Abstract
In order to provide information for further research and utilization of Erianthus arundinaceus (Retz.) Jesw., the phenotypic variation and genetic diversity in the collection of E. arundinaceus (Retz.) Jesw. germplasm in Guangxi, China were investigated. The coefficient of variation (CV) in four quantitative traits, plant height, stalk diameter, spikelet length and number of panicle nodes, of 50 E. arundinaceus (Retz.) Jesw. clones ranged from 12.71 to 16.14 %, stalk diameter had the highest coefficient of variation, followed by spikelet length, and number of panicle nodes showed the lowest. Fifteen start condon targeted polymorphism (SCoT) marker primers were used to assess the genetic diversity of the 50 collections of E. arundinaceus (Retz.) Jesw. germplasm. A total of 336 genotypes were investigated for SCoT polymorphism, which produced 284 amplicons with 83.97 % polymorphism, indicating that E. arundinaceus (Retz.) Jesw. germplasm had abundant diversity in Guangxi. The Jaccard’s similarity coefficients among these accessions varied from 0.61 to 0.88 with an average of 0.71. The UPGMA clustering analysis showed the 50 E. arundinaceus (Retz.) Jesw. accessions could be divided into four major groups at the similarity coefficient level 0.73. The UPGMA dendrogram reflected the geographic distribution patterns of these accessions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sugarcane is the most important sugar crop in tropical and subtropical regions of the world, and it has also become a very important bio-energy crop recently (Li 2010; Grivet and Arruda 2001). The cane sugar production accounts for over 90 % of total consumable sugar in China (Que et al. 2014). However, narrow genetic background is now the main restrictive factor in sugarcane breeding. Modern sugarcane cultivars have complex polyploidy genomes resulted from the crosses among several species of the Saccharum genus, namely S. officinarum, S. spontaneum and S. barberi (Li 2010). Since it is difficult to produce advantageous breakthrough through crossing between modern sugarcane varieties, and thus, widening the genetic basis of sugarcane with more wild germplasm is regarded as one of the key approaches for making a significant progress in sugarcane breeding. Erianthus arundinaceus (Retz.) Jesw. is the important wild germplasm of sugarcane with many useful characteristics for breeding, including strong vegetative vigor, good tillering ability and strong resistance to abiotic and biotic stresses such as drought and mosaic virus (Nair et al. 1993; Li 2010). However, research on the main quantitative traits variation and genetic diversity of E. arundinaceus (Retz.) Jesw. in China is limited.
In recent years, molecular markers have been widely used in crop genetic diversity analysis, offspring identification and gene mapping, etc. (Chen et al. 2010; Cai et al. 2012; Liu et al. 2012a, b; Su et al. 2012; Luo et al. 2013; Song et al. 2013; Zhang et al. 2013; Kalwade and Devarumath 2014; Sharma et al. 2014; Hu et al. 2015; Nerkar et al. 2015; Priya et al. 2005; Rajesh et al. 2015; You et al. 2015; Satya et al. 2015; Zan et al. 2015; Feng et al. 2016). Among the molecular markers reported, start condon targeted polymorphism (SCoT) has been proven to be effective for analyzing crop genetic diversity and clustering relationship (Collard and Mackill 2009; Xiong et al. 2010; Wu et al. 2010, 2013; Chen et al. 2012; Zhao et al. 2012; Wei et al. 2012). In the present study, we analyzed the phenotypic variation, genetic diversity and clustering relationship of fifty accessions of E. arundinaceus (Retz.) Jesw. collected in Guangxi, China, in order to provide a reference for further research and utilization of the wild germplasm in sugarcane improvement.
Materials and Methods
Plant Materials
The experiment was conducted at Sugarcane Research Institute, Guangxi Academy of Agricultural Sciences/Sugarcane Research Center, Chinese Academy of Agricultural Sciences, Nanning, China in 2015. Fifty E. arundinaceus (Retz.) Jesw. accessions were collected from its originally distributed areas in Guangxi, China, and preserved at the nursery of Sugarcane Research Institute, Guangxi Academy of Agricultural Sciences. All the 50 accessions could be distinguished morphologically. The detailed information of the studied accessions is summarized and presented in Table 1.
Traits Survey
The experiment was laid out in a randomized block design with three replications. Each accession was accommodated in a plot having three rows of 5 m in length with spacing 1.2 m between rows. The field management was done as for commercial sugarcane production. Observations were recorded for four quantitative traits viz. plant height, stalk diameter, spikelet length and number of panicle nodes.
DNA Extraction
Genomic DNA was extracted from approximately 0.5 g of fresh leaf samples using a modified SDS method as described by Liu et al. (2012b). The quantity and quality of extracted DNA were verified using the spectrophotometer and agarose gel electrophoresis and then stored at −20 °C for PCR amplification.
SCoT Marker Analysis
SCoT reaction condition and protocol were adapted from the previous reports with minor modifications (Wu et al. 2010; You et al. 2015). All primers were synthesized by Shanghai Sangon Biotech (Shanghai, China) (Table 2). PCR amplification of these primers was carried out in a thermal cycle in 20 μL volume, with the reaction mixture containing 2 μL of 10 × PCR buffer, 0.4 μL dNTPs (10 mM), 2 μL primer (10 μM), 0.2 μL Taq DNA polymerase (5 U/μL) and 0.5 μL template DNA (50 ng/μL). Cycling conditions were: initial denaturation at 94 °C for 4 min followed by 36 cycles of 94 °C for 50 s, 50 °C for 40 s, 72 °C for 2 min and final extension at 72 °C for 5 min. After amplification, PCR products were run on a polyacrylamide gel as described for the SCoT products.
Data Analysis
Only clearly amplified products from the SCoT primers were used for further analysis. The bands were scored as absent (0) or present (1). Software package NTSYS-pc version 2.0 was used for further analysis using the scored results. Genetic similarity analysis between the accessions was estimated using similarity matrix, generated by calculating Jaccard’s similarity coefficient. These similarity coefficients were used for cluster analysis, and a dendrogram was constructed by the unweighted pair group method (UPGMA) and the sequential hierarchical and nested clustering (SHAN) routine in the NTSYS program. Four quantitative traits viz. plant height, stalk diameter, spikelet length and number of panicle nodes were analyzed with the coefficients of variation using software SAS.
Results
Phenotypic Variation Analysis
The variation range of four quantitative traits, plant height, stalk diameter, spikelet length and number of panicle nodes, of the 50 E. arundinaceus (Retz.) Jesw. clones was 12.71–16.14 % (Table 3). The range of plant height was 219–404 cm, and the average was 337 cm; the range of stalk diameter was 0.56–1.07 cm, and the average was 0.82 cm; the range of spikelet length was 40.2–80.4 cm, and the average was 59.7 cm; and the number of panicle nodes was 11–21, and the average was 17. There were obvious morphological differences among the different materials. Stalk diameter showed the largest variation range and the variation coefficient reached 16.14 %, followed by spikelet length whose variation coefficient was 15.97 %, and the variation coefficients of plant height and number of panicle nodes were 13.12 % and 12.71 %, respectively.
Polymorphism Detected Using SCoT Markers
In the 50 E. arundinaceus (Retz.) Jesw. accessions, a total of 46 SCoT primers were screened, and 15 primers were successfully produced, which showed scorable, reproducible with clear high-intensity and highly polymorphic bands (Fig. 1). The total number of bands scored was 336, of which 284 were polymorphic (83.93 %) with an average of 19.0 per primer (Table 4). For each primer, the fragment sizes ranged from 250 to 1000 bp and the total number of bands varied from 17 (SCoT2) to 30 (SCoT27) with a mean 22, and the polymorphic rate ranged from 69.57 to 100 % and the total umber of polymorphic bands varied from 12 (SCoT2) to 26 (SCoT27) with a mean 19.
Similarity Coefficient Analysis
The scored bands were used to calculate the genetic diversity among the 50 E. arundinaceus (Retz.) Jesw. accessions. The Jaccard’s similarity coefficients varied from 0.61 to 0.88 with an average of 0.71. The accessions GUX09 and GUX10 had the highest genetic similarity coefficient of 0.88, while GUX10 and GUX30 had the lowest genetic similarity coefficient of 0.61. In terms of the genetic similarity coefficient of the accessions, the majority of the accession from the same area showed high similarity coefficient. For example, GUX01, GUX02, GUX03 and GUX04 coming from Lingchuan County, Guilin City, and their similarity coefficients were over 0.78; GUX12, GUX13, GUX14, GUX15 and GUX16 coming from Wuming County, Nanning City, and their similarity coefficients were over 0.75; GUX25, GUX26, GUX27, GUX28 and GUX29 coming from Liusanjie Township, Yizhou City, and their similarity coefficients were over 0.76. These results indicated that the accessions of E. arundinaceus (Retz.) Jesw. clones from the same area have relatively lower genetic differences and closer genetic relationships compared with those from different areas.
Clustering Analysis
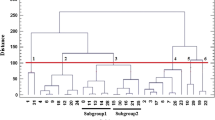
A UPGMA dendrogram based on the similarity coefficients was constructed to estimate the genetic relationships among the 50 E. arundinaceus (Retz.) Jesw. accessions (Fig. 2). As shown in Fig. 2, these accessions were distinctly clustered into four groups, which shared a common node at similarity coefficient of 0.73. The accessions GUX45, GUX49 and GUX50 belonged to cluster I, GUX48 belonged to cluster II, GUX38 belonged to cluster III, and the rest 45 clones belonged to cluster IV. The cluster IV was further subdivided into six subgroups, that is, eight were in subcluster A, 21 in subcluster B, six in subcluster C, six in subcluster D, two in subcluster E and two in subcluster F, which shared a common node at similarity coefficient of 0.76. The result shows that most of the accessions collected from the same area are gathered in the same class. For example, the accessions GUX01, GUX02, GUX03 and GUX04 from Lingchuan County, and GUX31, GUX32 and GUX33 from Hechi City were classified into subcluster A; GUX5, GUX6, GUX7, GUX8, GUX9 and GUX10 from Rongan County and neighbored Rongshui County, and GUX12, GUX13, GUX14, GUX15, GUX16, GUX21, GUX22, GUX23 and GUX24 from Wuming County were classified into subcluster B; GUX25, GUX26, GUX27, GUX28 and GUX29 from Liusanjie Village, Yizhou City were classified into subcluster C; and GUX40, GUX41 GUX42 and GUX43 from different areas of Hechi City are classified into subcluster D.
Discussion
The coefficient of variance (CV) is an index of population variation. According to Wang et al. (2012), CV 0—10 % is on the lower variation level, 10–20 % is on the medium variation level and over 20 % is on the high variation level. In this study, the CVs of four quantitative traits, plant height, stalk diameter, spikelet length and number of panicle nodes, in the 50 E. arundinaceus (Retz.) Jesw. clones collected from Guangxi ranged from 12.71 to 16.14 % which were on the medium variation level, lower than those reported by Liu et al. (2013) who found that the CVs for the quantitative traits of E. arundinaceus (Retz.) Jesw. accessions from Hainan Province were on the high level. In this study, the CV of stalk diameter was the highest; therefore, this trait has the greatest potential for genetic improvement.
The SCoT markers are expected to be linked to functional genes and corresponding traits; thus, the amplicons can be converted to gene targeted marker systems (Xiong et al. 2011). SCoT markers have been used for analyses of genetic diversity and clustering relation in several crops (Collard and Mackill 2009; Xiong et al. 2010; Wu et al. 2010; Chen et al. 2012; Zhao et al. 2012; Wei et al. 2012; Wu et al. 2013). Compared with RAPD, ISSR, AFLP and SSR, SCoT marker analysis has the advantages of simpleness, low cost and good repeatability. Zhang et al. (2015) analyzed the genetic diversity of E. arundinaceus (Retz.) Jesw. collections from Sichuan using SRAP markers and the polymorphic bands ratio was 74.63 %. Rao et al. (2014) analyzed the genetic diversity of E. arundinaceus (Retz.) Jesw. collections using RAPD markers and the polymorphic bands ratio was 78.85 %. In this study, the polymorphism of 50 E. arundinaceus (Retz.) Jesw. collections from Guangxi was analyzed using 15 SCoT primers and the total polymorphic bands ratio reached up to 83.93 %, which was higher than the previously reported results. The Jaccard’s similarity coefficients ranged from 0.61 to 0.88, and the UPGMA dendrogram reflected the geographic distribution patterns of the 50 accessions of E. arundinaceus (Retz.) Jesw. collected from different areas of Guangxi, indicating that the collections of E. arundinaceus (Retz.) Jesw. from Guangxi have rich genetic diversity. Collection, identification, rational conservation and exploitation of the E. arundinaceus (Retz.) Jesw. germplasm in Guangxi might be potentially useful in sugarcane breeding.
Conclusion
The CVs of four quantitative traits, plant height, stalk diameter, spikelet length and number of panicle nodes, of 50 E. arundinaceus (Retz.) Jesw. collection from Guangxi ranged from 12.71 to 16.14 %. The polymorphism and similarity analyses based on 15 SCoT primers reflected the rich diversity and geographic distribution patterns of these accessions.
References
Cai, Q., K.S. Aitken, Y.H. Fan, G. Piperidis, X.L. Liu, C.L. McIntyre, X.Q. Huang, and P. Jackson. 2012. Assessment of the genetic diversity in a collection of Erianthus arundinaceus. Genetic Resources and Crop Evolution 59: 1483–1491.
Chen, H., X.H. He, G.X. Huang, F. Li, and J.C. Jiang. 2012. Comparison and analysis of SCoT and ISSR markers for genetic diversity of Longan. Guihaia 32(4): 536–541.
Chen, X.L., Y.R. Li, L.T. Yang, J.M. Wu, C. Luo, F.Q. Xiong, and L. Yang. 2010. cDNA-SCoT differential display of cold resistance related genes in sugarcane under low temperature stress. Biotechnology Bulletin 8: 120–124.
Collard, B.C.Y., and D.J. Mackill. 2009. Start codon targeted (SCoT) polymorphism: A simple, novel DNA marker technique for generating gene targeted markers in plants. Plant Molecular Biology Reporter 27: 86–93.
Feng, S.G., R.F. He, M.Y. Jiang, J.J. Lu, X.X. Shen, J.J. Liu, Z.A. Wang, and H.Z. Wang. 2016. Genetic diversity and relationships of medicinal Chrysanthemum morifolium revealed by start codon targeted (SCoT) markers. Scientia Horticuturae 201: 118–123.
Grivet, L., and P. Arruda. 2001. Sugarcane genomics: depicting the complex genome of an important tropical crop. Current Opinion in Plant Biology 5: 122–127.
Hu, R., M.Z. Bao, X.Q. Wu, H.S. Tan, and X.P. Fu. 2015. Analyses and utilization of the phenotypic diversity of carnation. Journal of Huangzhong Agricultural University 34(2): 16–23.
Kalwade, S.B., and R.M. Devarumath. 2014. Single strand conformation polymorphism of genomic and EST-SSRs marker and its utility in genetic evaluation of sugarcane. Physiol Mol Biol Plants 20(3): 313–321.
Li, Y.R. 2010. Modern Sugarcane Science, 61–125. Beijing: China Agriculture Press.
Liu, X.H., F.X. Fang, R.H. Zhang, H.Z. Song, R.Z. Yang, Y.J. Gao, H.P. Ou, J.C. Lei, T. Luo, W.X. Duan, G.M. Zhang, and Y.R. Li. 2012a. Identification of progenies from sugarcane × Narenga porphyrocoma (Hance) Bor. by SSR marker. Southwest China. Journal of Agricultural Science 25(1): 38–42.
Liu, X.H., F.X. Feng, R.H. Zhang, H.Z. Song, R.Z. Yang, Y.J. Gao, J.C. Lei, T. Luo, W.X. Duan, and G.M. Zhang. 2012b. Identification and genetic analysis of hybrid from cross between Erianthus arundinaceum (Retz.) Jesw. and Saccharum spontaneum L. Acta Agronomica Sinica 32(6): 11–15.
Liu, Y., X.L. Liu, H.B. Liu, Y.L. Yao, and J.B. Su. 2013. Collection and genetic diversity of wild germplasm resource in sugarcane from Hainan Province. Chinese Agricultural Science Bulletin 29(1): 199–208.
Luo, T., H.X. Yang, H.F. Qin, X.H. Xu, Y.J. Gao, W.X. Duan, R.H. Zhang, X.H. Liu, H.Z. Song, Y.X. Huang, and G.M. Zhang. 2013. Application of SCoT molecular marker in construction of molecular genetic linkage mao of Saccharum spontaneun L. Journal of Plant Genetic Resources 14(4): 704–710.
Nair, N.V., A.W. Jebadhas, and T.V. Sreenivasan. 1993. Saccharum germplasm collections in Arunachal Pradesh. Indian Journal of Plant Genetic Resources 6(1): 21–26.
Nerkar, G., F. Farsangi, and R. Devarumath. 2015. Organellar genome diversity in Saccharum and Erianthus spp. Revealed by PCR-RFLP. Molecular Plant Breeding 6(11): 1–11.
Priya, M.S., K.H.P. Reddy, M.H. Kumat, V. Rajarjeswari, G.M. Naidu, R. Narasimhulu, B. Reddy, and R. Kumar. 2005. Genetic diversity and character aassociation among sugarcane (Saccharum Spp.) clones. Bioinfolet 12: 444–451.
Que, Y.X., Y.B. Pan, Y.H. Lu, C. Yang, Y.T. Yang, N. Huang, and L.P. Xu. 2014. Genetic analysis of diversity within a Chinese local sugarcane germplasm based on start codon targeted polymorphism. BioMed Research International 350: 1–10.
Rajesh, M.K., A.A. Sabna, K.E. Rachana, S. Rahman, B.A. Jerard, and A. Karun. 2015. Genetic relationship and diversity among coconut (Cocos nucifera L.) accessions revealed through SCoT analysis. Biotechnology 5: 999–1006.
Rao, V.P., R. Chaudhary, S. Singh, R.S. Sengar, and V. Sharma. 2014. Assessment of genetic diversity analysis in contrasting sugarcane varieties using random amplified polymorphic DNA (RAPD) markers. African Journal of Biotechnology 13(37): 3736–3741.
Satya, P., M. Karan, S. Jana, S. Mitra, A. Sharma, P.G. Karmakar, and D.P. Ray. 2015. Start codon targeted (SCoT) polymorphism reveal genetic diversity in wild and domesticated populations of ramie (Boehmeria nivea L. Gaudich.), a premium textile fible producting species. Meta Gene 3: 62–70.
Sharma, M.D., U. Dobhal, P. Singh, S. Kumar, A.K. Gaur, S.P. Singh, A.S. Jeena, E.P. Koshy, and S. Kumar. 2014. Assessment of genetic diversity among sugarcane cultivars using novel microsatellite marker. African Journal of Biotechnology 13(13): 1444–1451.
Song, S., Y.W. Wu, X.B. Ji, L.Q. Song, and X.P. Wen. 2013. Genetic analysis of apple rootstock germplasm from Guizhou province using ISSR markers. Journal of Huazhong Agricultural University 23(1): 19–24.
Su, Y.C., H. Lin, H.B. Wang, Y.X. Que, Q.B. Wu, S.S. Chen, and L.P. Xu. 2012. Optimization of SCoT-PCR reaction system, and screening and utilization of polymorphic primers in sugarcane. Chinese Journal of Applied and Environmental Biology 18(5): 810–818.
Wang, J.S., Z.X. Liu, F. Fan, L.P. Han, and G.H. Xie. 2012. Analysis of genetic diversity and inheritability of agronomic traits and chemical compositions in sweet sorghun (sorghum bicolour). Journal of China Agricultural University 17(6): 83–91.
Wei, Y.L., X.H. He, C. Luo, and H. Chen. 2012. Genetic diversity of podocarpus by SCoT markers. Guihaia 32(1): 90–93.
Wu, J.M., Y.R. Li, A.Q. Wang, L. Yang, and L.T. Yang. 2010. Different expression of gibberellin-induced genes for stalk elongation of sugarcane analysis with cDNA–ScoT. Acta Agronomica Sinica 36(11): 1883–1890.
Wu, K.C., Y.R. Li, L.T. Yang, and J.M. Wu. 2013. Establishment, optimization and application of cDNA-SCoT reaction system in sugarcane. Chinese Journal of Tropical Crops 34(5): 892–898.
Xiong, F.Q., J. Jiang, R.C. Zhong, Z.H. Qiang, L.Q. He, Z. Li, W.J. Zhuang, and R.H. Tang. 2010. Application SCoT molecular marker in Genus arachis. Acta Agronomica Sinica 36(12): 2055–2061.
Xiong, F.Q., R.C. Zhong, Z.Q. Han, J. Jiang, L.Q. He, W.J. Zhuang, and R.H. Tang. 2011. Start codon targeted polymorphism for evaluation of functional genetic variation and relationships in cultivated peanut (Arachis hypogaea L.) genotypes. Molecular Biology Reports 38: 3487–3494.
You, Q., Y.B. Pan, L.P. Xu, S.W. Gao, Q.N. Wang, Y.C. Su, Y.Q. Yang, Q.B. Wu, D.G. Zhou, and Y.X. Que. 2015. Genetic diversity analysis of sugarcane germplasm based on fluorescence-labeled simple sequence repeat markers and a capillary electrophoresis-based genotyping platform. Sugar Tech. doi:10.1007/s12355-015-0395-9.
Zan, F.G., X.M. Ying, C.W. Wu, P.F. Zhao, X.K. Chen, L. Ma, H.S. Su, and J.Y. Liu. 2015. Genetic diversity of 98 collections of sugarcane germplasm with AFLP markers. Scientia Agricultura Sinica 48(5): 1002–1010.
Zhang, J.B., J.J. Yan, S.Q. Bai, Y.W. Zhang, Z.X. Dao, D.X. Li, C.B. Zhang, M.H. You, Y. Zhang, J. Zhang, and F.Y. Yang. 2013. Genetic diversity of native Erianthus arundinaceus germplasm detected by SRAP markers. Jourmal of Agricultural Biotechnology 21(10): 1193–1202.
Zhang, Y., J.J. Yan, S.Q. Bai, Y. Peng, X.Z. Liang, J.B. Zhang, J. Zhang, and C. Hu. 2015. SRAP analysis of genetic structure of wide Erianthus arundinaceum populations in Sichuan China. Chinese Journal of Grassland 37(1): 15–21.
Zhao, R.Q., Y.H. Gao, X.L. Zhang, and J.P. Si. 2012. Establishment and optimization of SCoT-PCR reaction system for Dendrobium officinale. Acta Agriculturae Nucleatae Sinica 26(4): 648–655.
Funding
This research was financially supported by the National High Technology Research and Development Program of China (863 Program) (2013AA102604), National Natural Science Foundation of China (31101195), Guangxi Special Funds for Bagui Scholars and Distinguished Experts (2013), Funds of Guangxi (2014GXNSFBA118118, 2012GXNSFBA053048, 14123001-1-4, 15-140-13), Fund for the Guangxi Innovation Teams of Modern Agriculture Technology (gjnytxgxcxtd-03) and Fund of Guangxi Academy of Agricultural Sciences (2016JM07).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Xi-Hui Liu and Huan-Zhong Song have contributed equally in preparation of this manuscript.
Rights and permissions
About this article
Cite this article
Liu, XH., Song, HZ., Zhang, GM. et al. Phenotypic Variation and Genetic Diversity in the Collections of Erianthus Arundinaceus (Retz.) Jesw.. Sugar Tech 19, 359–367 (2017). https://doi.org/10.1007/s12355-016-0475-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12355-016-0475-5