, Saccharina seaweeds not only are used as food for humans and animals but also have become an important raw material of the marine chemical industry (Bixler and Porse 2011; Jensen 1993).
In China, many breeding technologies such as selection breeding in the early 1960s (Fang et al. 1962), radioactive mutation in the 1970s (IOIMF Institute of Oceanology, Institute of Marine Fisheries 1976), interspecies hybridization in the 1980s (Fang et al. 1985), intra-species hybridization breeding in the 1980s and 1990s (Zhang et al. 2007), and direct use of hybrids in the 1990s (Li et al. 2007a) were established and more than ten cultivation varieties have been bred and used successfully. Since the mid-1990s, some Saccharina varieties have been approved by the Chinese Approving Committee of Aquacultural Elite Varieties and Stock Seeds including “901” (approval no. GS01001-1997), “Dongfang No. 2” (approval no. GS02-001-2004), “Rongfu” (approval no. GS02-002-2004), “Dongfang No. 3” (approval no. GS02-002-2007), and “Dongfang No. 6” (approval no. GS02-004-2013). The yield of these varieties has been much improved, but the genetic stability has not been dealt properly with. Therefore, these improved varieties can lose their economic characteristics in the course of cultivation. In addition, since the early twenty-first century, there have been quite limited new improved varieties in China. Breeding and cultivation of the coverage ratio for improved varieties are still insufficient. Aiming to obtain more stable, elite Saccharina varieties for cultivation, we used hybridization of gametophytes, continuous self-crossing, and targeted selection to develop the high-yield variety “Ailunwan” and employed Grey relational analysis as well as molecular marker to evaluate its genetic stability.
Although molecular markers, such as RAPD (Billot et al. 1999; Yotsukura et al. 2001), AFLP (Zhang et al. 2008), SSR (Shi et al. 2008), and ISSR (Wang et al. 2005), have been applied to assess population genetic diversity in Saccharina, they have not yet been widely used in evaluating the genetic stability evaluation. Current stability evaluation of Saccharina breeding only depends on measurement, weight, and component detection of traits. Based on traditional methods, sequence-related amplified polymorphism (SRAP) was conducted to evaluate genetic stability of breeding varieties for the first time in this study, thus providing the molecular evidence during the “Ailunwan” breeding process. SRAP is a novel molecular marker technique without using restriction endonuclease. A single step with PCR can detect polymorphism of DNA sites and requires less labor. In addition, SRAP aims at amplifying open reading frames (ORFs). Therefore, it is more stable, reproducible, and informative than other molecular markers (Li and Quiros 2001). Moreover, Grey relational analysis is considered to be able to overcome the controversy caused by evaluation of single index (Lai et al. 2009); it was also first used to study the correlation between fresh weight and related traits in order to confirm the genetic stability of new variety.
After 5-year selection breeding, the new Saccharina variety “Ailunwan” was obtained. As shown in a series of evaluations, it was found to be high in yield and algin. More importantly, variety “Ailunwan” had acquired higher genetic stability compared to the other improved varieties.
Materials and methods
Sources of the parents and selection process
“Yuanza No. 10,” a high-yield variety selected from Saccharina japonica and Saccharina latissima, was used as female parent, while “Fujian,” a later maturing variety of S. japonica, was chosen as male parent. The new variety bred in this study was selected in the open sea near Rongcheng city, China. The most widely used commercial variety of S. japonica without any systematic selection was chosen as the control.
Hybridization of gametophytes, continuous self-crossing, and targeted selection have been adopted to breed the new variety since the year of 2005. Culture of gametophyte clones, crossing of gametophytes, and raising of young sporophytes (sporelings) used the method developed by our team and reported by Zhang et al (2011). The female and male gametophytes isolated by sterilized glass pipette one by one were cultured separately in nutrient-supplemented seawater (NaNO3-N 4 mg L−1, KH2PO4-P 0.4 mg L−1) at 10 °C and under 30 μmol photons m−2 s−1 irradiance. After the isolated gametophytes reached the diameter of 2 mm, they were cut into fragments (eight to ten cells). Male and female gametophyte clones were mixed 1:1 with final concentration of 105 cells mL−1. Then, mixed gametophyte clones (about 3000-mL mixture) were sprinkled onto palm rope and were cultured in filtered seawater enriched with nutrients. Sporelings generated from the mixture of gametophyte clones were taken out to the circulating seawater, when they grew up to 0.8–1 cm in length. Then, they were transferred from the laboratory to the open sea in the first or second 10 days of October.
During the targeted selection process, individuals with high-yield trait were selected, based on the highest means of blade length, width, and fresh weight. In addition, phenotypic traits including blade color, longitudinal groove, and blade base shape were taken into consideration. The 20 best integrated individuals with the largest values of blade length, width, and fresh weight were selected from 1000 individuals of the first filial generation (F1) and used to make ten new self lines. Five cycles of self-crossing and selection were carried out, and 1/50 best individuals were selected during each generation. Forty individuals with best biological traits each from the generation F2 (total 2000 individuals) and F3 (total 2000 individuals) were selected. For generation F4 and F5, 200 individuals were selected from 10,000 individuals, respectively. Moreover, large-scale reproduction and cultivation were tested.
Traits evaluation and Grey relational analysis
Blade length, width, fresh weight, and ratio of fresh to dry weight of each generation of “Ailunwan” (F1–F5) were consecutively measured and were compared with those of the control. In addition, frequency distributions and coefficient of variations of blade length, width, and fresh weight were analyzed between “Ailunwan” (F5) and the control. Fifty complete individuals used for evaluation were selected randomly, and all of the above traits were measured using previously described methods (Zhang et al. 2011). The quantitative traits are influenced by the seawater condition. Hence, “Ailunwan” and the control were all cultured in the same sea area. In the meantime, all indicators were measured at the same designated time. Five individuals each from “Ailunwan” and the control were used to analyze the content of algin, crude protein, fat, and ash and 12 amino acids following the methods reported by Ji (1997). SPSS20.0 software was used for statistical analysis. Differences between two groups were considered as significant if P < 0.05 with t test.
Using DPS7.05 software, Grey relational analysis for “Ailunwan,” “Pingbancai” (widely cultured in the open sea near Rongcheng city), and the control was applied to illustrate the correlation between fresh weight and other related traits including blade length, width, thickness, and fascia width.
Molecular marker analysis
To evaluate the genetic stability, SRAP analysis was performed for six varieties including “Ailunwan,” “Zaohoucheng,” “Dongfang No. 2,” “Dongfang No. 3,” “Pingbancai” and the control according to the procedure described by Li and Quiros (2001) with minor modification. Twenty-four individuals from each variety were selected randomly for the analysis, and all were provided by the Culture Collection of Seaweed at the Ocean University of China. Genomic DNAs were isolated with improved CTAB method (Guillemaut and Drouard 1992).
Primers synthesized at Invitrogen, Shanghai, China, with clearly separated bands, stable amplification, and rich polymorphism were selected from 88 primer combinations which were reported by Ding et al. (2010). Approximately 50 ng DNA was amplified in a final PCR volume of 20 μL which contained 0.5 μM primer, 125 μM dNTPs, 1 U Taq polymerase (TaKaRa Biotech, Japan), and 2.0 μL 10× buffer (2 mM MgCl2) by means of Thermal Cyclers (ABI 9700, USA). The amplification was performed for 5 min of denaturing at 95 °C; then 5 cycles of 1 min at 94 °C, 1 min at 35 °C, and 2 min at 72 °C; and then 35 cycles of 1 min at 94 °C, 1 min at 48 °C, and 2 min at 72 °C, with a final extension step of 10 min at 72 °C.
All PCR products were separated on 6 % denaturing polyacrylamide gel (1× TBE buffer) at 60 W for 2 h and visualized by silver staining (Bassam et al. 1991). A 20-bp DNA ladder was used as a reference marker for allele size determination.
The silver-stained bands in each gel were scored as presence (1) or absence (0) manually, and only reproducible bands were used to construct the original binary data matrix. The percentage of polymorphic loci among populations (P), Nei’s genetic diversity (H), Shannon’s information index (I), and effective number of alleles (Ne) were calculated from the SRAP data using POPGENE version 1.31 (Yeh et al. 1999). The phylogenetic dendrogram was constructed based on the Unweighted Pair Group Method with Arithmetic Mean (UPGMA) by using TFPGA software (Miller 1997) and the reliability of the aggregating method checked by bootstrap analysis (1000 replicates).
Results
After five-generation self-crossing and selection, variety “Ailunwan” was obtained in 2010. It has a dark brown blade with oblate stipe, round base, and well-developed rhizoid (Fig. 1). As to quantitative traits, the blade length, width, thickness, and fresh weight of variety “Ailunwan” have reached 285.1 ± 38.4 cm, 39.4 ± 6.3 cm, 4.17 ± 0.49 mm, and 1614.0 ± 399.2 g, respectively (Table 1). Compared with the control, they have increased by 20.2, 11.0, 49.5, and 27.3 %, respectively, and the dry matter content increased by 11.8 %, showing obvious advantages of the variety “Ailunwan.”
For different generations of “Ailunwan,” the blade length ranged from 285.1 to 524.2 cm and was always significantly larger than that of the control (p < 0.05) (Fig. 2). Blade width ranged from 36.8 to 41.6 cm, while the control ranged from 31.1 to 37.4 cm. “Ailunwan” was also significantly larger than that of the control (p < 0.05) except in 2008 (Fig. 3). The average fresh weights and fresh to dry weight ratios of “Ailunwan” and the control are shown in Fig. 4. The average fresh weight of each “Ailunwan” generation was always greater than that of the control (p < 0.05). At the same time, ratios of fresh to dry weight for five consecutive years were all significantly better than that of the control (p < 0.05).
Frequency distributions and coefficients of variation of variety “Ailunwan” and the control were analyzed. Blade length, width, and fresh weight of “Ailunwan” were mainly concentrated in 240–320 cm, 34–50 cm, and 1.2–2.0 kg respectively, while the control was mainly 200–280 cm, 26–42 cm, and 0.8–1.2 kg, indicating that these traits of variety “Ailunwan” were all better than the control. The results are illustrated in Figs. 5, 6, and 7. Furthermore, coefficients of variation of above traits of “Ailunwan” were 0.135, 0.171, and 0.247 while those of the control were 0.182, 0.203, and 0.313. Coefficients of variation of these traits in “Ailunwan” were all lower than those of the control, which showed that “Ailunwan” as a new variety was not significantly different and the genetic background had been sufficiently homogenized.
Grey relational degrees of F4 and F5 were calculated between fresh weight and related traits. They were both ranked as follows: fascia width > blade width > blade length > blade thickness. However, the order for “Pingbancai” was blade width > blade length > fascia width > blade thickness in 2009 and fascia width > blade width > blade length > blade thickness in 2010. For the control, the order was blade width > blade thickness > fascia width > blade length in 2009 and blade width > fascia width > blade thickness > blade length in 2010. This also showed that “Ailunwan” with five-generation breeding possessed higher genetic stability. The Grey relational coefficients between fresh weight and other related traits in three Saccharina varieties are shown in Table 2.
Contents of major economical components were different between “Ailunwan” and the control (Table 3). Except for crude protein, fat, and ash, variety “Ailunwan” was close to, or higher than, the control in the content of algin and 12 amino acids. The content of algin was 17.84 % in “Ailunwan,” which was 43.29 % higher than the control. It is clear that the biochemical traits of variety “Ailunwan” were satisfactory. In addition, “Ailunwan” contained seven essential amino acids for humans including lysine, threonine, valine, methionine, isoleucine, leucine, and phenylalanine. The contents of five of these were higher in “Ailunwan” than in the control. “Ailunwan” showed the excellent economic value.
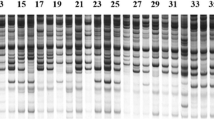
Fifteen of 88 primer combinations which produced stable and reproducible amplification patterns (Table 4) were chosen to generate all SRAP profiles for genetic structure research among “Ailunwan” and the other five Saccharina varieties. Using the selected 15 SRAP primer combinations, 252 amplified fragments ranging from 80 to 700 bp were identified from 144 individuals, 249 of which were polymorphic bands. The percentage of polymorphic loci (P) per variety ranged from 64.68 % (“Ailunwan”) to 82.14 % (“Zaohoucheng”). Nei’s genetic diversity (H) ranged from 0.234 (“Ailunwan”) to 0.324 (“Zaohoucheng”) and Shannon’s information index (I) from 0.347 (“Ailunwan”) to 0.472 (“Zaohoucheng”) (Table 5). The results showed that variety “Ailunwan” had the lowest genetic diversity compared to the other five widely used Saccharina varieties.
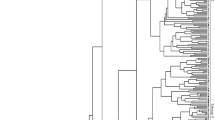
A phylogenetic dendrogram was constructed by UPGMA (Fig. 8). “Ailunwan,” “Zaohoucheng,” “Dongfang No. 3” and “Pingbancai” clustered together first and then “Dongfang No. 2.” The control formed an independent cluster. Different individuals of the same varieties were clustered together, indicating that the genetic structure was relatively stable and had genetic differences among varieties.
Discussion
After continuous selection breeding, variety “Ailunwan” was provided with stable excellent traits. It has an advantage over the control not only in the quantitative traits, but also in major economical components. Due to its high yield and high content of algin, “Ailunwan” could be used as an important chemical raw material. Based on its excellent traits, “Ailunwan” has been approved by the Chinese Approving Committee of Aquacultural Elite Varieties and Stock Seeds in 2011 with approval number GS01-010-2010.
Fresh weight is a complex biological trait and is influenced by many quantitative traits such as blade length, width, thickness, and so on. In the present evaluation system of fresh weight for Saccharina, the different traits were compared and analyzed (Li et al. 2007a; Zhang et al. 2007; 2011), which separated the comprehensive influence of each trait to fresh weight. Grey relational analysis is considered as an effective method to evaluate materials fully combined with the different factors and overcomes the controversy caused by the evaluation of one factor (Li and Shang 2007b). Since its development, it has been widely used in crop breeding, especially in the comprehensive evaluation of agronomic traits (Lai et al. 2009; Wang et al. 2013; Yin et al. 2006). In this study, Grey relational analysis was adopted for the first time to examine the correlation coefficient between fresh weight and the related traits (blade length, width, thickness, and fascia width) in “Ailunwan,” “Pingbancai,” and the control. The correlation order of variety “Ailunwan” in 2009 was the same as that in 2010, while they are different for “Pingbancai” and the control. “Pingbancai” and the control were both the cultivars without genetic improvement. Artificial breeding via the strategy of this research improved the genetic stability. Results of Grey relational analysis showed that fascia width and frond width were the main factors which influenced fresh weight. Therefore, we should focus on the selection and identification of these traits in order to obtain high-yield varieties in the future. Meanwhile, other traits should be combined in the process of breeding.
Molecular marker analysis is an effective method to reveal the genetic difference and genetic structure of the population. SRAP is a novel molecular marker technique aimed at amplifying ORFs which are the essential components of genes and relate to functional genes (Li and Quiros 2001). The observed polymorphism originates from the length variation of extrons, introns, promoters, and spacers among individuals or species (Ferriol et al. 2003). Compared to other molecular markers, SRAP is more efficient and can disclose more accurate genetic information. It has been applied to investigate genetic structure in many plant species including Brassica (Li and Quiros 2001), cotton (Lin et al. 2004), alfalfa (Vandemark et al. 2006), and Pinus koraiensis (Feng et al. 2009). However, it has only been used to explore the genetic diversity of Porphyra/Pyropia in seaweeds (Qiao et al. 2007). The SRAP analytical system was successfully adapted for the genetic analysis of Saccharina in this study. The diversity index showed that “Ailunwan” had acquired higher genetic stability through 5-year continuous selection than the other five varieties. “Dongfang No. 2” (S. japonica × Saccharina longissima) and “Dongfang No. 3” (S. japonica × S. longissima) were both the direct hybrids of the first filial generation from the female gametophytes of S. japonica and the male of S. longissima. “Pingbancai” (S. japonica × S. longissima) were hybridized by the S. japonica (female) and S. longissima (male). Genetic diversities of these three Saccharina varieties lie at medium level of the six varieties in this work. However, although variety “Zaohoucheng” (S. japonica) was an inbred line of S. japonica, it had the highest genetic diversity indicating poor genetic stability. The data demonstrated that the breeding strategy might affect the genetic structure. Cluster analysis based on SRAP marker suggested that “Ailunwan” can be distinguished from the other five varieties and forms an independent variety.
The cultivation area of “Ailunwan” has reached more than 2800 ha in Shandong province so far. The breeding and promotion of improved varieties have promoted the industry of Saccharina cultivation effectively in China. Breeding and application of variety “Ailunwan” demonstrated that it is feasible to breed elite Saccharina varieties by hybridization of gametophytes, continuous self-crossing, and targeted selection. Actually, this is similar to the interspecies crossing of crops (for example, rice). Kelp (Saccharina) are the only macroalgae that are comparable with rice in terms of cultivation, pure lines (gametophyte clones to rice varieties), and breeding strategies (selection, hybridization, and selection and direct use of hybrid) (Zhang et al. 2007). Although it is feasible, breeding through this strategy is labor intensive and time-consuming. Genetic breeding of rice by utilizing natural resistance or tolerance genes is the most economic and efficient way to combat or adapt to these stresses (Luo and Yin 2013). Unfortunately, basic research in Saccharina lags far behind that in rice. If the gap between rice and Saccharina was narrowed, a desirable trait of Saccharina can be designed before initiating a breeding program according to closely linked molecular markers and can then be selected with the assistance of these markers. This means that Saccharina breeding will achieve the expected efficiency easily in the future.
References
Bassam BJ, Caetano-Anolles G, Gresshoff PM (1991) Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal Biochem 196:80–83
Billot C, Boury S, Benet H, Kloareg B (1999) Development of RAPD markers for parentage analysis in Laminaria digitata. Bot Mar 42:307–314
Bixler HJ, Porse H (2011) A decade of change in the seaweed hydrocolloids industry. J Appl Phycol 23:321–335
Ding WD, Cao ZM, Cao LP (2010) Molecular analysis of grass carp (Ctenopharyngodon idella) by SRAP and SCAR molecular markers. Aquac Int 18:575–587
Fang TC, Wu CY, Jiang BY, Li JJ, Ren KJ (1962) The breeding of a new breed of Haidai (Laminaria japonica) and its preliminary genetic analysis. Acta Bot Sin 10:197–209 (in Chinese with English abstract)
Fang TC, Ou YL, Cui JJ (1985) Breeding of hybrid Laminaria “Danza No. 10”—an application of the Laminaria haploid cell clones. J Shandong Coll Oceanol 15:64–72 (in Chinese with English abstract)
Feng FJ, Chen MM, Zhang DD, Sui X, Han SJ (2009) Application of SRAP in the genetic diversity of Pinus koraiensis of different provenances. Afr J Biotechnol 8:1000–1008
Ferriol M, Pico B, Nuez F (2003) Genetic diversity of a germplasm collection of Cucurbita pepo using SRAP and AFLP markers. Theor Appl Genet 107:271–282
Guillemaut P, Drouard LM (1992) Isolation of plant DNA: a fast, inexpensive, and reliable method. Plant Mol Biol Report 10:60–65
IOIMF (Institute of Oceanology, Institute of Marine Fisheries) (1976) The breeding of new varieties of Haidai (Laminaria japonica) with high production and high iodine content. Sci Sin 19:243–252 (in Chinese with English abstract)
Jensen A (1993) Present and future needs for algae and algal products. Hydrobiologia 269:13–23
Ji MH (1997) Seaweed chemistry. Science, Beijing, p 777 (in Chinese)
Kain JM (1979) A view on the genus Laminaria. Oceanogr Mar Biol Annu Rev 17:101–161
Lai YP, Li J, Zhang ZQ, Dong XF, Liu XC, Wei HT, Hu XR, Peng ZS, Yang WY (2009) Grey correlation analysis of morphological traits related to drought tolerance of wheat at seedling stage. J Triticeae Crops 29(6):1055–1059 (in Chinese with English abstract)
Li G, Quiros CF (2001) Sequence related amplified polymorphism (SRAP), a new marker system based on a simple PCR reaction: its application to mapping and gene tagging in Brassica. Theor Appl Genet 103:455–461
Li ZB, Shang XW (2007b) Evaluating quality of flour for Lanzhou hand-extended noodles by Grey related degree analysis. J Triticeae Crops 27(1):59–62, in Chinese with English abstract
Li XJ, Cong YZ, Yang GP, Qu SC, Li ZL, Wang GW, Zhang ZZ, Luo SJ, Dai HL, Xie JZ, Jiang JL, Wang TY (2007a) Trait evaluation and trial cultivation of Dongfang No. 2, the hybrid of a male gametophyte clone of Laminaria longissima (Laminariales, Phaeophyta) and a female one of L. japonica. J Appl Phycol 19:139–151
Lin ZX, Zhang XL, Nie YC (2004) Evaluation of application of a new molecular marker SRAP on analysis of F2 segregation population and genetic diversity in cotton. Acta Genet Sin 31:622–626
Luo YC, Yin ZC (2013) Marker-assisted breeding of Thai fragrance rice for semi-dwarf phenotype, submergence tolerance and disease resistance to rice blast and bacterial blight. Mol Breed 32:709–721
Miller MP (1997) Tools for population genetic analysis (TFPGA) 1.3: a window program for the analysis of allozyme and molecular population genetic data. Computer software distributed by author
Qiao LX, Liu HY, Guo BT, Weng ML, Dai JX, Duan DL, Wang B (2007) Molecular identification of 16 Porphyra lines using sequence-related amplified polymorphism markers. Aquat Bot 87:203–208
Shi YY, Yang GP, Liao MJ, Liu YJ, Shang S, Li XJ, Cong YZ (2008) Comparative study on the microsatellite DNA polymorphism of the gametophytes of L. japonica and L. longissima. Period Ocean Univ China 38(2):303–308 (in Chinese with English abstract)
Vandemark GJ, Ariss JJ, Bauchan GA, Larsen RC, Hughes TJ (2006) Estimating genetic relationships among historical sources of alfalfa germplasm and selected cultivars with sequence related amplified polymorphisms. Euphytica 152:9–16
Wang XL, Liu CL, Li XJ, Cong YZ, Duan DL (2005) Assessment of genetic diversities of selected Laminaria (Laminarales, Phaeophyta) gametophytes by inter-simple sequence repeat analysis. J Integr Plant Biol 47(6):753–758
Wang ZK, Wang C, Guan FC (2013) Comprehensive evaluation of walnuts from different growing regions. Food Sci 34(15):100–103 (in Chinese with English abstract)
Yeh FC, Yang RC, Boyle T (1999) POPGENE, version 1.31. Microsoft window-based freeware for population genetic analysis. Quick user guide (Francis Yeh, University of Alberta, Canada). www.ualberta.ca/-fyeh
Yin L, Lu XP, Li MN, Guo J (2006) The Grey relation analysis and evaluation of hybrid Pacesetter. J Grassl 28(3):21–25 (in Chinese with English abstract)
Yotsukura N, Kawai T, Motomura T, Ichimura T (2001) Random amplified polymorphic DNA markers for three Japanese Laminaria species. Fish Sci 67:857–862
Zhang QS, Tang XX, Cong YZ, Qu SC, Luo SJ, Yang GP (2007) Breeding of an elite Laminaria variety 90-1 through interspecific gametophyte crossing. J Appl Phycol 19:303–311
Zhang QS, Shi YY, Cong YZ, Qu SC, Shang S, Yang GP (2008) AFLP analysis of the gametophyte clones derived from introduced Laminaria (Phaeophyta) and cultured varieties of China. Period Ocean Univ China 28:429–435 (in Chinese with English abstract)
Zhang J, Liu Y, Yu D, Song HZ, Cui JJ, Liu T (2011) Study on high-temperature-resistant and high-yield Laminaria variety “Rongfu”. J Appl Phycol 23:165–171
Acknowledgments
We thank Lichao Wang, Guojun Li, Yi Li, and Lei Wang from Xunshan Group Co., Ltd for their work in Saccharina cultivation. This work was supported by Special Fund for Agro-scientific Research in the Public Interest (201203063), Science and Technology Development Planning Project of Shandong Province (2014GGE29091), Agricultural Seed Project of Shandong Province, and TaiShan industrial Experts Programme.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhang, J., Liu, T., Bian, D. et al. Breeding and genetic stability evaluation of the new Saccharina variety “Ailunwan” with high yield. J Appl Phycol 28, 3413–3421 (2016). https://doi.org/10.1007/s10811-016-0810-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10811-016-0810-y