Abstract
Habitat structure and predation are major factors that influence the distribution of zooplankton species and functional traits. Here, we analyzed how the structure provided by macrophytes (richness, Shannon’s diversity, and biomass) and potential predation by small-bodied fishes (richness and abundance) determine the richness of species and functional traits of zooplankton, as well as the spatial dissimilarity (beta-diversity) of species and traits. Zooplankton, fish, and macrophytes were simultaneously sampled across a gradient of 30 multi-species macrophyte beds. We assessed spatial patterns of zooplankton under taxonomic and functional approaches, using linear regression models, Generalized Dissimilarity Models, a Structural Equation Model, and a Fourth-Corner Analysis. Zooplankton taxonomic beta-diversity was most represented by the turnover component and zooplankton functional beta-diversity by nestedness. Zooplankton taxonomic richness and taxonomic beta-diversity were positively related to macrophyte biomass, richness, and Shannon’s diversity, whereas zooplankton functional richness and functional beta-diversity were positively related to fish richness and abundance. Macrophyte biomass and diversity oppositely influenced fish structure, which had influence on zooplankton structure. Macrophytes also negatively influenced the zooplankton traits such as body size, reproduction type, habitat, lifespan, and predatory escape response and positively influenced the trait feeding type. Fish were negatively related to the trait body size. The spatial structure generated by macrophyte beds and fish community determined the distribution of zooplankton species and functional traits.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The recent loss of species caused by human activities and climate change (Loreau et al., 2001; Naeem et al., 2012) has motivated scientists to understand patterns that determine the variation of biodiversity between different localities. Biodiversity is directly related to ecosystem functioning and fundamental services for human beings, such as food production, water resource, and nutrient cycling (Gagic et al., 2015). Biodiversity can be assessed through taxonomic and/or functional diversity. The taxonomic diversity does not consider or indicate ecological functions (Magurran, 2004), while the functional diversity considers the variation of characteristics of species related to their function in ecosystems (Barnett et al., 2007; Lavorel et al., 2013; Mouillot et al., 2013). Studies indicate that the functional diversity rather than taxonomic is essential to understand ecosystems processes, such as resilience to perturbations and invasions, productivity, and matter flux (Bellwood et al., 2004; Petchey et al., 2004; Waldbusser et al., 2004).
The functional diversity is estimated through functional traits that represent the morphological and physiological characteristics of species, and describe responses or effects of environmental factors on species growth, reproduction, and survival (Suding et al., 2008; Hebert et al., 2016). To assess the functional trait values (weight) of each species in the assembly, the species are placed inside a functional space that determines their distribution, most of the time taking into account their numeric density (Villéger et al., 2008; Maire et al., 2015). With these procedures, it is possible to measure several functional aspects of biological communities, such as diversity, richness, evenness, divergence, dispersion, entropy, and beta-diversity (Mouillot et al., 2013).
Here, we will address two aspects of functional diversity: beta-diversity and richness. Beta-diversity represents the dissimilarity in species/traits composition between two communities and can be decomposed into turnover—the replacement between communities—and nestedness—the difference in the number of species/traits between communities (Villéger et al., 2013). Functional richness represents the amount of functional space filled by the community (Mouillot et al., 2013; Villéger et al., 2013), i.e., the difference between the maximum and minimum functional values of species in the community (Mason et al. 2005).
Several environmental filters influence the distribution of species and functional traits in space and time, reflecting in the ecosystem functioning (Heino et al., 2015a). Habitat structure is perhaps the most important environmental filter, since more structurally diverse habitats provide more niches opportunities, allowing species to track environmental variation among sites, potentially increasing the regional diversity (Wellborn et al., 1996; Tews et al., 2004; Heino et al., 2015b). In most subtropical aquatic environments, macrophytes represent the main habitat structure shaping the variation in community composition (Pinheiro et al., 2016; Brito et al., 2020). Macrophyte beds with different structure, i.e., total biomass and species diversity, may provide favorable microhabitats, increasing habitat diversity and niche availability (Choi et al., 2014). The greater the habitat structuration provided by this vegetation, the more organic particles, ciliates, flagellates, bacteria, and periphytic algae are attached or around its structures (Dabés & Velho, 2001), increasing food availability and creating refuges for different communities, such as zooplankton (Rossa & Bonecker, 2003; Brito et al., 2020) and small-bodied fishes (Meerhoff et al., 2007a; Lopes et al., 2015; Pinheiro et al., 2016; Cunha et al., 2019). Therefore, in the same way that macrophyte beds can offer refuge and food for zooplankton, they could also work as a trap because they can increase the density of zooplankton predators, such as macroinvertebrates and small-bodied fish (Meerhoff et al., 2007a; Sagrario et al., 2009). Moreover, the spatial distribution of macrophytes may lead to spatial differences in the zooplankton community composition, once certain zooplankton species are usually associated to certain macrophyte species, i.e., different macrophyte species can shelter different zooplankton species/traits (Zeng et al., 2017). Thus, different amount of macrophyte biomass and diversity might contribute to the increase of zooplankton diversity and dissimilarity (Debastiani-Júnior et al., 2016). Nevertheless, it could also increase the strength of the relationship between zooplankton and their predators, as macrophyte beds offer refuge and food for both groups.
Fish predation is recognized as a key factor determining taxonomic and functional zooplankton diversity (Iglesias et al., 2011; Sodré & Bozelli, 2019), reducing their density and changing traits such as fecundity and survival (Barnett et al., 2007; Santangelo et al., 2018). Predation can also alter competitive relationships among species, reducing dominant species or traits and allowing other less dominant zooplankton species to occur (Chase et al., 2002). Furthermore, predation may select some functional traits, such as large body size and lower predatory escape response (Barnett et al., 2007; Sodré & Bozelli, 2019), which affect the functional composition of communities. In this way, the effect of predation could potentially work by increasing or decreasing zooplankton taxonomic and functional diversity (Litchman et al., 2010).
Consequently, the habitat structure created by macrophyte beds can mediate complex trophic interactions (Meerhoff et al., 2007b; Sagrario et al., 2009; Akhurst et al., 2017; Carniatto et al., 2020) and provide an excellent system to analyze the site-by-site taxonomic and functional variation, especially as studies analyzing the interaction of macrophytes, fish, and functional structure of zooplankton are rare. Also, understanding the spatial filters that determine species diversity can assist in ecosystem conservation and management strategies (Heino et al., 2019).
Here we analyzed how the habitat structure provided by macrophytes (biomass, richness, and Shannon’s diversity) and potential predation by small-bodied fishes (richness and abundance) determine the richness and dissimilarity of zooplankton species and functional traits. We expected that the structure of macrophytes and fish community would be positively related to both taxonomic and functional diversity of zooplankton because macrophyte habitat structure might allow greater species and traits occurrence. Regarding taxonomic and functional beta-diversity of zooplankton, the macrophyte richness may promote spatial dissimilarity, as different macrophyte species can shelter different zooplankton species/traits. Additionally, an increase or decrease in macrophyte biomass may also contribute to increase the spatial dissimilarity of zooplankton, since denser macrophyte beds tend to support littoral species, while pelagic species would mostly occur in less dense beds (Sodré & Bozelli, 2019). Furthermore, fish predation might control the most dominant species, allowing more species and functional traits to co-occur.
Methods
Study area
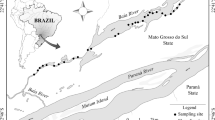
The Baía River presents slow water flow that ranges from 0.11 to 0.50 m/s (Bonecker et al., 1998). This tributary belongs to the Upper Paraná River Floodplain, and is located in the state of Mato Grosso do Sul/Brazil (Fig. 1). During the last 20 years, studies developed in the entire floodplain recorded 71 macrophyte species (Souza et al., 2017), 211 fish species (Lopes et al., 2015; Ota et al., 2018), and 617 zooplankton species (Bonecker et al., 2020). Baía River has approximately 18 m of width and 1 m of depth (Rodrigues et al., 2009), with the riverbanks covered mainly by herbaceous pasture vegetation (Lopes et al., 2006). This river also presents a pronounced littoral zone with aquatic macrophyte beds over almost all its perimeter, which allows a great diversity of species.
Sampling and community analyses
Zooplankton, fish, and macrophytes were simultaneously sampled during the dry season in August 2018. During this period, macrophyte beds are more intact and the other communities are less dispersed in floodplain environments. Sampling covered ~ 13.7 km, taking into account a minimum distance of ~ 350 m between each macrophyte bed, and following the river flow (Fig. 1). To sample a gradient of low to high macrophyte bed habitat structure, multi-species beds with different biomass were visually selected, totalizing 30 macrophyte beds considered here as sampled sites.
Zooplankton samples were taken at the subsurface (approximately 30 cm below the water–air interface), in the central region of the macrophyte beds (near the traps used for fish sampling). At each sampled site, 100 L of water were filtered through plankton net (68 µm) with buckets, and fixed with 4% formalin solution buffered with calcium carbonate. Specialized bibliographies (Koste, 1978; Reid, 1985; El-moor-Loureiro, 1997; Orlova-Bienkoswskaja, 2001; Perbiche-Neves et al., 2015) were used for species identification. To determine the zooplankton abundance (individuals m−3), we used an optical microscope (Olympus CX-41) and a Sedgewick-Rafter camera. At least 50 individuals of each zooplankton group (rotifers, cladocerans, and copepods) were counted in each sample. When the samples did not reach this minimum number of individuals, they were fully counted. The identification and classification of functional traits for each zooplankton species were made according to data available in the specialized literature (Braghin et al., 2018). The traits body size; habitat; lifespan; type of feeding; type of reproduction; and predatory escape response were selected (Table 1). These traits represent how zooplankton respond to environmental conditions, and directly influence ecosystem processes (Barnett et al., 2007; Rizo et al., 2017; Obertegger & Flaim, 2018). Predation might alter the traits body size and type of reproduction by selecting large-bodied zooplankton, including ovate females (Iglesias et al., 2011), predatory escape response by selecting the slower swimmers, and habitat by selecting pelagic species, whereas habitat structure provided by macrophytes might influence the zooplankton habitat, by favoring littoral species, as well as different type of feeding, lifespan, and type of repoduction, by offering more food resources and development conditions. For more details about the importance of each trait chosen, please see Braghin et al. (2018).
We used floating minnow traps for fish sampling. Minnow traps are acrylic side traps with a 1 cm opening for fish entry. Its upper side is composed by pieces of polyethylene to allow its placement on the surface layer of the water, while its down portion has a plastic plate with small fixed holes to drain the water and remove the fishes (Cunha et al., 2011). Three traps were installed in each macrophyte bed, 4 m from the riverbank, to minimize the effect of fish species that transit between the open area and the area structured by macrophytes. The traps were installed at 7 a.m. and checked for fish every 8 h, accounting for a sampling effort of 24 h. Following ethical practice, the captured fishes were anesthetized with clove oil and fixed in 4% formalin. Subsequently, they were identified at the species level (Ota et al., 2018), and quantified (individuals m−2). Fish sampling was approved by the Ethics Committee on the Use of Animals of the State University of Maringá (CEUA/UEM - No. 5980040618). We identified 20 fish species distributed into six families (supplementary material A). Characidae (13 species), and Cichlidae (3 species) were the families with the greatest species richness. The richness varied from 1 to 11 species among sampling locations, and the abundance from 1 to 228 ind. m−2 (Supplementary Material B).
Macrophytes were also sampled near the fish traps. A quadrat (0.5 m × 0.5 m) was used, and all plants material contained to a depth of 0.5 m depth were sampled. In the laboratory, macrophytes were identified, washed, and dried (at 60°C) to obtain the dry weight (DW) (adapted from Wetzel & Likens, 1991). The habitat structure provided by macrophytes can be expressed by various characteristics, so we calculated total biomass (g DW m−3), taxonomic richness [TRic—number of species (S) in each bed, where rare and common species have the same weight), and Shannon index (H′—in which the biomass of each species was used to evaluate the relative abundance values and it is sensitive to rare species (Magurran, 2004)]. Through macrophyte biomass, we can evaluate the quantity of roots, branch, and leaves that might be available to zooplankton as refuge, and shelves for food resources. Species richness can represent different biotypes of macrophytes and also influence the availability of refuge and food resource for zooplankton community, while Shannon’s diversity combines quantity and quality, i.e., biomass and richness. A total of 18 macrophyte species were identified, belonging to 10 families (Supplementary Material A). Salviniaceae (four species), Polygonaceae (three species), and Pontederiaceae (three species) were the families with the greatest species richness. The macrophyte richness varied from 1 to 13 species; the lowest biomass was 45.77 g DW m−3 and the highest was 883.29 g DW m−3, and the Shannon’s diversity (H′) varied between 2.2 and 0 (Supplementary Material B).
Functional analyses
All analyses in our study were performed in R software (R Core Team, 2020). We calculated functional richness (FRic) following the methodology proposed by Villéger et al. (2008) [R function ‘dbFD’, package “FD” (Laliberté et al., 2014)]. FRic represents the total functional space that the species occupy in a given community. High values indicate that species have distinct combinations of traits, whereas lower values indicate that the species have more similar traits (functionally redundant) (Villéger et al., 2008). We calculated FRic using one matrix of species density by site, and one matrix of functional traits by species. We applied Gower distance as we have categorical and numerical traits (Gower, 1966). The functional space was calculated through a Principal Coordinate Analysis (PCoA) based on the combination of traits per species of each site. The first four axes of PCoA were retained, which presented the high quality of functional space occupied by the zooplankton species. Regarding the rule or process used to determine how many axes to retain, according to the method proposed by Maire et al. (2015), a high-quality functional space needs to show congruence between the initial functional distance (Gower distance) and the standardized distances in the new space (generated by PCoA).
To analyze the distribution of zooplankton species and functional traits, we calculated the beta-diversity and its components (turnover and nestedness). We followed the methodology proposed by Baselga (2010) for taxonomic beta-diversity [R function ‘beta.pair’, package “betapart” (Baselga & Orme, 2012)], whereas to determine functional beta-diversity, we used the methodology proposed by Villéger et al. (2013) [R function ‘functional.beta.pair’, package “betapart” (Baselga & Orme, 2012)]. We calculated three metrics, the Jaccard dissimilarity index (‘jac’), which represents the total variation of species and traits between sites; the nestedness component (‘jne’) that corresponds to the loss of species or traits; and the turnover component (‘jtu’) that represents the replacement of species and traits between sites. The pairwise distance between sites was computed using the Gower distance, and from this matrix, the functional traits were summarized through a PCoA, following the same procedure described in the previously paragraph.
Statistical analyses
To analyze how the habitat structure provided by macrophytes and potential predation by fish determine the richness of zooplankton species and functional traits, we performed linear regression models [R function ‘lm’; package ‘stats’ (Bolar, 2019)] for taxonomic richness and beta regressions for functional richness [R function ‘betareg’, package ‘betareg’ (Cribari-Neto & Zeileis, 2010)]. We used five predictors: macrophyte species richness, Shannon’s diversity, and biomass, and fish species richness and abundance of individuals. Beta regression is linked to generalized linear models (GLMs) and is used for response variables that vary between 0 and 1 (such as functional richness) (Cribari-Neto & Zeileis, 2010).
We performed Generalized Dissimilarity Models (GDM) (Ferrier et al., 2007) controlling for the effect of space [R function ‘gdm.varImp’; package “gdm” (Fitzpatrick et al., 2020)] to analyze which of the five predictors related to macrophytes (species richness, Shannon’s diversity, and biomass) and fish (species richness and abundance of individuals) determine the distribution of zooplankton species and functional traits. GDM is based on matrices of regression, a technique that allows controlling spatial variation in the composition of sets between pairs of geographic locations, and it can be based on any dissimilarity matrix as a response (Ferrier et al., 2007). We used the three dissimilarities described above (total beta-diversity, and its components turnover and nestedness) as response variables to calculate GDM analysis. Model assumptions were tested for all analyses.
We performed piecewise Structural Equation Models (SEM; Lefcheck, 2016) to analyze the effects of macrophytes on fishes and how this may influence the zooplankton structure. We tested several pathways (Fig. 2). Before performing SEM analysis, we prepared and summarized the response variables (zooplankton functional and taxonomic diversity) by calculating the local contribution of each site to total beta-diversity (LCBD—Legendre & De Cáceres, 2013; R function ‘LCBD.comp’ of package “adespatial”). LCBD can be performed through a dissimilarity matrix or beta-diversity component matrix. Here, we used the beta-diversity component matrix previously calculated by ‘jac’ (described in the above subsection), which represents the total variation of species and traits between sites. Piecewise Structural Equation Models were created using GLMs—generalized linear models (R function ‘glm’ of package “stats”), and fitted using the function ‘psem’ (R package “piecewiseSEM”). Fish abundance of individuals and richness, and macrophyte richness, Shannon’s diversity, and biomass, were used as predictors in the models. To choose the best model, we reduced the number of variables using Akaike Information Criteria corrected for small sample size (AICc), and checked correlations between the predictors by tools implemented in the “piecewiseSEM” R package (Lefcheck, 2016).
Finally, we analyzed which zooplankton functional traits were influenced by macrophytes structure and potential fish predation by performing a Fourth-Corner Analysis [R function ‘fourthcorner’, package “ade4” (Bougeard & Dray, 2018)]. To obtain a more accurate model, we first transformed the predictor variables (biomass of macrophytes per species in each site, and abundance of fish per species in each site) into distances through two PCoAs [R function ‘pcoa’, package “ape” (Paradis & Schliep, 2018)] and retained the first axis of each predictor. In this way, three matrices were used in the Fourth-Corner Analysis, the zooplankton species distribution (L), the zooplankton functional traits per species (Q), and the predictor variables (R—composed by the first axis of macrophyte and fish from the PCoAs). We used the model type 6, which is more robust because permute the rows and columns of the matrix L (Dray & Legendre, 2008; ter Braak et al., 2012). We tested the model significance through 999 permutations, and applied p-adjusted for multiple comparisons (“Bonferroni” correction).
Results
Description of the zooplankton community
A total of 142 species of zooplankton were identified, most of them belonging to Rotifera (75 species), followed by Cladocera (52 species), and Copepoda (15 species) (Supplementary Material A). The species were distributed into 25 families, among them, Lecanidae, Trichocercidae, and Brachionidae were the families with the greatest species richness of rotifers (21, 14, and 12 species, respectively), Chydoridae was the family with the greatest species richness of cladocerans (38 species), and Cyclopidae the family with the greatest species richness of copepods (12 species). The zooplankton richness varied from 23 to 71 species among sampling locations (for more details please see Supplementary Material B). We found that the zooplankton total taxonomic beta-diversity (Tjac = 0.58) was slightly higher than the total functional beta-diversity (Fjac = 0.47). The turnover component had a greater contribution to the total taxonomic beta-diversity (Tjtu = 0.48), and the nestedness to the total functional beta-diversity (Fjne = 0.29) (Fig. 3).
Relationship between zooplankton taxonomic and functional diversity and the predictors (macrophyte and fish)
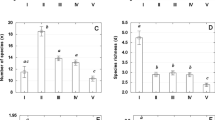
The taxonomic richness of zooplankton was positively related to macrophyte richness (lm-model: F = 5.48, P = 0.02, Std. Error: 10.21, R2 adj.: 0.13; Degrees of Freedom: 28) and macrophyte diversity (Shannon’s diversity) (lm-model: F = 5.90, P = 0.02, Std. Error: 10.14, R2 adj.: 0.14; Degrees of Freedom: 28) (Fig. 4a, b), whereas zooplankton functional richness was positively related to fish richness (P = 0.02, z-value = 2.30, pseudo-R2: 0.001, Std. Error: 0.04; Log-likelihood: 265.7 on 5 Df) and macrophyte richness (P = 0.007, z-value = 2.67, pseudo-R2: 0.1783, std. error: 0.03, Log-likelihood: 266.3 on 5 Df) (Fig. 4c, d). The other predictors were not related to zooplankton taxonomic and functional richness (Table 2).
Significant regressions (considering P < 0.05) between taxonomic richness of zooplankton (= species richness) and macrophyte richness (a), and macrophyte diversity (Shannon’s diversity) (b). And significant regressions between the functional richness of zooplankton and macrophyte richness (c) and fish richness (d)
Macrophyte structure (biomass, Shannon’s diversity, and richness) determined most of the zooplankton taxonomic beta-diversity variation. We observed that a small increase in these variables was sufficient to determine the increase in zooplankton taxonomic beta-diversity, and after that, there was a stabilization trend (Fig. 5a, c, e). All predictors explained 32.58% (model-P < 0.001) of the zooplankton total taxonomic beta-diversity. Concerning the beta components, all predictors explained 19.24% (model-P = 0.01) of the zooplankton taxonomic turnover, and the macrophyte biomass was the most important predictor for this component (24.65%) (Table 3). Zooplankton taxonomic nestedness was not explained by the predictors (Table 3).
GDM regressions to zooplankton total taxonomic beta-diversity (y-axis). On the x-axis are the predictor variables: macrophyte biomass (a), macrophyte diversity (Shannon’s diversity) (b), macrophyte richness (c), fish abundance (d), and fish richness (e). The percentages of explanation (%) of each preditor for the model are shown
We found that 9.98% of zooplankton total functional beta-diversity was explained by the global GDM model (model-P = 0.02). Fish abundance contributed to 62.21% of this total variation, with an exponential increase in zooplankton total functional beta-diversity from a high fish density (around 100; Fig. 6b). Macrophyte richness (4.75%) was the second most important predictor of zooplankton functional beta-diversity, followed by macrophyte Shannon’s diversity (4.59%); the slopes increased after high values of these predictors (Fig. 6c, e). The predictor variables were important in explaining only the variation in functional nestedness (total explanation: 17.82%; model-P = 0.02), and the abundance of fish was the most important predictor for this component (54.06%) (Table 3).
GDM regressions to zooplankton total functional beta-diversity (y-axis). On the x-axis are the predictor variables: Fish abundance (a), macrophyte richness (b), macrophyte diversity (Shannon’s diversity) (c), macrophyte biomass (d), and fish richness (e). The percentages of explanation (%) of each preditor for the model are shown
We observed through the Structural Equation Model that macrophyte biomass and Shannon’s diversity affected the fish richness in opposite ways. Macrophyte biomass had a negative effect on fish richness (SEM std. estimate: − 0.47, P = 0.006), while Shannon’s diversity had a positive effect (SEM std. estimate: 0.33, P = 0.04). Zooplankton taxonomic LCBD was negatively affected by macrophyte Shannon’s diversity (SEM std. estimate: − 0.41, P = 0.03); the other relationships were not significant (Fig. 7). Fourth-Corner Analysis showed several negative relationships, with habitat structure provided by macrophytes influencing all zooplankton traits. The only positive relationship was between macrophytes and feeding type (P = 0.03). Fish predation was negatively related only to body size (Fig. 8).
Path diagram representing final structural equation model pathways linking stream macrophyte, fish, and zooplankton communities. Boxes represent measured variables. Arrows represent unidirectional relationships among variables. Black arrows denote significant paths, and semi-transparent represents non-significant (P > 0.05). Pointed arrows represent negative relationships and arrows ending in circles represent positive relationships. R2, P value, and std. estimate are shown
Fourth-Corner significant results (P-adjusted for multiple comparisons) showing the interaction between the six functional traits and the predictor variables – (first axis of PCoA from macrophytes and fishes, please for more details see methods). The positive interaction is shown in red and negative interactions in blue
Discussion
The potential predation, represented by fish community structure (i.e., richness and abundance), explained most of the variation of zooplankton functional beta-diversity, while habitat structure, represented by macrophyte community structure (Shannon’s diversity, richness, and biomass), explained most of the variation of zooplankton taxonomic richness and taxonomic beta-diversity, and was related to six functional traits, in agreement with our predictions. Littoral regions of subtropical environments often support high richness of zooplankton species, owing to the presence of families morphologically adapted to explore resources in the macrophytes, such as Lecanidae (rotifers), Chydoridae (cladocerans), Trichocercidae (rotifers), and Cyclopidae (copepods) (Lansac-Tôha et al., 2009; Serafim-Júnior et al., 2016; Bomfim et al., 2017). Roots and submerged parts of aquatic vegetation allow zooplankton organisms to settle and attach their eggs (Debastiani-Júnior et al., 2016; Battauz et al., 2017), whereas the great richness and abundance of microorganisms associated with macrophytes provide a variety of food resource for zooplankton, such as omnivorous and carnivorous microcrustaceans (Thomaz & Cunha, 2010; Debastiani-Júnior et al., 2016), and rotifers (Rossa & Bonecker, 2003; Grzybkowska et al., 2018).
The refuge and food availability offered by macrophytes (Dabés & Velho, 2001) may allow a more diverse assembly with zooplankton organisms that explore several types of food resources, reflecting in the positive relationships observed between macrophytes and the zooplankton trait feeding type, as well, zooplankton taxonomic richness and taxonomic beta-diversity. Nevertheless, this strong habitat structure provided by macrophytes also negatively influenced other zooplankton traits. High food availability is linked to small-bodied zooplankton organisms (Bomfim et al., 2018). Small-bodied zooplankton, in turn, is represented by organisms with asexual reproduction instead sexual, short lifespan instead long, and low predatory escape response instead medium, high, or maximum, as observed in our data. Therefore, the high food availability offered by macrophyte structure might have favored these specific sub-traits (cited above), which lead to the negative relationships observed in the Fourth-Corner Analysis between macrophytes and the traits body size, reproduction type, habitat, lifespan, and predatory escape response. In other words, macrophytes structure may have favored the prevalence of small-bodied zooplankton organisms that is linked to other specific traits, which were also selected by this environmental filter.
On the other hand, we observed that macrophyte diversity was positively related to fish diversity, which probably increased predation pressure on zooplankton organisms and lead to a positive effect on zooplankton functional diversity. Due to nutritional advantages and easy detection, fish usually predate large-bodied zooplankton species, reducing their numerical density and favoring smaller-bodied species (Bonecker et al., 2011; Iglesias et al., 2017). Our findings support this viewpoint through the negative relationship observed between fish and the zooplankton trait body size. The size-selective predation by fish on large-bodied zooplankton species may also reflect in other zooplankton traits (Litchman et al., 2010; Hébert et al., 2017), once small-bodied zooplankton present particular sub-traits, as exemplified in the previously paragraph. Fish richness may have promoted high zooplankton functional richness, as well as fish abundance may have promoted high zooplankton functional beta-diversity. These findings indicate that certain levels of predation had a positive effect on zooplankton functional diversity by balancing the distribution of numerical density within species and functional traits (i.e., promoting a more equal distribution of density; as functional richness weigh species density). Also, predation is well recognized for reducing competition between groups (Chase et al., 2002); sight-feeding fish generally prey on large-bodied species and on most abundant zooplankton species that normally are better competitors and dominate the environments. Therefore, predation may provide space and conditions for other species to establish and grow (Brooks & Dodson, 1965; Litchman et al., 2010), and consequently increase functional richness and functional beta-diversity of zooplankton.
Macrophyte biomass and diversity had different effects on fish richness, which also influenced the zooplankton beta-diversity in opposite ways. Macrophytes can mediate the relationship between zooplankton and their predators (Meerhoff et al., 2006; Santos et al., 2020), because more complex macrophyte beds provide shelter for small organisms that occupy microhabitats predators cannot access (Meerhoff et al., 2007b; Iglesias et al., 2017; Strzałek & Koperski, 2019). Therefore, once again, this expected relationship reinforces the link between macrophytes and small-bodied zooplankton. Macrophyte diversity promoted high fish richness, but macrophyte biomass reduced fish richness. Dense macrophyte beds limit the predators’ movement and prey detection (Diehl, 1988), thus, normally higher fish densities are found in intermediate levels of macrophyte biomass (Warfe & Barmuta, 2006; Cunha et al., 2019). These relationships between macrophytes and fish confounded the patterns and may have led to a negative effect of macrophyte diversity on zooplankton taxonomic beta-diversity (by favoring fishes).
The spatial relationship between macrophyte structure and fish also influenced the zooplankton turnover and nestedness. Zooplankton taxonomic beta-diversity was most explained by turnover component and influenced by macrophyte diversity and biomass, which means that the spatial distribution of macrophyte beds with different characteristics determined the species replacement among sites. Meanwhile, the increase in fish abundance promoted the development of functional trait sub-sets formed from a large set (nestedness). Fewer structured macrophyte habitats usually host fewer zooplankton and fish species (Braghin et al., 2016; Stephan et al., 2019). Conversely, highly structured macrophyte habitats host greater zooplankton and fish species (Lopes et al., 2015; Brito et al., 2020). Therefore, the difference between macrophyte habitat structure created a spatial environmental heterogeneity (Choi et al., 2014) that selectively filtered different zooplankton species and functional traits, consequently increasing zooplankton taxonomic and functional beta-diversity. Our results have direct implication for guiding biomonitoring and conservation, since areas that contain high macrophytes richness and diversity may assure high fish and zooplankton richness, which in turn, will continue to ensure ecosystem functions offered by these species.
Our results showed complex interactions between different metrics of macrophytes, fish, and zooplankton. We demonstrated how these abiotic and biotic filters working together affected zooplankton taxonomic and functional diversity. Moreover, our findings revealed that zooplankton species and functional traits may respond differently to these filters, reaffirming the importance to analyze as many factors as possible to have a realistic view of the species interaction and ecosystem functioning in natural environments.
References
Akhurst, D. J., G. B. Jones, M. Clark & A. Reichelt-Brushett, 2017. Effects of fish and macrophytes on phytoplankton and zooplankton community structure in a subtropical freshwater reservoir. Limnologica 62: 5–18.
Barnett, A. J., K. Finlay & B. E. Beisner, 2007. Functional diversity of crustacean zooplankton communities: towards a trait-based classification. Freshwater Biology 52: 796–813.
Baselga, A., 2010. Partitioning the turnover and nestedness components of beta diversity. Global Ecology and Biogeography 19: 134–143.
Baselga, A. & C. D. L. Orme, 2012. Betapart: an R package for the study of beta diversity. Methods in Ecology and Evolution 3: 808–812.
Battauz, Y. S., S. B. J. De-Paggi & J. C. Paggi, 2017. Macrophytes as dispersal vectors of zooplankton resting stages in a subtropical riverine floodplain. Aquatic Ecology 51: 191–201.
Bellwood, D. R., T. P. Hughes, C. Folke & M. Nyström, 2004. Confronting the coral reef crisis. Nature 429: 827–833.
Bolar, K., 2019. STAT: interactive document for working with basic statistical analysis in R.
Bomfim, F. F., L. S. M. Braghin, C. C. Bonecker, & F. A. Lansac-Tôha, 2018. High food availability linked to the dominance of small zooplankton in a subtropical floodplain. International Review of Hydrobiology 103: 26–34.
Bomfim, F. F., T. Mantovano, D. C. Amaral, W. S. Palhiarini, C. C. Bonecker, & F. A. Lansac-Tôha, 2017. Adjacent environments contribute to the increase of zooplankton species in a neotropical river. Acta Limnologica Brasiliensia 29: 1–14
Bonecker, C. C., F. A. Lansac-Tôha & D. C. Rossa, 1998. Planktonic and non-planktonic rotifers in two environments of the Upper Paraná River floodplain, state of Mato Grosso do Sul, Brazil. Brazilian Archives of Biology and Technology 41: 447–456.
Bonecker, C. C., F. De Azevedo & N. R. Simões, 2011. Zooplankton body-size structure and biomass in tropical floodplain lakes: relationship with planktivorous fishes. Acta Limnologica Brasiliensia 23: 217–228.
Bonecker, C. C., L. P. Diniz, L. de S. M. Braghin, T. Mantovano, J. V. F. da Silva, F. de F. Bomfim, D. A. Moi, S. Deosti, G. N. T. Dos Santos, D. A. Das Candeias, A. J. de M. M. Mota, L. F. M. Velho & F. A. Lansac-Tôha, 2020. Synergistic effects of natural and anthropogenic impacts on zooplankton diversity in a subtropical floodplain: a long-term study. Oecologia Australis 24: 524–537.
Bougeard, S. & S. Dray, 2018. Supervised Multiblock Analysis in R with the ade4 Package. Journal of Statistical Software 86: 1–17.
Braghin, L. de S. M., B. de A. Almeida, D. C. Amaral, T. F. Canella, B. C. G. Gimenez & C. C. Bonecker, 2018. Effects of dams decrease zooplankton functional β-diversity in river-associated lakes. Freshwater Biology 63: 721–730.
Brito, M. T. da S., J. Heino, U. M. Pozzobom & V. L. Landeiro, 2020. Ecological uniqueness and species richness of zooplankton in subtropical floodplain lakes. Aquatic Sciences 82: 43–56.
Brooks, J. L. & S. I. Dodson, 1965. Predation, Body Size and Composition of Plankton. Science 150: 28–35.
Carniatto, N., E. R. Cunha, S. M. Thomaz, B. A. Quirino & R. Fugi, 2020. Feeding of fish inhabiting native and non-native macrophyte stands in a Neotropical reservoir. Hydrobiologia 847: 1553–1563.
Chase, J. M., P. A. Abrams, J. P. Grover, S. Diehl, P. Chesson, R. D. Holt, S. A. Richards, R. M. Nisbet & T. J. Case, 2002. The interaction between predation and competition: a review and synthesis. Ecology Letters 5: 302–315.
Choi, J. Y., K. S. Jeong, S. K. Kim, G. H. La, K. H. Chang & G. J. Joo, 2014. Role of macrophytes as microhabitats for zooplankton community in lentic freshwater ecosystems of South Korea. Ecological Informatics Elsevier B.V. 24: 177–185.
Cribari-Neto, F. & A. Zeileis, 2010. Beta Regression in R. Journal of Statistical Software 34: 1–24.
Cunha, E. R., S. M. Thomaz, H. B. Antoniazi Evangelista, J. Carniato, C. F. Souza & R. Fugi, 2011. Small-sized fish assemblages do not differ between a native and a recently established non-indigenous macrophyte in a neotropical ecosystem. Natureza a Conservacao 9: 61–66.
Cunha, E. R., K. O. Winemiller, J. C. da Silva, T. M. Lopes, L. C. Gomes, S. M. Thomaz & A. A. Agostinho, 2019. α and β diversity of fishes in relation to a gradient of habitat structural complexity supports the role of environmental filtering in community assembly. Aquatic Sciences 81: 38.
Dabés, M. B. G. e S. & L. F. M. Velho, 2001. Assemblage of testate amoebae (Protozoa, Rhizopoda) associated to aquatic macrophytes stands in a marginal lake of the São Francisco river floodplain, Brazil. Acta Scientiarum 23: 291–297.
de Sodré, E. O, & R. L. Bozelli, 2019. How planktonic microcrustaceans respond to environment and affect ecosystem: a functional trait perspective. International Aquatic Research 11: 207–223
Debastiani-Júnior, J. R., L. M. A. Elmoor-Loureiro & M. G. Nogueira, 2016. Habitat architecture influencing microcrustaceans composition: a case study on freshwater Cladocera (Crustacea Branchiopoda). Brazilian Journal of Biology 76: 93–100.
Diehl, S., 1988. Nordic Society Oikos foraging efficiency of three freshwater fishes: effects of structural complexity and light. Oikos 53: 207–214.
Dray, S. & P. Legendre, 2008. Testing the species traits-environment relationships: the fourth-corner problem revisited. Ecology 89: 3400–3412.
El-moor-Loreiro, L. M. A., 1997. Manual de Identificação de Cladóceros Límnicos do Brasil. Universa, Brasília.
Ferrier, S., G. Manion, J. Elith & K. Richardson, 2007. Using generalized dissimilarity modelling to analyse and predict patterns of beta diversity in regional biodiversity assessment. Diversity and Distributions 13: 252–264.
Fitzpatrick, M. C., K. Mokany, G. Manion, M. Lisk, S. Ferrier & D. Nieto-Lugilde, 2020. Generalized Dissimilarity Modeling (GDM) in R.
Gagic, V., I. Bartomeus, T. Jonsson, A. Taylor, C. Winqvist, C. Fischer, E. M. Slade, I. Steffan-Dewenter, M. Emmerson, S. G. Potts, T. Tscharntke, W. Weisser & R. Bommarco, 2015. Functional identity and diversity of animals predict ecosystem functioning better than species-based indices. Proceedings of the Royal Society B: Biological Sciences 282: 20142620.
Gower, J. C., 1966. Some distance properties of latent root and vector methods used in multivariate analysis. Biometrika 53: 325–338.
Grzybkowska, M., M. Dukowska, J. Leszczyńska, J. Lik, E. Szczerkowska-Majchrzak & M. Przybylski, 2018. The food resources exploitation by small-sized fish in a riverine macrophyte habitat. Ecological Indicators 90: 206–214.
Hebert, M.-P., B. E. Beisner & R. Maranger, 2016. A meta-analysis of zooplankton functional traits influencing ecosystem function A meta – analysis of zooplankton functional traits influencing – ecosystem function. Ecology 97: 1069–1080.
Hébert, M.-P. P., B. E. Beisner & R. Maranger, 2017. Linking zooplankton communities to ecosystem functioning: toward an effect-Trait framework. Journal of Plankton Research 39: 3–12.
Heino, J., A. S. Melo & L. M. Bini, 2015a. Reconceptualising the beta diversity-environmental heterogeneity relationship in running water systems. Freshwater Biology 60: 223–235.
Heino, J., A. S. Melo, T. Siqueira, J. Soininen, S. Valanko & L. M. Bini, 2015b. Metacommunity organisation, spatial extent and dispersal in aquatic systems: patterns, processes and prospects. Freshwater Biology 60: 845–869.
Heino, J., J. Alahuhta, S. Fattorini & D. Schmera, 2019. Predicting beta diversity of terrestrial and aquatic beetles using ecogeographical variables: insights from the replacement and richness difference components. Journal of Biogeography 46: 304–315.
Iglesias, C., N. Mazzeo, M. Meerhoff, G. Lacerot, J. M. Clemente, F. Scasso, C. Kruk, G. Goyenola, J. García-Alonso, S. L. Amsinck, J. C. Paggi, S. J. de Paggi & E. Jeppesen, 2011. High predation is of key importance for dominance of small-bodied zooplankton in warm shallow lakes: evidence from lakes, fish exclosures and surface sediments. Hydrobiologia 667: 133–147.
Iglesias, C., E. Jeppesen, N. Mazzeo, J. P. Pacheco, F. T. de Mello, F. Landkildehus, C. Fosalba, J. M. Clemente & M. Meerhoff, 2017. Fish but not macroinvertebrates promote trophic cascading effects in high density submersed plant experimental lake food webs in two contrasting climate regions. Water (Switzerland) 9: 514.
Koste, W., 1978. Rotatoria die Rädertiere Mitteleuropas begründet von Max Voight. Monogononta, Gebrüder Borntraeger, Berlin.
Laliberté, E., P. Legendre & B. Shipley, 2014. FD: measuring functional diversity from multiple traits, and other tools for functional ecology. R package version 1.0-12..
Lansac-Tôha, F., C. Bonecker, L. Velho, N. Simões, J. Dias, G. Alves & E. Takahashi, 2009. Biodiversity of zooplankton communities in the Upper Paraná River floodplain: interannual variation from long-term studies. Brazilian Journal of Biology 69: 539–549.
Lavorel, S., J. Storkey, R. D. Bardgett, F. De Bello, M. P. Berg, X. Le Roux, M. Moretti, C. Mulder, R. J. Pakeman, S. Díaz & R. Harrington, 2013. A novel framework for linking functional diversity of plants with other trophic levels for the quantification of ecosystem services. Journal of Vegetation Science 24: 942–948.
Lefcheck, J. S., 2016. piecewiseSEM: piecewise structural equation modelling in r for ecology, evolution, and systematics. Methods in Ecology and Evolution 7: 573–579.
Legendre, P. & M. De Cáceres, 2013. Beta diversity as the variance of community data: dissimilarity coefficients and partitioning. Ecology Letters 16: 951–963.
Litchman, E., P. de Tezanos Pinto, C. A. Klausmeier, M. K. Thomas & K. Yoshiyama, 2010. Linking traits to species diversity and community structure in phytoplankton. Hydrobiologia 653: 15–28.
Lopes, C. D. A., A. C. E. A. De Faria, G. I. Manetta & E. Benedito-Cecilio, 2006. Caloric density of aquatic macrophytes in different environments of the Baía river subsystem, Upper Paraná river floodplain, Brazil. Brazilian Archives of Biology and Technology 49: 835–842.
Lopes, T. M., E. R. Cunha, J. C. B. Silva, R. D. L. Behrend & L. C. Gomes, 2015. Dense macrophytes influence the horizontal distribution of fish in floodplain lakes. Environmental Biology of Fishes 98: 1741–1755.
Loreau, M., S. Naeem, P. Inchausti, J. Bengtsson, J. P. Grime, A. Hector, D. U. Hooper, M. A. Huston, D. Raffaelli, B. Schmid, D. Tilman & D. A. Wardle, 2001. Ecology: biodiversity and ecosystem functioning: current knowledge and future challenges. Science 294: 804–808.
Magurran, A., 2004. Measuring Biological Diversity. Blackwell Science Ltd, a Blackwell Publishing company, Malden.
Maire, E., G. Grenouillet, S. Brosse & S. Villéger, 2015. How many dimensions are needed to accurately assess functional diversity? A pragmatic approach for assessing the quality of functional spaces. Global Ecology and Biogeography 24: 728–740.
Mason, N. W. H., D. Mouillot, W. G. Lee & J. B. Wilson, 2005. Functional richness, functional evenness and functional divergence: the primary components of functional diversity. Oikos 111: 112–118.
Meerhoff, M., C. Fosalba, C. Bruzzone, N. Mazzeo, W. Noordoven & E. Jeppesen, 2006. An experimental study of habitat choice by Daphnia: plants signal danger more than refuge in subtropical lakes. Freshwater Biology 51: 1320–1330.
Meerhoff, M., J. M. Clemente, F. T. de Mello, C. Iglesias, A. R. Pedersen & E. Jeppesen, 2007a. Can warm climate-related structure of littoral predator assemblies weaken the clear water state in shallow lakes? Global Change Biology 13: 1888–1897.
Meerhoff, M., C. Iglesias, F. T. De Mello, J. M. Clemente, E. Jensen, T. L. Lauridsen & E. Jeppesen, 2007b. Effects of habitat complexity on community structure and predator avoidance behaviour of littoral zooplankton in temperate versus subtropical shallow lakes. Freshwater Biology 52: 1009–1021.
Mouillot, D., N. A. J. Graham, S. Villéger, N. W. H. Mason & D. R. Bellwood, 2013. A functional approach reveals community responses to disturbances. Trends in Ecology and Evolution 28: 167–177.
Naeem, S., J. E. Duffy & E. Zavaleta, 2012. The functions of biological diversity in an age of extinction. Science 336: 1401–1406.
Obertegger, U. & G. Flaim, 2018. Taxonomic and functional diversity of rotifers, what do they tell us about community assembly? Hydrobiologia 823: 79–91.
Orlova-Bienkoswskaja, M. J., 2001. Cladocera: Anomopoda - Daphniidae: genus Simocephalus. Guides to the identification of the Microinvertebrates of Continental waters of the world. Backhuys Publishers.
Ota, R. R., G. de C. Deprá, W. J. da Graça & C. S. Pavanelli, 2018. Peixes da planície de inundação do alto rio Paraná e áreas adjacentes: revised, annotated and updated. Neotropical Ichthyology 16: 1–111.
Paradis, E. & K. Schliep, 2018. ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 35: 526–528.
Perbiche-Neves, G., G. A. Boxshall, D. Previattelli, M. G. Nogueira & C. E. F. da Rocha, 2015. Identification guide to some Diaptomid species (Crustacea, Copepoda, Calanoida, Diaptomidae) of “de la Plata” River Basin (South America). ZooKeys 111: 1–111.
Petchey, O. L., A. Hector & K. J. Gaston, 2004. How do different measures of functional diversity perform? Ecology 85: 847–857.
Pinheiro, D. T., J. Max, S. Corrêa, C. S. Chaves, D. Patrick, F. Campos, S. Cristina & D. M. Zacardi, 2016. Distribution and ichthyofauna diversity associated with aquatic macrophytes bank of an Amazon floodplain lake, Pará State, Brazil. Acta of Fisheries and Aquatic Resources 4: 59–70.
R Core Team, 2020. A language and environment for statistical computing. Vienna, Austria. https://www.r-project.org.
Reid, J. W., 1985. Chave de identificação e lista de referências bibliográficas para as espécies continentais sulamericanas de vida livre da ordem Cyclopoida (Crustacea, Copepoda). Boletim de Zoologia 9: 17.
Rizo, E. Z. C., Y. Gu, R. D. S. Papa, H. J. Dumont & B. Han, 2017. Identifying functional groups and ecological roles of tropical and subtropical freshwater Cladocera in Asia. Hydrobiologia 799: 83–99.
Rodrigues, L., S. Train, V. Bovo-Scomparin, S. Jati, C. Borsalli & E. Marengoni, 2009. Interannual variability of phytoplankton in the main rivers of the Upper Paraná River floodplain, Brazil: influence of upstream reservoirs. Brazilian Journal of Biology 69: 501–516.
Rossa, D. C. & C. C. Bonecker, 2003. Abundance of planktonic and non-planktonic rotifers in floodplain lakes of the Upper Paraná River floodplain. Amazoniana 17: 567–581.
Sagrario, G., M. De Los Ángeles, E. Balseiro, R. Ituarte & E. Spivak, 2009. Macrophytes as refuge or risky area for zooplankton: a balance set by littoral predacious macroinvertebrates. Freshwater Biology 54: 1042–1053.
Santangelo, J. M., B. N. Soares, T. Paes, P. Maia-Barbosa, R. Tollrian & R. L. Bozelli, 2018. Effects of vertebrate and invertebrate predators on the life history of Daphnia similis and Moina macrocopa (Crustacea: Cladocera). Annales de Limnologie - International Journal of Limnology 54: 25.
Santos, N. G., L. R. Stephan, A. Otero, C. Iglesias & M. S. M. Castilho-Noll, 2020. How free-floating macrophytes influence interactions between planktivorous fish and zooplankton in tropical environments? An in-lake mesocosm approach. Hydrobiologia 847: 1357–1370.
Serafim-Júnior, M., F. A. Lansac-Tôha, R. M. Lopes & G. Perbiche-Neves, 2016. Continuity effects on rotifers and microcrustaceans caused by the construction of a downstream reservoir in a cascade series (Iguaçu River, Brazil). Brazilian Journal of Biology 76: 279–291.
Souza, D. C., E. R. Cunha, R. de A. Murillo, M. J. Silveira, M. M. Pulzatto, M. S. Dainez-Filho, L. A. Lolis, & S. M. Thomaz, 2017. Species inventory of aquatic macrophytes in the last undammed stretch of the Upper Parana River, Brazil. Acta Limnologica Brasiliensia 29: e115
Stephan, L. R., B. E. Beisner, S. G. M. Oliveira & M. S. M. Castilho-Noll, 2019. Influence of Eichhornia crassipes (Mart) solms on a tropical microcrustacean community based on taxonomic and functional trait diversity. Water (Switzerland) 11: 1–16.
Strzałek, M. & P. Koperski, 2019. The effect of dense patches of Stratiotes aloides L. on the spatial structure of microcrustacean assemblages in an oxbow lake. Ecohydrology and Hydrobiology European Regional Centre for Ecohydrology of the Polish Academy of Sciences 19: 75–82.
Suding, K. N., S. Lavorel, F. S. Chapin, J. H. C. Cornelissen, S. Díaz, E. Garnier, D. Goldberg, D. U. Hooper, S. T. Jackson & M. L. Navas, 2008. Scaling environmental change through the community-level: a trait-based response-and-effect framework for plants. Global Change Biology 14: 1125–1140.
ter Braak, C., A. Cormont & S. Dray, 2012. Improved testing of species traits-environment relationships in the fourth corner problem. Ecology 93: 1525–1526.
Tews, J., U. Brose, V. Grimm, K. Tielborger, M. C. Wichmann, M. Schwager & F. Jeltsch, 2004. Animal species diversity driven by habitat heterogeneity/diversity: the importance of keystone structures. Journal of Biogeography 31: 79–92.
Thomaz, S. M. & E. R. da Cunha, 2010. The role of macrophytes in habitat structuring in aquatic ecosystems: methods of measurement, causes and consequences on animal assemblages’ composition and biodiversity. Acta Limnologica Brasiliensia 22: 218–236.
Villéger, S., N. W. H. Mason & D. Mouillot, 2008. New Multidimensional Functional Diversity Indices for a Multifaceted Framework in Functional Ecology. Ecology 89: 2290–2301.
Villéger, S., G. Grenouillet & S. Brosse, 2013. Decomposing functional β-diversity reveals that low functional β-diversity is driven by low functional turnover in European fish assemblages. Global Ecology and Biogeography 22: 671–681.
Waldbusser, G. G., R. L. Marinelli, R. B. Whitlatch & P. T. Visscher, 2004. The effects of infaunal biodiversity on biogeochemistry of coastal marine sediments. Limnology and Oceanography 49: 1482–1492.
Warfe, D. M. & L. A. Barmuta, 2006. Habitat structural complexity mediates food web dynamics in a freshwater macrophyte community. Oecologia 150: 141–154.
Wellborn, G. A., D. K. Skelly & E. E. Werner, 1996. Mechanisms creating community structure across a freshwater habitat gradient. Annual Review of Ecology and Systematics 27: 337–363.
Wetzel, R. G. & G. E. Likens, 1991. Limnological Analyses. Springer, Verlag.
Zeng, L., B. Liu, Z. Dai, Q. Zhou, L. Kong, Y. Zhang, F. He & Z. Wu, 2017. Analyzing the effects of four submerged macrophytes with two contrasting architectures on zooplankton: a mesocosm experiment. Journal of Limnology 76: 581–590.
Acknowledgments
The authors thank the Brazilian Council of Research (CNPq) and the Coordination for the Improvement of Higher Education Personnel (CAPES) for financial support and infrastructure for developing this study. S. Deosti, F. F. Bomfim, F. M. Lansac-Tôha, B. A. Quirino, C. C. Bonecker, and F. A. Lansac-Tôha acknowledge the Brazilian Council of Research (CNPq and CAPES) for providing grants.
Author information
Authors and Affiliations
Corresponding author
Additional information
Handling editor: Eric R. Larson
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Deosti, S., de Fátima Bomfim, F., Lansac-Tôha, F.M. et al. Zooplankton taxonomic and functional structure is determined by macrophytes and fish predation in a Neotropical river. Hydrobiologia 848, 1475–1490 (2021). https://doi.org/10.1007/s10750-021-04527-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10750-021-04527-8