Abstract
Carbonic anhydrases (CAs) play important roles in the utilization of inorganic carbon and have been studied in many higher plants and algae. Herein, based on unigene sequences from Pyropia haitanensis, six full-length CA of P. haitanensis (PhCA) genes were obtained by rapid amplification of complementary DNA (cDNA) ends, or by direct polymerase chain reaction (PCR), and named PhαCA1, PhαCA2, PhβCA1, PhβCA2, PhβCA3, and PhγCA1. The full-length cDNAs of the six PhCA genes comprised 1156, 1151, 1146, 847, 1124, and 1013 nucleotides, respectively. The encoded proteins were 275, 287, 206, 245, 307, and 290 amino acids, with isoelectric points of 10.26, 5.40, 6.03, 6.42, 7.66, and 4.75, respectively. On the basis of conserved motifs and phylogenetic tree analysis, the PhCAs were divided into three CA classes: PhαCA1 and PhαCA2 are α-CAs; PhβCA1, PhβCA2, and PhβCA3 are β-CAs; and PhγCA1 is a γ-CA. In different CO2 concentrations, the expression levels of PhβCA2, PhβCA3, and PhγCA1 showed no significant changes; however, the expression levels of PhαCA1, PhαCA2, and PhβCA1 decreased significantly under high CO2. The expression level of each PhβCA gene was significantly higher in the sporophytes than in the gametophytes; however, the expression levels of PhαCA and PhγCA in the sporophytes were significantly lower than those in the gametophytes. The expression level of each PhCA under desiccation and high-temperature stress had a different pattern. These results suggested that PhCAs play important roles in the utilization of inorganic carbon of P. haitanensis. Additionally, the expressions of PhCAs were significantly affected by environmental stresses.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
For photosynthetic organisms, the marine environment is seriously CO2-limited, mainly because of the alkaline nature of seawater (currently pH 8.2). Most of the dissolved inorganic carbon is present as bicarbonate (HCO3 −), with only a small fraction (∼10–20 μmol kg−1) present as free CO2 (aqueous). Additionally, the diffusion rate of CO2 is slow: ∼10,000 times slower than that in air (Lee et al. 2013). To adapt to the unfavorable low-CO2 conditions, aquatic photosynthetic organisms have developed a panoply of carbon-concentrating mechanisms (CCMs) that elevate CO2 concentration intracellularly around the active site of ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) (Moroney et al. 2001). A number of studies have shown that carbonic anhydrase (CA) plays a crucial role in the CCM process (Moroney et al. 2001).
CA, which was first discovered in red blood cells, has since been found in most organisms, including animals, plants, archaebacteria, and eubacteria. CA catalyzes the reversible interconversion of CO2 and HCO3 −, a reaction fundamental to many biological processes, such as photosynthesis and respiration, pH homeostasis, ion transport, and catalysis of key steps in the pathways for the biosynthesis of physiologically important metabolites (Badger 2003). On the basis of their amino acid sequences, CA proteins are categorized into six distinct classes: α, β, γ, δ, ζ, and ε (Moroney et al. 2011). In animals, all enzymes so far discovered belong to the α class, while in plants and macroalgae, almost all known CAs belong to the α, β, and γ classes, with the β class predominating (Moroney et al. 2001, 2011). To date, the δ-CAs have only been described in some marine diatoms (Roberts et al. 1997; Soto et al. 2006), the representatives of the ζ class have only been discovered in marine cyanobacteria and some chemolithoautotrophs (Fabre et al. 2007), and the ε-CAs are limited to bacteria containing α-type carboxysomes (So et al. 2004; So and Espie 2005). Although all six types of CAs share no sequence similarity in their primary amino acid sequences and seem to have evolved independently, they all are zinc metalloenzymes and appear to share a similar catalytic mechanism (Moroney et al. 2011).
Over the last decade, data provided by various genome sequencing studies have revealed the multiplicity of CA isoforms in plants. For example, analysis of the Arabidopsis thaliana genome revealed the presence of 19 genes encoding CAs, including eight α, six β, and five γ-CAs (Fabre et al. 2007). In the model microalga Chlamydomonas reinhardtii, there are at least 12 genes that encode CA isoforms, including three α, six β, and three γ or γ-like CAs (Moroney et al. 2011). CA genes have also been identified in a number of macroalgae; for example, one β-CA and one α-CA were reported in Pyropia yezoensis (Zhang et al. 2010) and Laminaria/Saccharina japonica (Yu et al. 2011; Ye et al. 2014).
Pyropia is a model organism of the intertidal zone. Its gametophytes are subjected to profound environmental changes, including desiccation, osmotic shock, intense sunlight exposure, high and/or freezing temperatures, and other changes, on a continuing basis (Sahoo et al. 2002; Blouin et al. 2011). Several studies have shown that environmental stress can significantly inhibit the rate of carbon fixation and consequently the rate of photosynthesis (Ashraf and Harris 2013; Xu et al. 2014). Furthermore, the life cycle of Pyropia differs significantly from that of terrestrial plants, consisting of a macroscopic foliose haploid thallus phase (gametophyte) and a microscopic diploid filamentous conchocelis phase (sporophyte) (Sahoo et al. 2002). A number of studies have reported that the carbon-fixation mechanisms in Pyropia are different between the gametophyte and sporophyte phases: The transcripts of phosphoenolpyruvate carboxykinase (PEPCK), phosphoenolpyruvate carboxylase (PEPC), and aspartate aminotransferase (AST) were abundant in the sporophytes of Pyropia haitanensis, which suggested that C4-like characteristics existed in this species and phase (Fan et al. 2007; Xie et al. 2013). Thus, it is important to determine whether there are different CAs expressed in the different life phases of Pyropia, to study the roles of each CA during adaptation and to examine the tolerance mechanisms to environmental stresses associated with the carbon-fixation mechanism of Pyropia. Thus, a first step would be to isolate and sequence the CA genes from Pyropia genomic DNA or complementary DNA (cDNA). Therefore, the aims of the present study are to isolate and characterize the CA genes from P. haitanensis and to determine their expression levels in different CO2 concentrations, in different life phases, and under high-temperature and desiccation stress.
Materials and methods
Pyropia haitanensis strain Z-61, the seaweed material used in this study, is tolerant to high temperatures and produces a high yield (Chen et al. 2008). It was selected and purified by the Laboratory of Germplasm Improvements and Applications of Pyropia in Jimei University, Fujian Province, China.
Under normal conditions, the gametophytes and sporophytes of Z-61 were cultured in natural seawater with Provasoli’s enrichment solution (PES) medium at 21 °C and 50–60 μmol photons m−2 s−1 (10:14, L/D cycle). The culture medium was bubbled continuously with filter-sterilized air and refreshed every 2 days. When the gametophytes had grown to 15 ± 2 cm in length, they were collected for the further experimentation.
In the CO2 concentration experiment, 25 gametophytes were randomly selected and cultured in five aerated flasks (3000 mL) containing five blades each, at 21 °C (high temperature), 50–60 μmol photons m−2 s−1 (10:14, L/D) and bubbled continuously with filter-sterilized 10,000 ppm CO2 for 0, 30 min and 1, 2, and 4 h.
In the high-temperature experiments, 30 gametophytes were randomly selected and cultured in six aerated flasks (3000 mL) containing five blades each at 29 °C (high temperature) and 50–60 μmol photons m−2 s−1 (10:14, L/D) for 0, 3, 6, 12, 24, and 48 h.
In the desiccation experiments, 25 gametophytes were randomly selected and separated into five groups, each with five gametophytes. The gametophytes of each group were desiccated in an incubator at 50–60 μmol photons m−2 s−1 and 21 °C. The desired levels of desiccation (0, 15, 30, 45, 60, 75, and 90 %) were obtained by varying the duration of exposure. Degrees of desiccation were expressed as percentage of water loss from the samples. The water loss (W L , %) was estimated using the equation: W L = (W 0 − W t )/(W 0 − W d ) × 100, where W 0 is the initial wet weight measured after removing surface water drops by lightly blotting with tissue paper, W t is the desiccated weight after a known time interval, and W d is the dry weight (60 °C, 6 h).
In the rehydration experiment, five gametophytes were first desiccated to a water loss of 90 %, and then recultured in culture medium for 30 min.
Each treatment was carried out as three biological replicates from independent flasks that were cultivated in parallel.
RNA extraction and cDNA synthesis
Total RNA was isolated from each sample listed above. The collected samples were first cleaned for three times with sterilized seawater and then sterilized by soaking in 0.7 % KI solution for 10 min to remove the unwanted algae and contaminants. After drying with hygroscopic filter paper, the samples were ground into a powder in liquid nitrogen. An E.Z.N.A. Plant RNA Kit (Omega, Germany) was used to extract the RNA. The quality and quantity of the purified RNA were determined by measuring the absorbance at 260/280 nm (A 260/A 280) and 260/230 nm (A 260/A 230) using a Nanodrop ND-1000 spectrophotometer (LabTech, USA). Only RNA samples with an A 260/A 280 ratio between 1.9 and 2.1 and an A 260/A 230 ratio greater than 2.0 were used for subsequent analyses. Agarose gel electrophoresis (1.2 %) was used to assess the integrity of the RNA samples.
The cDNA used for 5′ rapid amplification of cDNA ends (RACE) and 3′ RACE was synthesized using a SMART RACE cDNA kit (Clontech, USA), according to the manufacturer’s protocol. The first-strand cDNA used for quantitative real-time PCR (qPCR) was synthesized from total RNA (1 μg) using a PrimeScript RT reagent kit (Takara, China) with 6-mer random primers, according to the manufacturer’s protocol. All cDNAs were diluted with nuclease-free water to 5 ng μL−1 for use as templates in qPCR.
Cloning of the full-length cDNA of P. haitanensis CA (PhCA) genes
Among the unigene database of P. haitanensis, which was obtained from whole transcriptome sequencing, five sequences (Unigene2306, Unigene8262, Unigene10445, CL1023, and Unigene235) were identified as homologous to the conserved domains of PhCA genes. The gene-specific primers that were used to amplify the full-length cDNA of the PhCA genes were designed based on these five sequences using Primer Premier 5.0 software (http://www.PremierBiosoft.com).
Ten gene-specific primers were designed and used to amplify the 5′ and 3′ ends of the PhαCA1, PhαCA2, PhβCA1, PhβCA3, and PhγCA1 genes via RACE using a SMART RACE cDNA kit (Clontech Lab., Inc.) (Table 1). Unigene CL1023 included the full-length cDNA of PhβCA2; therefore, two head-to-toe primers, HβCA2F and HβCA2R, were designed and used to amplify the full-length PhβCA2 cDNA via PCR. All the processes were performed according to the manufacturer’s protocol. The obtained PCR products were cloned into the pMD19-T simple vector (Takara) and transformed into competent Escherichia coli JM109 cells. The recombinants were identified through blue-white selection on ampicillin-containing Luria-Bertani plates; PCR verification of white colonies used primers RV-M and M13-20. An ABI Prism 3730 automated DNA sequencer was used to sequence the positive clones. The sequence of each primer used in this study is shown in Table 1.
Sequence analysis
DNAMAN 5.2.2 software (Lynnon BioSoft) was used to assemble the obtained 5′ and 3′ end sequences into the full-length cDNA of each PhCA gene. The obtained sequences were examined for identity with other known sequences using the BLAST program, available at the National Center for Biotechnology Information (NCBI) website (http://www.ncbi.nlm.nih.gov/blast). The deduced amino acid (AA) sequences were analyzed using the Conserved Domain of NCBI and the Expert Protein Analysis System (http://www.expasy.org/). Swiss-model (http://swissmodel.expasy.org/tools) and WoLFPSORT (http://wolfpsort.seq.cbrc.jp/) predicted the transmembrane helices, 3D structure, and subcellular localization. Pfam HMM (http://pfam.sanger.ac.uk/search) predicted their domains and SignalP (http://www.cbs.dtu.dk/services/signalP) predicted the potential signal sequences. Multiple sequence alignments were generated using BioEdit (http://www.mbio.ncsu.edu/bioedit/bioedit.html). The neighbor-joining algorithm of the MEGA5.10 software (Tamura et al. 2011) was used to construct a phylogenetic tree with Poisson correction and pairwise deletion parameters; all other parameters were set to default. A total of 1000 bootstrap replicates were performed.
qPCR analysis of PhCA gene expression
qPCR measured the expressions of the PhCA genes under different treatments. qPCR reactions were performed in 96-well plates using an ABI 7300 Real-time PCR Detection system. No-template control (NTC) and no-reverse transcriptase (NRC) reactions were included in the same 96-well plates to confirm no target contamination and negligible genomic DNA contamination. Reactions were performed in 25-μL volumes containing 12.5 μL of 2× SYBR green Master Mix (ToYoBo, Japan), 0.25 μL (20 mM) of each sense and antisense primers, 2 μL of the diluted template, and 10 μL of RNA-free water. The ubiquitin-conjugating enzyme (UBC) gene was used as an internal control gene. The sequence of each primer is shown in Table 1. The thermal profile for qPCR was 95 °C for 10 min, followed by 40 cycles of 95 °C for 10 s and 60 °C for 30 s. Dissociation curve analysis of the amplicons was performed at the end of each PCR reaction to confirm that only one specific PCR product was amplified and detected. qPCR was performed in triplicate for each sample. After the PCR program, the ABI optical system software analyzed the data. To maintain consistency, the baseline was set automatically by the software. All data are presented as means ± SE in terms of relative mRNA expression. Student’s t test analyzed the results, and P < 0.05 was set as the level of statistical significance.
Results
Identification of the PhCA unigenes
Previously, high-throughput sequencing was used to identify 24,575 unigenes from one cDNA library constructed with mixed samples, including the thallus and conchocelis, of P. haitanensis cultured under normal or different stress conditions (Xie et al. 2013). Functional annotation of each unigene allowed us to identify unigenes encoding putative CAs. Among the putative CA unigenes, five unigenes (Unigene2306, Unigene8262, Unigene10445, CL1023, and Unigene235) that had the longest sequences and the highest query scores were selected and used as core sequences to clone the full sequences of PhCA cDNAs. Homology searching using the BLASTX program showed that the five putative PhCA cDNAs encoded different PhCAs. The PhCA genes were named PhαCA1, PhαCA2, PhβCA1, PhβCA2, PhβCA3, and PhγCA1, respectively (Table 2). The unigene CL1023 included the full-length cDNA of PhβCA2 and was also the core sequence of PhβCA3.
Cloning and sequence analysis of PhCA cDNAs
PhαCA1
On the basis of the sequence of unigene2306, two gene-specific primers (RαCA1-5′ and RαCA1-3′) were designed to clone the partial cDNA of PhαCA1. Using SMART RACE technology, a fragment comprising 517 bp at the 5′ end (Fig. 1a) and a fragment comprising 666 bp at the 3′ end (Fig. 1b) of the PhαCA1 cDNA were obtained. The two fragments were then assembled into the full-length cDNA of PhαCA1, which comprised 1156 bp. The full-length sequence was submitted to GenBank under the accession number KJ778685. Open reading frame (ORF) finder analysis showed that the PhαCA1 cDNA comprised a 5′ untranslated region (UTR) of 130 bp, a 3′ UTR of 198 bp, and an ORF of 828 bp. The ORF encoded a polypeptide of 275 amino acids with a predicted molecular mass (MM) of 29.4 kDa and a theoretical isoelectric point (pI) of 10.26 (Table 2). Domain searching showed that amino acids 37–273 comprise the CA (score = 102.5), and the SignalP software revealed that the cleavage site of the signal peptide in this sequence was between amino acid 32 and 33; residues 9–32 were predicted to be a transmembrane helix (Fig. 2).
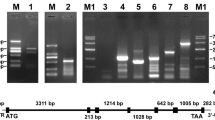
Agarose gel electrophoresis of RACE or head-to-toe PCR products of carbonic anhydrase of Pyropia haitanensis (PhCA) genes. M DL2000 DNA marker; lanes 1 and 2 are the biological replicates; lanes a, b, c, d, e, f, g, i, j, and k are the 5′ and 3′ RACE products of PhαCA1, PhαCA2, PhβCA1, PhβCA3, and PhγCA1, respectively; h shows the head-to-toe PCR products of PhβCA2. The arrows indicate the target fragments
Sequence alignment of the PhαCA proteins. Conserved residues are indicated with asterisks and the similar residues with colons. The three histidine residues involved in zinc binding are indicated by a gray background; residues that interact with the water molecule and help charge the zinc with a hydroxyl ion are indicated by boxes; residues that form the CO2 hydrophobic pocket are indicated by equal signs; the sequences between the two vertical lines are the CA domains; downward arrows are the cleavage sites of the signal peptide, and the transmembrane helix is indicated by en dash
PhαCA2
On the basis of the sequence of Unigene8262, two gene-specific primers (RαCA2-5′ and RαCA2-3′) were designed to clone the partial cDNA of PhαCA2. Using SMART RACE technology, a fragment comprising 698 bp at the 5′ end (Fig. 1c) and a fragment comprising 685 bp at the 3′ end (Fig. 1d) of the PhαCA1 cDNA were obtained. The two fragments were then assembled into the full-length cDNA of PhαCA2, which comprised 1151 bp. The full-length sequence was submitted to GenBank under the accession number KJ778686. ORF finder analysis showed that the PhαCA2 cDNA comprised a 5′ UTR of 99 bp, a 3′ UTR of 188 bp, and an ORF of 864 bp. The ORF encoded a polypeptide of 287 amino acids with a predicted MM of 29.7 kDa and a theoretical pI of 5.40 (Table 2). Domain searching showed that amino acids 42–281 comprise the CA (score = 95.7), and the SignalP software revealed that the cleavage site of the signal peptide in this sequence was between amino acids 28 and 29; residues 16–33 were predicted to be a transmembrane helix (Fig. 2).
PhβCA1
On the basis of the sequence of Unigene10445, two gene-specific primers (RβCA1-5′ and RβCA1-3′) were designed to clone the partial cDNA of PhβCA1. Using SMART RACE technology, a fragment comprising 331 bp at the 5′ end (Fig. 1e) and a fragment comprising 837 bp at the 3′ end (Fig. 1f) of the PhβCA1 cDNA were obtained. The two fragments were then assembled into the full-length cDNA of PhβCA1, which comprised 1146 bp. The full-length sequence was submitted to GenBank under the accession number KC895403. ORF finder analysis showed that the PhβCA1 cDNA comprised a 5′ UTR of 364 bp, a 3′ UTR of 161 bp, and an ORF of 621 bp. The ORF encoded a polypeptide of 206 amino acids with a predicted MM of 22.7 kDa and a theoretical pI of 6.03 (Table 2). Domain searching showed that amino acids 20–172 comprise the CA (score = 163.6); the SignalP software revealed that there is no signal peptide in this sequence (Fig. 3).
Multiple sequence alignment of the PhβCA proteins. The conserved residues are indicated with asterisks and similar residues with colons. Zinc ligands are marked by en dash. Thirteen residues are clustered on the concave surface of the active-site clefts and classified into two groups: Group I residues, indicated by boxes, are located around Zn; group II residues, marked with a gray background, are oriented toward Zn. The sequences between the two vertical lines are the carbonic anhydrase (CA) domains
PhβCA2
CL1023 included the full-length cDNA of PhβCA2; therefore, two head-to-toe primers (HβCA2F and HβCA2R) were used to amplify the full-length cDNAs of PhβCA2. The nucleotide sequence of PhβCA2 was 847 bp (Fig. 1g), comprising a 738-bp ORF encoding a protein of 245 amino acids with a predicted MM of approximately 24.6 kDa and a pI of 6.42 (Table 2). The sequence also contained a 47-bp 5′ UTR and a 62-bp 3′ UTR. The full-length sequence was submitted to GenBank under the accession number KJ778687. Domain searching showed that amino acids 84–242 comprise the CA (score = 126.2); the SignalP software revealed that there is no signal peptide in this sequence (Fig. 3).
PhβCA3
On the basis of the sequence of CL1023, two gene-specific primers (RβCA3-5′ and RβCA3-3′) were designed to clone the partial cDNA of PhβCA3. Using SMART RACE technology, a fragment comprising 621 bp at the 5′ end (Fig. 1h) and a fragment comprising 599 bp at the 3′ end (Fig. 1i) of the PhβCA3 cDNA were obtained. The two fragments were then assembled into the full-length cDNA of PhβCA3, which comprised 1124 bp. The full-length sequence was submitted to GenBank under the accession number KJ778688. ORF finder analysis showed that the PhβCA3 cDNA comprised a 5′ UTR of 25 bp, a 3′ UTR of 175 bp, and an ORF of 924 bp. The ORF encoded a polypeptide of 307 amino acids with a predicted MM of 32.0 kDa and a theoretical pI of 7.66 (Table 2). Domain searching showed that amino acids 147–303 comprise the CA (score = 115.0); the SignalP software revealed that there is no signal peptide in this sequence (Fig. 3).
PhβCA1
On the basis of the sequence of Unigene235, two gene-specific primers (RγCA1-5′ and RγCA1-3′) were designed to clone the partial cDNA of PhγCA1. Using SMART RACE technology, a fragment comprising 484 bp at the 5′ end (Fig. 1j) and a fragment comprising 622 bp at the 3′ end (Fig. 1k) of the PhγCA1 cDNA were obtained. The two fragments were then assembled into the full-length cDNA of PhγCA1, which comprised 1013 bp. The full-length sequence was submitted to GenBank under the accession number KJ778689. ORF finder analysis showed that the PhγCA1 cDNA comprised a 5′ UTR of 71 bp, a 3′ UTR of 69 bp, and an ORF of 783 bp. The ORF encoded a polypeptide of 290 amino acids with a predicted MM of 28.9 kDa and a theoretical pI of 4.75 (Table 2). Domain searching showed that amino acids 194–227 form the left-handed parallel beta-helix (LbH) structure in CA (score = 22.2); the SignalP software revealed that there is no signal peptide in this sequence. In addition, residues 202–221 were predicted to be a transmembrane helix (Fig. 4).
Multiple sequence alignment of γ-CA proteins. The conserved residues are indicated with asterisks and similar residues with colons. The left-handed parallel beta-helix structures are indicated by a gray background, residues that are strictly conserved in γ-CAs are marked with boxes, and conserved Zn ligands are marked by equal signs. The transmembrane helix is indicated by en dash. AtγCA γ-CA of Arabidopsis thaliana, CrγCA γ-CA of Chlamydomonas reinhardtii, EhγCA γ-CA of Emiliania huxleyi
Multiple sequence alignment and phylogenetic analysis of PhCA genes
Pairwise sequence alignment of the deduced amino acid sequences of the PhαCA1 and PhαCA2 indicated that the sequence similarity of the two PhαCAs was only 49 % (Fig. 2); however, the alignment revealed that the amino acid sequences of PhαCA1 (His-126, His-128, and His-145) and PhαCA2 (His-132, His-134, and His-139) contain three histidine residues that were predicted to be the ligands for the active-site zinc. The alignment also showed that other amino acid residues near the Zn active site were highly conserved: Most of the residues were predicted to form the hydrogen bond network to Zn-bound solvent molecules in α-CAs (Fig. 2).
The multiple sequence alignment of the deduced amino acid sequences of the PhβCA1, PhβCA2, and PhβCA3 indicated that the sequence similarities (%) among the three PhβCAs were 31, 25, and 67 %, respectively (Fig. 3). The alignment also revealed that several sequence features present in other β-CAs could be identified in the three sequences. The active-site zinc of PhβCA1 and PhβCA2 was all coordinated by a Cys-Asp-His-Cys tetrad, and the active-site zinc of PhβCA3 was Cys-His-Cys (Fig. 3). These active sites are strictly conserved among β-CAs and conform to the active site in higher plant β-CAs, based on extended X-ray absorption fine structures.
Multiple sequence alignment of the deduced amino acid sequence of PhγCA1 and other γ-CAs indicated that several sequence features present in other γ-CAs could also be identified in the PhγCA1 sequence, such as amino acid residues Arg-123 and Asp-125, which are strictly conserved in γ-CAs; however, amino-acid residue Gln-139, which is also conserved in other γ-CAs, was absent in PhγCA1 (Fig. 4). Especially, the conserved Zn ligands His-144, His-177, and His-182 present in γ-CAs were completely absent from the PhγCA1, but these residues were also missing from the γ-CA in Emiliania huxleyi, a single-celled planktonic alga (Fig. 4).
The phylogenetic tree of PhCAs (Fig. 5) can be divided into three different clusters with the different CA classes (α-CAs, β-CAs, and γ-CAs). As expected, PhαCA1 and PhαCA2 clustered together with other α-CAs, PhβCA1, PhβCA2, and PhβCA3 were grouped with the β-CAs, and PhγCA1 was grouped with the other γ-CAs. Thus, the phylogenetic tree and the analysis of conserved motifs in the PhCAs indicated that PhαCA1 and PhαCA2 are α-CAs, PhβCA1, PhβCA2, and PhβCA3 are β-CAs, and PhγCA1 is a γ-CA.
Phylogenetic tree constructed using CA amino acid sequences. The accession numbers were as follows: Arabidopsis thaliana (NP_193831), Chlamydomonas reinhardtii (AAC49983), Dunaliella salina (1Y7W), Nostoc sp. PCC 7120 (BAB74628), Saccharomyces cerevisiae (CAA95901), Coccomyxa sp. PA (AAC33484), Phaeodactylum tricornutum (AAL07493), Nostoc sp. PCC 7120(NP_486950), Synechocystis sp. PCC 6803 (NP_442201), Acinetobacter sp. ADP1 (CAG67944), C. reinhardtii (AAC49888), Heterocapsa triquetra chl (AAW79300), Porphyridium purpureum (BAA12981), Pyropia yezoensis (ACI01451), Oryza sativa (AAA86943), A. thaliana (NP_974782), Zea mays (AAA86945), C. reinhardtii (EDO97352), A. thaliana (AAK28403), Thermoplasma acidophilum DSM 1728 (NP_394340), Pyrococcus horikoshii Ot3 (1V67), Selaginella moellendorffii (XP_002960200), Methanosarcina (1827571), C. reinhardtii (AAS48197), Candidatus koribacter versatilis Ellin345 (YP_590283), and Emiliania huxleyi (DQ644551)
Expression of PhCAs in different life phases and under different levels of stress
Information on the expression of the PhCAs would promote a better understanding of their physiological functions. Therefore, qPCR was used to measure the relative expressions of the six PhCAs in different life phases and under different temperatures and levels of desiccation stress. In the qPCR, NTC and NRC reactions consistently demonstrated a lack of target contamination and negligible genomic DNA contamination. The amplification specificity for each PhCA and the UBC gene was determined by analyzing the dissociation curves of the PCR products. There was only one peak in the dissociation curve for the PhCAs and UBC gene (data not shown).
First, we tested if changes in environmental CO2 concentration could alter PhCA gene expression. After bubbling with filter-sterilized 10,000 ppm CO2 for 15 min, the pH of culture medium decreased from 8.24 to 6.53; the pH remained at a constant 6.53 as the CO2 bubbled continuously. The relative expressions of each PhCA gene in the thallus of P. haitanensis at different time points were measured. The dynamic changes of expression levels of PhαCA1, PhαCA2, and PhβCA1 were similar: Their expression levels decreased significantly after bubbling with filter-sterilized 10,000 ppm CO2 for 30 min or 1 h (P < 0.05); thereafter, their expression levels did not change significantly (P > 0.05) (Fig. 6a, b, c). During the whole bubbling with filter-sterilized 10,000 ppm CO2 process, the expression levels of PhβCA2, PhβCA3, and PhγCA1 showed no significant changes (P > 0.05) (Fig. 6d, e, f).
Second, the relative expressions of each PhCA gene in the sporophytes and gametophytes of P. haitanensis were measured. The results showed that the expression level of each PhβCA gene was significantly higher in the sporophytes than in the gametophytes (P < 0.01): The expression levels of PhβCA1, PhβCA2, and PhβCA3 were 20-fold, 110-fold, and 7-fold higher, respectively, in the sporophytes than in the gametophytes (Fig. 7). However, the expression levels of the PhαCAs and PhγCA in the sporophytes were significantly lower than those in the gametophytes (P < 0.01): the expression levels of PhαCA1, PhαCA2, and PhγCA1 were 12-fold, 230-fold, and 5-fold lower, respectively, in the sporophytes than in the gametophytes (Fig. 7). These results indicated that the CA genes were expressed differently in the different phases of the P. haitanensis life cycle.
Third, the relative expressions levels of each PhCA gene in gametophytes of P. haitanensis under different levels of desiccation and in rehydration for 30 min were also measured. The expression level changes of the six PhCA genes were different. During desiccation, the expression level of PhαCA1 was not significantly different from the normal level when the water loss was 10 % (P > 0.05); however, when the water loss was 40–90 %, the expression level increased significantly (P < 0.05), reaching its maximum level when the water loss was 90 %. In addition, the expression level decreased significantly, but did not return to the normal level, after rehydration in culture medium for 30 min (Fig. 8a) (P < 0.05). For PhαCA2, the expression level was not significantly different from the normal level when the water loss was ≤40 % (P > 0.05); however, when the water loss was 60 %, the expression level increased significantly (P < 0.05), but the expression level returned to the normal level when the water loss was 90 % and after rehydration in culture medium for 30 min (Fig. 8b). For PhβCA1, the expression level was significantly decreased after 10 % water loss (P < 0.05). However, during subsequent desiccation and rehydration, the expression level did not change significantly (P > 0.05) (Fig. 8c). For PhβCA2 and PhβCA3, when the water loss was 10 and 40 %, respectively, their expression levels increased significantly (P < 0.05) and reached the maximum levels (P < 0.05). However, during subsequent desiccation and rehydration, the expression levels did not change significantly (P > 0.05) (Fig. 8d, e). For PhγCA1, its expression level was not significantly different from the normal level when the water loss was ≤60 % (P > 0.05); however, when the water loss was 90 %, the expression level increased significantly (P < 0.05). In addition, the expression level did not return to the normal level after rehydration in culture medium for 30 min (Fig. 8f).
Fourth, the relative expressions of each PhCA gene in the gametophytes of P. haitanensis were measured at different time points of high-temperature stress (29 °C). During high-temperature stress, the dynamic changes in the expression level of PhαCA1 and PhγCA1 were similar: Their expression levels increased significantly after 3 h of stress, reaching their maximum levels (P < 0.05), and then decreased to the normal level as the high-temperature stress continued (P > 0.05) (Fig. 9a, f). However, the dynamic changes in expression levels of other PhCA genes were different. The expression level of PhαCA2 decreased significantly and steadily under prolonged high-temperature stress (P < 0.05) and reached its minimum level after 24 h of high-temperature stress (Fig. 9b). By contrast, the expression level of PhβCA1 did not change significantly during the whole high-temperature stress process. The expression level of PhβCA2 increased significantly after 3 h of stress and reached its maximum level at 6 h (P < 0.05) and then significantly decreased; however, its expression level was still significantly higher than the normal level after 12 and 24 h of high-temperature stress (P < 0.05), but as the high-temperature stress continued to 48 h, its expression level decreased to the normal level (P > 0.05) (Fig. 9d). The expression level of PhβCA3 increased significantly after 3 h of stress and reached its maximum level at 6 h (P < 0.05); its expression level did not change significantly under further high-temperature stress (P > 0.05) (Fig. 9e).
Discussion
A number of studies have shown that CA or CA-like activity plays an important role in photosynthesis and in the operation of the CCM in algae (Moroney et al. 2001; Badger 2003). In this study, based on the unigene sequences of P. haitanensis, six full-length PhCA genes, which could be divided into three CA classes, were cloned. The results of expression analysis suggested that PhCAs play important roles in the utilization of inorganic carbon of P. haitanensis and their expressions were significantly affected by environmental stresses.
Over the last decade, data provided by various genome or transcriptome sequencing projects have revealed the multiplicity of CA isoforms in plants and algae; for example, C. reinhardtii has at least 12 CA genes (Moroney et al. 2011) and Arabidopsis has 19 CA genes (Fabre et al. 2007). To understand the function of each CA, obtaining the full-length sequence of each gene or cDNA is the first step. Thus, in the present study, based on the sequencing of the whole transcriptome of P. haitanensis, five CA unigenes were identified (Xie et al. 2013). Using RACE or direct PCR with PhCA unigene-based primer sequences, six full-length genes were cloned. The six PhCA genes share little primary sequence similarity and appear to have evolved independently; however, the essential residues appear well conserved (Figs. 2, 3, and 4). To the best of our knowledge, this is the first report of CA genes in P. haitanensis.
Based on the amino acid sequences and differences of the active site, the known CAs in eukaryotes can generally be divided into six classes, of which α-, β-, and γ-CAs are distributed widely in algae and higher plants (Moroney et al. 2011). Most α-CAs are active as monomers of about 30 kDa with three histidines coordinating the zinc atom (Moroney et al. 2001). However, for β-CAs, the deduced molecular mass of a single polypeptide is 24–30 kDa, and Zn2+ is coordinated by two cysteine and one histidine residues (Moroney et al. 2001). By contrast, the amino acid sequence and active site of γ-CAs are strikingly different from either the α-CAs or β-CAs; they function as a trimer of identical subunits, and the structure of each monomer is dominated by a left-handed β-helix (Kisker et al. 1996; Ferry 2010). In this study, the putative molecular masses of the six PhCAs were 29.4, 29.7, 22.7, 24.6, 32.0, and 28.9 kDa, respectively; the deduced protein sequences of PhαCA1and PhαCA2 all contain three conserved histidine residues, one signal peptide cleavage site and a transmembrane helix (Fig. 2). The deduced protein sequences of PhβCA1, PhβCA2, and PhβCA3 all contain one zinc active site of Cys-His-Cys (Fig. 3); however, the sequence of PhγCA1 contains one left-handed β-helix domain (Fig. 4). These characteristics and the phylogenetic tree of the CAs (Fig. 5) indicated that the six PhCAs could be divided into three classes: PhαCA1 and PhαCA2 are α-CAs, PhβCA1, PhβCA2, and PhβCA3 are β-CAs, and PhγCA1 is a γ-CA.
Generally, CAs all contain one Zn per enzyme unit and likely have a common Zn-hydroxide mechanism for catalysis (Moroney et al. 2001); however, the conserved amino acid residues of Zn2+ ligand in PhγCA1 are absent (Fig. 5), which is similar to the γ-CA in E. huxleyi (Fig. 4). This suggests that alternative residues may coordinate the Zn2+ or that another metal cofactor may be employed in the P. haitanensis enzyme (e.g., Co or Cd), requiring a different ligand coordination motif. It is important to note that there are relatively few reports of γ-CAs in eukaryotes, and variations in the γ-family metal ligand motif will arise as more of these enzymes are identified and characterized in eukaryotes. For example, the His-122 residue component is also absent in the γ-CA of C. reinhardtii but is present in the Arabidopsis sequence (Fig. 4).
The presence of so many distinct CAs in P. haitanensis underscores the importance of catalyzing the interconversion of CO2 and HCO3 − in this organism and even suggests that new functions for this enzyme may emerge from in-depth investigations. In C. reinhardtii, CA biochemistry and function have been explored extensively, leading to a better understanding of the function of each CA gene: The physiological functions of each CA correlate with its localization in the cell (Moroney et al. 2011). Although the localization of the PhCAs is unknown at this time, the genes are sufficiently different to indicate that their protein products may be located in different organelles or compartments within the P. haitanensis cell. Perhaps, some of the genes are expressed only under certain growth conditions or environmental stresses.
CA has been proven to fluctuate in activity in a number of species with changes in environmental CO2 concentration; its activity rapidly decreases on bubbling air from low CO2 to high CO2 and vice versa (Tiwari et al. 2005). Therefore, we examined the effect of exposure to a high CO2 concentration on the expression of CA genes in P. haitanensis thallus. We detected no change in the expression levels of PhβCA2, PhβCA3, and PhγCA1; however, the expression levels of PhαCA1, PhαCA2, and PhβCA1 significantly decreased under high CO2 concentrations (Fig. 6), suggesting that the enzymes of PhαCA1, PhαCA2, and PhβCA1 are important for the acclimation of P. haitanensis under low CO2 conditions. This result was the same as that for CAs in C. reinhardtii, where CAH1, CAH4, and CAH5 showed strong upregulation under low CO2 growth conditions; however, the other CA genes appear to be constitutively expressed and do not show such a strong response to changes in the CO2 level (Moroney et al. 2011). CAH1 has been proposed as a periplasmic α-CA that can promote the equilibrium between CO2 and HCO3 −, so that CO2 at the cell surface can diffuse across the plasma membrane (Moroney et al. 1985; Van and Spalding 1999). Therefore, CAH1 has been postulated to be part of the CCM in C. reinhardtii (Moroney et al. 2001, 2011). However, CAH4 and CAH5 are β-CAs (Eriksson et al. 1996, 1998), and their proposed roles were in retaining CO2 for photosynthesis by converting the CO2 generated by photorespiration to HCO3 −, thus preventing the CO2 from leaking out of the cell, which is important for algal CCM (Raven 2001; Moroney et al. 2011). Based on these observations, we hypothesized that PhαCA1, PhαCA2, and PhβCA1 must play important roles in the CCM of P. haitanensis.
Pyropia has a unique heteromorphic digenetic life cycle, with a macroscopic gametophyte phase and a microscopic sporophyte phase (Sahoo et al. 2002). Several studies have reported that the carbon-fixation mechanisms in Pyropia are different between the sporophytes and gametophytes and that a special C4-like carbon-fixation pathway might exist in the sporophytes (Fan et al. 2007; Zhang et al. 2010; Xie et al. 2013). Given the important roles of CAs in carbon-fixation pathways of plant photosynthesis, we investigated the expression level of each PhCA gene in the different life phases of P. haitanensis. The results showed that the expression level of each PhβCA gene was significantly higher in the sporophytes than in the gametophytes; however, the expression levels of PhαCA and PhγCA in the sporophytes were significantly lower than in the gametophytes (Fig. 7). The difference of expression level of each PhCA gene in different life phases of P. haitanensis may correlate with differences in physiology. In C3 plants, CA is thought to maintain the supply of CO2 to the enzyme Rubisco by speeding up the dehydration of HCO3 −; however, in C4 plants, CA is thought to catalyze the first critical step of C4 photosynthesis, the hydration of CO2 to bicarbonate, which phosphoenolpyruvate carboxylase uses as the substrate for carboxylation of phosphoenolpyruvate to oxaloacetate (Tiwari et al. 2005). Luo et al. (2002) and Zou and Gao (2002a) reported that external CA dehydration of HCO3 − to CO2 was the main pathway of inorganic carbon utilization by the thallus of P. haitanensis, but the conchocelis of P. haitanensis mainly takes up HCO3 − directly; these manners of inorganic carbon utilization correlate with the characteristics of C3 and C4-like pathways, respectively. The expression analysis of one β-CA (CA3) in Flaveria bidentis using an antisense strategy showed that the CA3 activity is essential to maximize C4 photosynthesis (Von Caemmerer et al. 2004). Another α-CA (CAH3) localized in the thylakoid lumen has been proposed to increase generation of CO2 from bicarbonate and consequently increase CO2 availability at the catalytic site of Rubisco in the pyrenoid (Raven 2001).
Expression of CA may also be regulated by some environmental factors, such as light, temperature, and salt content (Moskvin et al. 2000; Karlsson et al. 1998; Yu et al. 2007). For example, expression of a rice carbonic anhydrase (OsCA1) gene and CA activity were upregulated by some environmental stresses, such as salts (NaCl, NaHCO3, and Na2CO3) and osmotic stress (10 %, w/v, PEG 6000) (Yu et al. 2007). A transgenic Arabidopsis overexpressing OsCA1 had a greater salt tolerance at the seedling stage than wild-type plants (Yu et al. 2007).
Pyropia, a sessile organism that inhabits the intertidal zone, an environment of rapidly changing physical conditions because of the turning tides, have high levels of tolerance to various abiotic stressors, such as desiccation, osmotic shock, temperature, and light (Blouin et al. 2011). In particular, under desiccation stress, the gametophytes of Pyropia can lose about 95 % of their water at low tides (Liu 2009). In recent years, high temperatures associated with global warming have markedly affected the cultivation of P. haitanensis and reduced its yield along the coasts of Fujian and Zhejiang Province of China, which comprise one of the two primary cultivation areas (Xu et al. 2014). Therefore, the relative expression levels of each PhCA gene in the gametophytes of P. haitanensis under different levels of desiccation and different times of high-temperature stress were measured. During desiccation stress, except for PhβCA1, which was significantly downregulated, the other five PhCA genes were significantly upregulated, although the water loss points at which they were upregulated were different (Fig. 8). During high-temperature stress, the dynamic changes of the expression levels of the six PhCA genes were different: PhαCA1, PhβCA2, and PhγCA1 were initially significantly upregulated and then significantly downregulated; PhβCA1 was not significantly changed; PhαCA1 was continuously and significantly downregulated, but PhβCA2 was significantly upregulated (Fig. 9). These results suggested that PhCA expression responds to environmental stresses and is related to stress tolerance.
Among the metabolic processes of plants, photosynthetic assimilation of CO2 is more sensitive to environmental stress (Ashraf and Harris 2013), as it is in P. haitanensis. During emersion at low tides, the enhanced desiccation results in decreased net photosynthesis, photosynthetic efficiency, apparent carboxylating efficiency, and the light saturation point in P. haitanensis (Zou and Gao 2002b). In addition, high-temperature stress can inhibit the utilization of inorganic carbon by P. haitanensis, and the higher the temperature, the lower the utilization (Wang et al. 2013). As the key enzyme of inorganic carbon utilization, the activity of CA must be upregulated or downregulated to maintain the equilibrium between CO2 and HCO3 − during stress. However, the role of each PhCA during environmental stress is unclear at present and should be further studied using transgenic technology or an antisense strategy.
References
Ashraf M, Harris PJC (2013) Photosynthesis under stressful environments: an overview. Photosynthetica 51:163–190
Badger M (2003) The roles of carbonic anhydrases in photosynthetic CO2 concentrating mechanisms. Photosynth Res 77:83–94
Blouin NA, Brodie JA, Grossman AC, Xu P, Brawley SH (2011) Porphyra: a marine crop shaped by stress. Trends Plant Sci 16:29–37
Chen CS, Ji DH, Xie CT, Xu Y, Liang Y, Zhen YJ, Shi XZ, Wang FX, Zhao LM (2008) Preliminary study on selecting the high temperature resistance strains and economic traits of Porphyra haitanensis. Acta Oceanol Sin 30:100–106 (in Chinese with English abstract)
Eriksson M, Karlsson J, Ramazanov Z, Gardestrom P, Samuelsson G (1996) Discovery of an algal mitochondrial carbonic anhydrase: molecular cloning and characterization of a low-CO2-induced polypeptide in Chlamydomonas reinhardtii. Proc Natl Acad Sci U S A 93:12031–12034
Eriksson M, Villand P, Gardestrom P, Samuelsson G (1998) Induction and regulation of expression of a low-CO2-induced mitochondrial carbonic anhydrase in Chlamydomonas reinhardtii. Plant Physiol 116:637–641
Fabre N, Reiter IM, Becuwe-Linka N, Genty B, Rumeau D (2007) Characterization and expression analysis of genes encoding α and β carbonic anhydrases in Arabidopsis. Plant Cell Environ 30:617–629
Fan XL, Fang YJ, Hu SN, Wang GC (2007) Generation and analysis of 5318 expressed sequence tags from the filamentous sporophyte of Porphyra haitanensis (Rhodophyta). J Phycol 43:1287–1294
Ferry JG (2010) The γ class of carbonic anhydrases. Biochim Biophys Acta 1804:374–381
Karlsson J, Clarke AK, Chen ZY, Hugghins SY, Park YI, Husic HD, Moroney JV, Samuelsson G (1998) A novel a-type carbonic anhydrase associated with the thylakoid membrane in Chlamydomonas reinhardtii is required for growth at ambient CO2. EMBO J 17:1208–1216
Kisker C, Schindelin H, Alber BE, Ferry JG, Rees DC (1996) A left-handed beta-helix revealed by the crystal structure of a carbonic anhydrase from the archaeon Methanosarcina thermophila. EMBO J 15:2323–2330
Lee RBY, Smith JAC, Rickaby REM (2013) Cloning, expression and characterization of the δ-carbonic anhydrase of Thalassiosira weissflogii (Bacillariophyceae). J Phycol 49:170–177
Liu YC (2009) Mechanism for differential desiccation tolerance in Porphyra species. PhD Thesis, Northeastern University
Luo QJ, Pei LQ, Pan SY (2002) Utilization of inorganic carbon in free-living conchocelis of Porphyra haitanensis. J Fish 26:477–480 (in Chinese with English abstract)
Moroney JV, Husic HD, Tolbert NE (1985) Effect of carbonic anhydrase inhibitors on inorganic carbon accumulation by Chlamydomonas reinhardtii. Plant Physiol 79:177–183
Moroney JV, Bartlett SG, Samuelsson G (2001) Carbonic anhydrases in plants and algae. Plant Cell Environ 24:141–153
Moroney JV, Ma YB, Frey WD, Fusilier KA, Pham TT, Simms TA, DiMario RJ, Yang J, Mukherjee B (2011) The carbonic anhydrase isoforms of Chlamydomonas reinhardtii: intracellular location, expression, and physiological roles. Photosynth Res 109:133–149
Moskvin OV, Ivanov BN, Ignatova LK, Kollmeier MA (2000) Light-induced stimulation of carbonic anhydrase activity in pea thylakoids. FEBS Lett 470:375–377
Raven JA (2001) A role for mitochondrial carbonic anhydrase in limiting CO2 leakage from low CO2-grown cells of Chlamydomonas reinhardtii. Plant Cell Environ 24:261–265
Roberts SB, Lane TW, Morel FMM (1997) Carbonic anhydrase in the marine diatom Thalassiosira weissflogii (Bacillariophyceae). J Phycol 33:845–850
Sahoo D, Tang XR, Yarish C (2002) Porphyra-the economic seaweed as a new experimental system. Curr Sci 83:1313–1316
So AK, Espie GS (2005) Cyanobacterial carbonic anhydrases. Can J Bot 83:721–734
So AK, Espie GS, Williams EB, Shively JM, Heinhorst S, Cannon GC (2004) A novel evolutionary lineage of carbonic anhydrase (epsilon class) is a component of the carboxysome shell. J Bacteriol 186:623–630
Soto AR, Zheng H, Shoemaker D, Rodriguez J, Read BA, Wahlund TM (2006) Identification and preliminary characterization of two cDNAs encoding unique carbonic anhydrases from the marine alga Emiliania huxleyi. Appl Environ Microbiol 72:5500–5511
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Tiwari A, Kumar P, Singh S, Ansari SA (2005) Carbonic anhydrase in relation to higher plants. Photosynthetica 43:1–11
Van K, Spalding MH (1999) Periplasmic carbonic anhydrase structural gene (Cah1) mutant in Chlamydomonas reinhardtii. Plant Physiol 120:757–764
Von Caemmerer S, Quinn V, Hancock NC, Price GD, Furbank RT, Ludwig M (2004) Carbonic anhydrase and C4 photosynthesis: a transgenic analysis. Plant Cell Environ 27:697–703
Wang SG, Yang R, Zhou XQ, Song DD, Sun X, Luo QJ (2013) Utilization of inorganic in Pyropia haitanensis (Rhodophyta) under heat stress. Oceanology Limnol Sin 44:1378–1385 (in Chinese with English abstract)
Xie C, Li B, Xu Y, Ji D, Chen C (2013) Characterization of the global transcriptome for Pyropia haitanensis (Bangiales, Rhodophyta) and development of cSSR markers. BMC Genomics 14:107
Xu Y, Chen CS, Ji DH, Hang N, Xie CT (2014) Proteomic profile analysis of Pyropia haitanensis in response to high-temperature stress. J Appl Phycol 26:607–618
Ye RX, Yu Z, Shi WW, Gao HJ, Bi HY, Zhou ZG (2014) Characterization of α-type carbonic anhydrase (CA) gene and subcellular localization of α-CA in the gametophytes of Saccharina japonica. J Appl Phycol 26:881–890
Yu S, Zhang XX, Guan QJ, Takano T, Liu SK (2007) Expression of a carbonic anhydrase gene is induced by environmental stresses in rice (Oryza sativa L.). Biotechnol Lett 29:89–94
Yu Z, Bi YH, Zhou ZG (2011) Cloning and characterization of carbonic anhydrase (CA) gene from Laminaria japonica gametophytes. J Fish China 35:1343–1353 (in Chinese with English abstract)
Zhang BY, Yang F, Wang GC, Peng G (2010) Cloning and quantitative analysis of the carbonic anhydrase gene from Porphyra yezoensis. J Phycol 46:290–296
Zou DH, Gao KS (2002a) Photosynthetic bicarbonate utilization in Porphyra haitanensis (Bangiales, Rhodophyta). Chin Sci Bull 47:1629–1633
Zou DH, Gao KS (2002b) Effects of desiccation and CO2 concentrations on emersed photosynthesis in Porphyra haitanensis (Bangiales, Rhodophyta), a species farmed in China. Eur J Phycol 37:587–592
Acknowledgments
This research was supported in part by the 863 Project of China (Grant No: 2012AA10A411), the National Natural Science Foundation of China (Grant Nos.: 41176151 and 41276177), and the National Marine Public Welfare Research Project (Grant Nos.: 201105008 and 201105023).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chen, C., Dai, Z., Xu, Y. et al. Cloning, expression, and characterization of carbonic anhydrase genes from Pyropia haitanensis (Bangiales, Rhodophyta). J Appl Phycol 28, 1403–1417 (2016). https://doi.org/10.1007/s10811-015-0646-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10811-015-0646-x