Abstract
South-western Australia is isolated from other forested regions of Australia by desert and bounded on southern and western sides by the Southern and Indian Oceans, respectively, with Westralunio carteri (Iredale, 1934) as the sole endemic freshwater mussel. Its conservation status is vulnerable. This species has a history of nomenclatural change and its systematic placement and population genetic history are largely unknown. We sampled 46 individuals from 13 sites across W. carteri’s distribution and sequenced two mitochondrial genes (16S rDNA and cytochrome c oxidase subunit I) and one nuclear gene (28S rDNA). The mitochondrial haplotype networks and COI phylogenies revealed three evolutionarily significant units (ESUs): “W. carteri” I including the west coast populations, “W. carteri” II from the south and south-eastern range, and “W. carteri” III only occurring in the south-western tip of Australia. Four species delimitation methods identified two molecular operational taxonomic units supporting two distinct species (“W. carteri” I and “W. carteri” II + III). Phylogeographic patterns revealed herein confirm the historical separation of Western and Southern paleo-basins, also highlighting the isolation of the south-western extremity of the region. This underlines the need for taxonomic revision and will require a re-evaluation of W. carteri’s conservation status.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Freshwater mussels (Bivalvia: Unionida) are among the most threatened groups of animals in the world with around 45% of assessed species being threatened or near threatened according to the IUCN Red List of Threatened Species (Lopes-Lima et al., 2018). Freshwater mussels provide a wealth of ecosystem services and given their importance to freshwater ecosystems and local economies, their conservation is crucial (Vaughn, 2018).
Species delimitation is essential because government biodiversity legislation is dependent on species’ names as core conservation units (Prié et al., 2012; Ferreira-Rodriguez et al., 2019). Yet, conservation assessments of freshwater mussels are hindered by phylogenetic and taxonomic uncertainties (Walker et al., 2014a; Lopes-Lima et al., 2017; Ferreira-Rodriguez et al., 2019). Many freshwater mussel species are difficult to distinguish based on morphological characters alone (e.g. due to homoplasy, morphological convergence and lack of distinctive synapomorphies) and molecular species delimitation methods have been used with success to identify molecular operational taxonomic units (MOTUs) within cryptic species (Araujo et al., 2018; Johnson et al., 2018; Lopes-Lima et al., 2019).
As adults, freshwater mussels have a limited ability to disperse, owing to their slow movement and relatively confined habitat. However, the larval stage (glochidia in the Margaritiferidae, Unionidae and Hyriidae) is parasitic and attaches to a vertebrate host, generally a fish. Using this strategy, freshwater mussels can disperse their offspring using the host as a vector (Kat, 1984; Strayer, 2008). Not surprisingly, species’ distribution among the Unionida closely follows the phylogeographic distribution of their host fishes (Haag, 2012).
Despite their larval dispersal strategy, rates of dispersal remain low among freshwater mussels, leading to substantial genetic structuring within species (Berg et al., 2007; Geist, 2010). To effectively manage conservation of genetic diversity within a freshwater mussel species, it is important to identify evolutionarily significant units (ESUs), because these represent groups of populations with distinct evolutionary trajectories (de Queiroz 2005, 2008; Froufe et al., 2016; Lopes-Lima et al., 2016). The analysis of current biogeographic distribution and phylogenetic structure, along with knowledge of paleo basin formation, tectonic events, and other past landscape modifications can help to explain the evolutionary history of freshwater mussels and thereby contribute to the identification of ESUs (Wilson, 1995; Araujo et al., 2018).
The freshwater mussels of Australia are all members of the Hyriidae, a family with representatives in South America, Australia, New Zealand and New Guinea comprising 90–94 species from 13 to 17 genera (Graf & Cummings, 2007; Marshall et al., 2014; Pereira et al., 2014; Walker et al., 2014a; Graf et al., 2015), with evidence of ‘cryptic speciation’ having been identified for several undescribed species complexes within the last two decades (Baker et al., 2003; Sheldon 2017). Furthermore, recent molecular phylogenetic analysis has revealed inconsistencies with systematic relationships constructed using traditional morphological characters (Graf et al., 2015). Molecular data are thus necessary as a complement to shell characters, larval forms and internal anatomy to build a solid taxonomic framework for the conservation of Australian freshwater mussels (Walker et al., 2014a). Two Australian hyriids have been assessed as threatened within the last 10 years. Hyridella glenelgensis (Dennant, 1898) is listed as critically endangered under State and Commonwealth legislation and is the IUCN Red List (Playford & Walker 2008; Walker et al., 2014a, b). Similarly, Westralunio carteri (Iredale, 1934) was assessed as vulnerable by the IUCN Red List of Threatened Species (Klunzinger & Walker, 2014) and subsequently listed as threatened (vulnerable) under State and Commonwealth legislation (Klunzinger et al., 2015).
Westralunio carteri is a regional endemic, restricted to south-western Western Australia. It is listed as vulnerable due to a contraction of its former range primarily from secondary salinization of its freshwater habitats (Klunzinger et al., 2015). The genus Westralunio was described by Iredale (1934) and included Westralunio ambiguus and the subspecies Westralunio ambiguus carteri. McMichael & Hisock (1958) later consolidated these names to a single species, W. carteri. The taxonomy of the species was based almost exclusively on shell morphology. Given that shells can vary with habitat and locality (Balla & Walker, 1991; Baker et al., 2003) and the conflicting taxonomy between Iredale (1934) and McMichael & Hiscock (1958), we herein examine the patterns of genetic diversity and phylogeographic structure of W. carteri to identify ESUs for conservation and delineate MOTUs to help clarify the taxonomy.
In this study, we aim to (a) describe the genetic structure of Westralunio carteri across the endpoints of its distribution, (b) reveal phylogeographic patterns and evolutionary history, (c) evaluate hidden cryptic diversity using a combination of species delimitation methods and (d) discuss the conservation implications of these results.
Materials and methods
Tissue sampling, DNA extraction, sequencing and alignment
A total of 46 individual specimens of W. carteri were collected from 13 sites (hydrology follows AWRC, 1976) across the distribution of the species (Table 1). In the field, a small sample from the foot was collected (following Naimo et al., 1998) and placed in 99% ethanol. Genomic DNA was extracted from the tissue samples, using a standard high-salt protocol (Sambrook et al., 1989) and amplified for one nuclear and two mitochondrial markers: the F-type mtDNA cytochrome oxidase subunit 1 gene (COI; ca. 700 bp fragment), with LCO_22me and HCO_700dy primers (Walker et al., 2006, 2007); the mtDNA 16Sr DNA (16S rRNA; ca. 500 bp fragment), with 16SL and 16SH primers (Palumbi et al., 1991); and the nuclear 28SrDNA (ca. 800 bp fragment), with 28S-RD1.3f and 28S-rD4b primers (Whiting, 2002). PCR conditions followed Froufe et al. (2014), with annealing temperatures ranging from 48 °C (16S and COI) to 54 °C (28S). Sequences were obtained by Macrogen Inc., Korea. Individual gene alignments were built for each marker with ClustalW, in Bioedit 7.2.5 (Hall, 1999), including sequences from GenBank (Table 1, phylogeny specimens).
Genetic diversity of Westralunio carteri
COI and 16S individual alignments were joined in unrooted networks using TCS 1.21 (Clement et al., 2000). Uncorrected p-distances among and within haplogroups were calculated using MEGAX (Kumar et al., 2018). For COI, genetic diversity indices (i.e. haplotype and nucleotide diversity) and indices of neutrality (Tjima’s and Fu’s) were calculated using DnaSp6 v6.11 (Rozas et al., 2017) and pairwise Fst values were calculated in Arlequin 3.5.2.2 (Excoffier & Lischer, 2010).
Phylogeny and species delimitation in Westralunio carteri
A COI alignment was constructed with GUIDANCE2 (Sela et al., 2015) following Fonseca et al. (2016). It included all newly sequenced individuals in addition to outgroups. Outgroups included two other Australian Hyriidae species which Graf et al. (2015) confirmed as non-Westralunio (Velesunioninae: Velesunio ambiguus (Philippi, 1847) and Hyriinae: Hyridellini: Cucumerunio novaehollandiae (Gray, 1834)) and one representative each from each of the other two freshwater mussel families which possess glochidia: one Unionidae (Unio pictorum (Linnaeus, 1758)) and one Margaritiferidae (Margaritifera margaritifera Linnaeus, 1758) (Table 1). The COI alignment was then analysed with maximum likelihood (ML) and Bayesian Inference (BI) methods using IQ-TREE v 1.6.10 (Nguyen et al., 2015) and MrBayes 3.2.7a (Ronquist et al., 2012), respectively. For the BI analyses, the best-fit models of nucleotide substitution and partition scheme were selected using PartitionFinder 2 (Lanfear et al., 2016) under the Bayesian Information Criterion. Two independent runs of 10 × 106 generations were sampled at intervals of 1,000 generations producing a total of 10,000 trees. Burn-in was determined upon convergence of log likelihood and parameter values using Tracer 1.7.1 (Rambaut et al., 2018). For the ML analysis, the best-fit models of nucleotide substitution and partition scheme were selected using ModelFinder (Kalyaanamoorthy et al., 2017). Maximum-likelihood searches were then conducted with an initial tree search followed by 10 independent runs and 10,000 ultrafast bootstrap replicates.
Four species delimitation methods were applied to the COI dataset (excluding outgroups) to determine the number of molecular operational taxonomic units (MOTUs). Two distance-based methods were implemented; the BOLD BIN system (Ratnasingham & Hebert, 2013) and the Automatic Barcode Gap Discovery (ABGD) (Puillandre et al., 2012). For BOLD, the COI dataset was analysed with an online cluster sequences tool implemented in BOLD4 (Ratnasingham & Hebert, 2013). The ABGD method was also applied (COI and 16S) using its online version (http://wwwabi.snv.jussieu.fr/public/abgd/abgdweb.html) with default settings and the Kimura‐2‐parameter distance matrix (Puillandre et al., 2012). An additional statistical parsimony method was implemented with TCS 1.21 (Clement et al., 2000), with a 95% connection limit following Lopes-Lima et al. (2019). Finally, we applied a coalescent‐based molecular species delimitation method, using Bayesian implementation of the Poisson Tree Processes model (bPTP) (Zhang et al., 2013). A BI phylogenetic tree (data not shown) was obtained (COI Codons Models: HKY+I; F81; HKY) and used as an input tree in the bPTP Web server (available at: http://species.h-its.org/) with 1 × 106 iterations of MCMC and 20% burn‐in.
Divergence time estimates
At present, no internally calibrated molecular clock is available for the family Hyriidae (using the fossil record). Therefore, divergence times among lineages were estimated from COI sequences using BEAST2 v2.6.1 (Bouckaert et al., 2014), and the substitution rate of 0.265 ± 0.06% per million years recently estimated for Unio spp. (Froufe et al., 2016) was applied with normal distribution prior. The dataset was run under the HKY+I substitution model according to results from PartitionFinder2 v2.1.1 (Lanfear et al., 2016). An uncorrelated lognormal relaxed clock (Drummond et al., 2006) and the two-parameter birth–death model (Nee et al., 1994; Gernhard, 2008) were used. Other parameters used default settings. The random seed was 1,572,354,798,200. The analysis ran for 107 generations, sampling every 1000 generations. The quality of the runs was assessed through parameter convergence using Tracer 1.7 (Rambaut et al., 2018). The maximum credibility tree of mean heights was constructed using TreeAnnotator and discarding 2000 trees as burn-in.
Results
Dataset parameters
From the COI alignment with 559 nucleotides (nt) in length, 46 sequences, 24 haplotypes, and 47 polymorphic and 40 parsimony-informative sites were retrieved. No insertions, deletions or stop codons were observed after translating all sequences to amino acids. From the 16S rRNA alignment with 491 nt in length, 11 sequences, 6 haplotypes, and 18 polymorphic and 16 parsimony-informative sites were retrieved. A single haplotype was detected in 28S rDNA with no variation across the sampling range. Full genetic datasets were deposited in GenBank (see Table 1 for accession numbers and specimen provenance details).
Genetic diversity, phylogeny and species delimitation
A single haplotype network based on COI mtDNA was obtained and three haplogroups (red, green and blue: Fig. 1) were retrieved (corresponding to the three obtained MOTUs, see below) separated by a minimum of 12 mutations. The haplogroup “W. carteri” I (red) presents the higher number of haplotypes (12) followed by “W. carteri” II (blue) with 9 and “W. carteri” III (green) with 3 (Fig. 1). “Westralunio carteri” I is separated from “W. carteri” II by a minimum of 24 mutations and from “W. carteri” III by at least 26 mutations, with a minimum of 10 mutations separating “W. carteri” II and “W. carteri” III (Fig. 1). The haplotype network of the 16S rRNA fragment also shows the same number of haplogroups, with “W. carteri” I separated from “W. carteri” II by 13 mutations and from “W. carteri” III by 16 mutations, and two mutations separating “W. carteri” II and “W. carteri” III.
Haplotype (TCS) networks showing the relationships of “Westralunio carteri” individuals sequenced for COI and 16S (Table 1). Circle size is proportional to the observed haplotype frequencies and black points represent unobserved haplotypes and potential intermediates. Colours represent the three lineages detected in the obtained phylogeny; Westralunio carteri I (red), “Westralunio carteri” II (blue) and “Westralunio carteri” III (green)
Genetic distances among haplogroups are shown in Table 2. For the COI gene, the mean pairwise genetic distances varied from 2.5% between “W. carteri” II and “W. carteri” III, to 5.4% between “W. carteri” I and “W. carteri” III. For the 16S rRNA gene, genetic distances varied from 0.6% between “W. carteri” II and “W. carteri” III, to 3.4% between “W. carteri” I and “W. carteri” III.
Nucleotide and haplotype diversity were greatest for “W. carteri” II and least for “W. carteri” III (Table 3). Neutrality indices were negative for all haplogroups (suggesting an excess number of alleles at low frequency), although this was significant only for Fu’s F in “W. carteri” I. All pairwise Fst values among haplogroups were similar and high (Table 4).
Both phylogenetic trees showed similar topologies in the main nodes, with the BI topology shown in Fig. 2. Within the ingroup, three clades were obtained, corresponding to the same three haplogroups, i.e. “W. carteri” I, II and III, with “W. carteri” I being sister to another clade formed by “W. carteri” II + W. carteri III (Fig. 2).
Phylogenetic tree obtained by Bayesian Inference (BI) analysis of “Westralunio carteri” individuals (COI + 16S + 28S). For the major nodes support values (%) are given as Bayesian posterior probability/maximum likelihood bootstrap support. Species delimitation methods applied in this study are represented by colour-coded bars to the right of each “W. carteri” lineage: BOLD-red, ABGD-green, bPTP-blue and TCS-black. Species delimitation separation between “W. carteri” II and III was only observed by bPTP, which is represented here as a white line
Molecular species delimitation methods (BOLD, ABGD, TCS (95%) and bPTP) applied in our study recovered, by consensus, two of these lineages as molecular operational taxonomic units (MOTUs), i.e. “W. carteri” I and “W. carteri” II + “W. carteri” III. Also, bPTP recovered “W. carteri” III as an additional MOTU, separate from “W. carteri” II. (Figure 2). The geographical distributions of these three lineages are shown in Fig. 3, colour-coded by the three lineages, as follows:
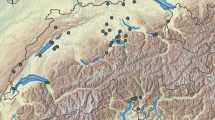
Map of freshwater mussel populations sampled for phylogenetic analysis: a red triangles “Westralunio carteri” I—(1) Gin Gin Brook, (2) Marbling Brook, (3) Lake Leschenaultia, (4) Canning River, (5) Neerigen Brook, (6) Wungong Brook, (7) Collie River, (8) Preston River; b blue diamonds “Westralunio carteri” II—(9) Blackwood River, (11) Kent River, (12) Goodga River, (13) Waychinicup River; c green circle “Westralunio carteri” III—(10) Margaret River. Refer to Table 1 for river basin and sample site details
-
(A)
“W. carteri” I found in north-westerly hydrographic basins to the north of and including the Preston River Basin, with a northern limit of Gin Gin Brook in the Moore-Hill Basin;
-
(B)
“W. carteri” II in south-westerly and southerly flowing river basins extending from the Blackwood Basin, eastward to the Albany Coast Basin, with an eastern limit of Waychinicup River;
-
(C)
“W. carteri” III from Margaret River in the Busselton Coast Basin.
Divergence time estimates
All effective sample size (ESS) values accessed in Tracer v.1.7 were above 1000. The average estimated time divergence for the crown ages for the three “W. carteri” lineages was between 0.98 and 1.96 Mya (Fig. 4). The estimated age of the most recent common ancestor (MRCA) of all three “W. carteri” lineages was around the mid-Miocene, 11.0 Mya (Fig. 4), while that of “W. carteri” lineages II and III was during the late Miocene, 4.9 Mya (Fig. 4).
Discussion
Although W. carteri has been included in a broader phylogeny of the Hyriidae (Graf et al., 2015), this is the first study to investigate the genetic diversity within this nominal taxon. The “W. carteri” phylogenies reveal two major allopatric clades: one (“W. carteri” I) in the drainages of the west coast, draining to the Indian Ocean, and the other (“W. carteri” II + “W. carteri” III) in the south coast, draining to the Southern Ocean of south-western Australia. This Southern Ocean clade is further divided into two subclades: one (“W. carteri” III) occurring in the Margaret River in the south-west and the other (“W. carteri” II) in southern drainages to the south and east of Blackwood River to Waychinicup River in the Albany Coast Basin. Moreover, these two groups are also evident in both 16S and COI networks and supported by the large pairwise FST and p-distance values. A signal of demographic expansion was only observed in “W. carteri” I as shown by the star-shaped topology of the COI network and the low, significant values of Fu’s F. No variation was detected for the nuclear marker 28S. This lack of 28S diversity has been reported for other congeneric species of freshwater mussels, due to its low substitution rate (Froufe et al., 2016; Araujo et al., 2018).
According to Bayesian analysis, “W. carteri” II + III populations diverged during the mid-late Miocene, which is consistent with divergence timing for a number of south-west Australian terrestrial taxa (Rix et al., 2014) and other freshwater taxa (Gouws et al., 2006, 2010; Unmack et al., 2011; Morgan et al., 2014). These studies suggest that vicariant events due to increased aridity periods acted as the main driving force reducing genetic connectivity and dispersion across river basins.
Furthermore, the separation of the “W. carteri” clades mirrors previous phylogenetic patterns of freshwater taxa in the region, including fishes (e.g. Unmack et al., 2011; Galeotti et al., 2015) and crayfishes (e.g. Gouws et al., 2006, 2010), and is likely a consequence of geological division of the south-west coast drainage division during the Eocene. Beard (1999) shows that geological formation appeared to create two distinct watersheds, or drainage subdivisions in south-western Australia: one on the south coast to the south of the “Jarrahwood Axis” and one on the west coast, to the west of the Yilgarn Craton and the Darling Scarp.
Our study estimates the separation of the “W. carteri” II and “W. carteri” III lineages took place between during the late Miocene/Pliocene. During these periods, Western Australian palaeodrainages suffered from cycles of intense aridification (Unmack, 2001; Hopper & Gioia, 2004) that might have confined the common ancestral populations to refuge areas promoting their separation. We speculate that increasing aridity would have decreased host fish movement among drainages, particularly given that freshwater fishes of south-western Australia do not undertake ocean migration and rely on flooding events for basin connectivity and dispersal (Morgan et al., 2014).
The south and western Australian drainages were free from considerable readjustments since the late Pliocene (Unmack, 2001; Murphy & Austin, 2004), justifying the separation of the three lineages. However, the presumed wet and humid climate over the past two million years (Hopper, 1979; Hopper & Gioia, 2004) may have provided more favourable conditions for regional inter-basin dispersal within each lineage. In fact, although unique haplotypes were found in some populations, we could not find a clear geographic structure within each lineage, indicating recent gene-flow or connectivity events. However, a higher number of individuals per population are required to confirm this apparent lack of structure.
Nevertheless, in the “W. carteri” I lineage, we identified signs of a bottleneck followed by a fast demographic expansion by the mid Pleistocene, as mirrored by the star-shaped topology of the “W. carteri” I lineage COI network, corroborated by its negative Fu’s F and Tajimas D values. Conversely, W. carteri II does not seem to show any evident demographic process.
Our results support the separation of “W. carteri” into two species: “W. carteri” I and “W. carteri” II + “W. carteri” III based on most of the employed species delimitation methods. The bPTP method further separates “W. carteri” II and “W. carteri” III, which is not surprising given that this method has been shown to overestimate the number of MOTUs (e.g. Dellicour & Flot, 2018). Future morphometric analyses combined with the present genetic results might support putative new species.
The third MOTU lineage (“W. carteri” III) revealed only by bPTP modelling suggests that a subspecies rank may be warranted, although further population sampling within the Busselton Coast Basin is required to determine the extent of this apparent MOTU. We hypothesize that populations within Busselton Coast to the north and west of Blackwood River are “W. carteri” I and that populations to the west of the Naturaliste Ridge are “W. carteri” III. Populations to the east of Blackwood River within the Donnelly, Warren and Shannon Basins, as well as other populations along the South Coast are likely “W. carteri” II.
The present results have major conservation implications for the Westralunio taxa in south-west Western Australia. The high divergence level revealed between “W. carteri” I and “W. carteri” II + “W. carteri” III supported by all species delimitation methods supports the separation into two species. Given that species are generally the taxonomic units used in conservation status assessment and legislation/policy, the taxonomic status of these two taxa needs urgent confirmation. Moreover, the three lineages “W. carteri” I, “W. carteri” II and “W. carteri” III correspond to distinct ESUs (as defined by Moritz, 1994); similarly, ESUs are recurrently observed in a number of other freshwater mussels (e.g. Froufe et al., 2016; Lopes-Lima et al., 2016; Sousa et al., 2018). These ESUs should be conserved and managed independently, given that they represent genetically unique populations and are geographically isolated.
Klunzinger et al. (2015) elucidated the conservation status of W. carteri by modelling historical and contemporary distributional records with environmental data. The authors revealed that salinity, flow permanency and total nitrogen were the variables most critical in limiting the species’ occurrence. Reduction in the species’ extent of occurrence was due primarily to secondary salinization of formerly freshwater habitats, which resulted in the species being assessed as vulnerable by the IUCN Red List (Klunzinger & Walker, 2014). Therefore, the taxonomic split, suggested here, implies that the individual conservation status of each MOTU, i.e. “W. carteri” I and “W. carteri” II + “W. carteri” III, should be re-assessed by the IUCN to inform protection management under State and Commonwealth conservation legislation.
Using distribution data from Klunzinger et al. (2015), the extent of occurrence (EOO) for “W. carteri” I is estimated to be 8814 km2, with an historic EOO of 31,559 km2, a reduction in EOO of approximately 72% which might qualify the MOTU as endangered under criterion A2c of the IUCN Red List. The EOO for “W. carteri” II is estimated to be 8660 km2 with an historic EOO of 10,070 km2, a reduction of approximately 14%, not qualifying for any threatened category under Criterion A. Due to an EOO of < 20,000 km2, “W. carteri” II is close to qualifying as vulnerable status under Criterion B. However, because there is no available evidence for severe habitat fragmentation, or number of locations ≤ 10, or extreme fluctuations, this rank cannot be attributed. Although some populations of “W. carteri” II are suggested to be increasing in size (Benson et al., 2017, 2019) and a large proportion of habitats and populations occur in national parks and specially protected areas for conservation (Klunzinger et al., 2015), it is unknown whether similar trends are true across this MOTU’s range. In the absence of these data, we suggest that “W. carteri” II + “W. carteri” III be listed as near threatened on the IUCN Red List but is likely to qualify for the vulnerable category in the near future.
Besides the two MOTUs or putative species here described, the three lineages or ESUs warrant independent conservation actions and management. For example, the brood stock for potential propagation programs on each ESU should consider the original geographic distribution. The same is true when considering eventual translocations and reintroductions. Biological traits important for conservation planning, such as the reproductive physiology and habitat requirements, as well as biotic interactions, should also be investigated independently for each ESU, especially the range of host fish use (and see Klunzinger et al., 2012), that is critical for the maintenance and viability of freshwater mussel populations.
Given the threatening processes outlined in recent publications (Klunzinger et al., 2015; Benson et al., 2017, 2019; Ferreira-Rodriguez et al., 2019), maintaining riparian vegetation and habitats for host fishes, environmental flows in regulated rivers, the flow of freshwater (and mitigation of salinity) and reductions in nutrient pollution should be conservation priorities for both species/MOTUs revealed in this study.
References
Araujo, R. D., K.-O. Buckley, R. García-Jiménez Nagel & A. Machordom, 2018. Species boundaries, geographic distribution and evolutionary history of the Western Palaearctic freshwater mussels Unio (Bivalvia: Unionidae). Zoological Journal of the Linnean Society 182: 275–299.
AWRC, 1976. Review of Australia’s Water Resources 1975. Canberra, Australian Water Resources Council (AWRC), Department of Natural Resources.
Baker, A. M., C. Bartlett, S. E. Bunn, K. Goudkamp, F. Sheldon & J. M. Hughes, 2003. Cryptic species and morphological plasticity in long-lived bivalves (Unionoida: Hyriidae) from inland Australia. Molecular Ecology 12: 2707–2717.
Balla, S. A. & K. F. Walker, 1991. Shape variation in the Australian freshwater mussel Alathyria jacksoni Iredale (Bivalvia, Hyriidae). Hydrobiologia 220: 89–98.
Beard, J. S., 1999. Evolution of the river systems of the south-west drainage division, Western Australia. Journal of the Royal Society of Western Australia 82: 147–164.
Benson, J. A., P. G. Close, B. A. Stewart & A. J. Lymbery, 2017. Upstream recolonization by freshwater mussels (Unionoida: Hyriidae) following installation of a fishway. Aquatic Conservation: Marine and Freshwater Ecosystems 28: 512–517.
Benson, J. A., P. G. Close, B. A. Stewart & A. J. Lymbery, 2019. Freshwater tributaries provide refuge and recolonization opportunities for mussels following salinity reversal. Science of the Total Environment 683: 231–239.
Berg, D. J., A. D. Christian & S. I. Guttman, 2007. Population genetic structure of three freshwater mussel (Unionidae) species within a small stream system: significant variation at local spatial scales. Freshwater Biology 52: 1427–1439.
Bouckaert, R., J. Heled, D. Kühnert, T. Vaughan, C.-H. Wu, et al., 2014. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Computational Biology 10: e1003537.
Clement, M., D. Posada & K. A. Crandall, 2000. TCS: a computer program to estimate gene genealogies. Molecular Ecology 9: 1657–1659.
de Queiroz, K., 2005. A unified concept of species and its consequences for the future of taxonomy. Proceedings of the California Academy of Sciences 56(Suppl. 1): 196–215.
de Queiroz, K., 2008. Species concepts and species delimitation. Systematic Biology 56: 879–886.
Dellicour, S. & J. Flot, 2018. The hitchhiker’s guide to single-locus species delimitation. Molecular Ecology Resources 2018: 1–13.
Drummond, A. J., S. Y. Ho, M. J. Phillips & A. Rambaut, 2006. Relaxed phylogenetics and dating with confidence. PLoS Biology 4: 699.
Excoffier, L. & H. E. L. Lischer, 2010. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources 10: 564–567.
Ferreira-Rodriguez, N., Y. B. Akiyama, O. V. Aksenova, R. Araujo, C. M. Barnhart, Y. V. Bespalaya, A. E. Bogan, I. N. Bolotov, P. B. Budha, C. Clavijo, S. J. Clearwater, G. Darrifran, V. T. Do, K. Douda, E. Froufe, C. Gumpinger, L. Henrikson, C. L. Humphrey, N. A. Johnson, O. Klishko, M. W. Klunzinger, S. Kovitvadhi, U. Kovitvadhi, J. Lajtner, M. Lopes-Lima, E. A. Moorkens, S. Nagayama, K. Nagel, M. Nakano, J. N. Negishi, P. Ondina, P. Oulasvirta, V. Prié, N. Riccardi, M. Rudzīte, F. Sheldon, R. Sousa, D. L. Strayer, M. Takeuchi, J. Taskinen, A. Teixeira, J. S. Tiemann, M. Urbańska, S. Varandas, M. V. Vinarski, B. J. Wicklow, T. Zając & C. C. Vaughn, 2019. Research priorities for freshwater mussel conservation assessment. Biological Conservation 231: 77–87.
Fonseca, M. M., M. Lopes-Lima, M. S. Eackles, T. L. King & E. Froufe, 2016. The female and male mitochondrial genomes of Unio delphinus and the phylogeny of freshwater mussels (Bivalvia: Unionida). Mitochondrial DNA Part B Resources 1: 954–957.
Froufe, E., C. Sobral, A. Teiseira, R. Sousa, S. Varandas, D. C. Aldridge & M. Lopes-Lima, 2014. Genetic diversity of the pan-European freshwater mussel Anodonta anatina (Bivalvia: Unionoida) based on CO1: new phylogenetic insights and implications for conservation. Aquatic Conservation Marine and Freshwater Ecosystems 24: 561–574.
Froufe, E., V. Prié, J. Faria, M. Ghamizi, D. V. Gonçalves, M. E. Gürlek, I. Karaouzas, Ü. Kebapçi, H. Şereflişan, C. Sobral, R. Sousa, A. Teixeira, S. Varandas, S. Zogaris & M. Lopes-Lima, 2016. Phylogeny, phylogeography, and evolution in the Mediterranean region: news from a freshwater mussel (Potomida, Unionida). Molecular Phylogenetics and Evolution 100: 322–332.
Galeotti, D. M., M. A. Castalanelli, D. M. Groth, C. McCullough & M. Lund, 2015. Genotypic and morphological variation between Galaxiella nigrostriata (Galaxiidae) populations: implications for conservation. Marine and Freshwater Research 66: 187–194.
Geist, J., 2010. Strategies for the conservation of endangered freshwater pearl mussels (Margaritifera margaritifera L.): a synthesis of conservation genetics and ecology. Hydrobiologia 644: 69–88.
Gernhard, T., 2008. The conditioned reconstructed process. Journal of Theoretical Biology 253: 769–778.
Gouws, G., B. A. Stewart & S. R. Daniels, 2006. Phylogeographic structure of a freshwater crayfish (Decapoda: Parastacidae: Cherax preissii) in south-western Australia. Marine and Freshwater Research 57: 837–848.
Gouws, G., B. A. Stewart & S. R. Daniels, 2010. Phylogeographic structure in the gilgie (Decapoda: Parastacidae: Cherax quinquecarinatus): a south-western Australian freshwater crayfish. Biological Journal of the Linnean Society 101: 385–402.
Graf, D. L. & D. Ó. Foighil, 2000a. The evolution of brooding characters among the freshwater pearly mussels (Bivalvia: Unionoidea) of North America. Journal of Molluscan Studies 66: 157–170.
Graf, D. L. & D. Ó. Foighil, 2000b. Molecular phylogenetic analysis of 28S rDNA supports a Gondwanan origin for Australasian Hyriidae (Mollusca: Bivalvia: Unionoida). Vie et Milieu 50: 245–254.
Graf, D. L. & K. S. Cummings, 2007. Review of the systematics and global diversity of freshwater mussel species (Bivalvia: Unionoida). Journal of Molluscan Studies 73: 291–314.
Graf, D. L., H. Jones, A. J. Geneva, J. M. Pfeiffer III & M. W. Klunzinger, 2015. Molecular phylogenetic analysis supports a Gondwanan origin of the Hyriidae (Mollusca: Bivalvia: Unionida) and the paraphyly of Australasian taxa. Molecular Phylogenetics and Evolution 85: 1–9.
Hall, T. A., 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Window 95/98/NT. Nucleic Acids Symposium Series 41: 95–98.
Haag, W. R., 2012. North American Freshwater Mussels: Natural History, Ecology and Conservation. Cambridge University Press, Cambridge.
Hopper, S. D., 1979. Biogeographical aspects of speciation in the southwest Australian flora. Annual Review of Ecology and Systematics 10: 399–422.
Hopper, S. D. & P. Gioia, 2004. The southwest floristic region: evolution and conservation of a global hot spot of biodiversity. Annual Review of Ecology Evolution and Systematics 35: 623–650.
Iredale, T., 1934. The freshwater mussels of Australia. Australian Zoologist 8: 57–78.
Johnson, N. A., C. H. Smith, J. M. Pfeiffer, C. R. Randklev, J. D. Williams & J. D. Austin, 2018. Integrative taxonomy resolves taxonomic uncertainty for freshwater mussels being considered for protection under the US Endangered Species Act. Scientific Reports 8: 15892.
Källersjö, M., T. von Proschwitz, S. Lundberg, P. Eldenas & C. Erseus, 2005. Evaluation of ITS rDNA as a complement to mitochondrial gene sequences for phylogenetic studies in freshwater mussels: an example using Unionidae from north-western Europe. Zoolica Scripta 34: 415–424.
Kalyaanamoorthy, S., B. Q. Minh, T. K. F. Wong, A. von Haeseler & L. J. Jermiin, 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nature Methods 14: 587–589.
Kat, P. W., 1984. Parasitism and the Unionacea (Bivalvia). Biological Reviews 59: 189–207.
Klunzinger, M. & K. F. Walker, 2014. Westralunio carteri. The IUCN Red List of Threatened Species 2014: e.T23073A58526341.
Klunzinger, M. W., S. J. Beatty, D. L. Morgan, G. J. Thomson & A. J. Lymbery, 2012. Glochidia ecology in wild fish populations and laboratory determination of competent host fishes for an endemic freshwater mussel of south-western Australia. Australian Journal of Zoology 60: 26–36.
Klunzinger, M. W., S. J. Beatty, D. L. Morgan, A. M. Pinder & A. J. Lymbery, 2015. Range decline and conservation status of Westralunio carteri Iredale, 1934 (Bivalvia: Hyriidae) from south-western Australia. Australian Journal of Zoology 63: 127–135.
Kumar, S., G. Stecher, M. Li, C. Knyaz & K. Tamura, 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution 35: 1547–1549.
Lanfear, R., P. B. Frandsen, A. M. Wright, T. Senfeld & B. Walcott, 2016. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Molecular Biology and Evolution 34: 772–773.
Lopes-Lima, M., R. Sousa, A. Teixeira, S. Varandas, N. Riccardi, D. C. Aldridge & E. Froufe, 2016. Newly developed microsatellite markers for the pan-European duck mussel, Anodonta anatina: revisiting the main mitochondrial lineages. Aquatic Conservation Marine and Freshwater Ecosystems 26: 307–318.
Lopes-Lima, M., E. Froufe, V. T. Do, M. Ghamizi, K. E. Mock, Ü. Kebapçı, O. Klishko, S. Kovitvadhi, U. Kovitvadhi, O. S. Paulo, J. M. Pfeiffer III, M. Raley, N. Riccardi, H. Şereflişan, R. Sousa, A. Teixeira, S. Varandas, X. Wu, D. T. Zanatta, A. Zieritz & A. E. Bogan, 2017. Phylogeny of the most species-rich freshwater bivalve family (Bivalvia: Unionida: Unionidae): defining modern subfamilies and tribes. Molecular Phylogenetics and Evolution 106: 174–191.
Lopes-Lima, M., L. E. Bulakova, A. Y. Kuratayev, K. Mehler, M. Seddon & R. Sousa, 2018. Conservation of freshwater bivalves at the global scale: diversity, threats and research needs. Hydrobiologia 810: 1–14.
Lopes-Lima, M., L. E. Bulakova, A. Y. Kuratayev, A. Gomes-dos-Santos, A. Zieritz, E. Froufe & A. E. Bogan, 2019. Revisiting the North American freshwater mussel genus Quadrula sensu lato (Bivalvia Unionidae): phylogeny, taxonomy and species delineation. Zoologica Scripta 48: 1–24.
Machordom, A., R. Araujo, D. Erpenbeck & M. A. Ramos, 2003. Phylogeography and conservation genetics of endangered European Margaritiferidae (Bivalvia: Unionoidea). Biological Journal of the Linnean Society London 78: 235–252.
Marshall, B. A., M. C. Fenwick & P. A. Ritchie, 2014. New Zealand recent Hyriidae (Mollusca: Bivalvia: Unionida). Molluscan Research 34: 181–200.
McMichael, D. F. & I. D. Hiscock, 1958. A monograph of the freshwater mussels (Mollusca: Pelecypoda) of the Australian region. Australian Journal of Marine and Freshwater Research 9: 372–508.
Morgan, D. L., P. J. Unmack, S. J. Beatty, B. C. Ebner, M. G. Allen, J. J. Keleher, J. A. Donaldson & J. Murphy, 2014. An overview of the ‘freshwater fishes’ of Western Australia. Journal of the Royal Society of Western Australia 97: 263–278.
Moritz, C., 1994. Defining ‘evolutionarily significant units’ for conservation. Trends in Ecology and Evolution 9: 373–375.
Murphy, N. P. & C. M. Austin, 2004. Phylogeography of the widespread Australian freshwater prawn, Macrobrachium australiense (Decapoda, Palaemonidae). Journal of Biogeography 31: 1065–1072.
Naimo, T. J., E. D. Damschen, R. G. Rada & E. M. Monroe, 1998. Nonlethal evaluation of the physiological health of unionid mussels: methods for biopsy and glycogen analysis. Journal of the North American Benthological Society 17: 121–128.
Nee, S., E. C. Holmes, R. M. May & P. H. Harvey, 1994. Extinction rates can be estimated from molecular phylogenies. Philosophical Transactions of the Royal Society of London B Biological Science. 344: 77–82.
Nguyen, L. T., H. A. Schmidt, A. von Haeseler & B. Q. Minh, 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molcular Biology and Evolution 32: 268–274.
Palumbi, S. R., A. P. Martin, S. Romano, W. O. McMillan, L. Stice & G. Grabowski, 1991. The Simple Fool’s Guide to PCR, Version 2.0. Published and distributed by the authors: Honolulu.
Pereira, D., M. C. D. Mansur, L. D. S. Duarte, A. S. de Oliveira, D. M. Pimpão, C. T. Callil, C. Ituarte, C. Parada, S. Peredo, G. Darrigran, F. Scarabino, C. Clavijo, G. Lara, I. C. Miyahira, M. T. R. Rodriguez & C. Lasso, 2014. Bivalve distribution in hydrographic regions in South America: historical overview and conservation. Hydrobiologia 735: 15–44.
Playford, T. & K. F. Walker, 2008. Status of the endangered Glenelg River Mussel Hyridella glenelgensis (Unionoida: Hyriidae) in Australia. Aquatic Conservation: Marine and Freshwater Ecosystems 18: 679–691.
Prié, V., N. Puillandre & P. Bouchet, 2012. Bad taxonomy can kill: molecular reevaluation of Unio mancus Lamarck, 1819 (Bivalvia: Unionidae) and its accepted subspecies. Knowledge and Management of Aquatic Ecosystems 8: 1–18.
Puillandre, N., M. V. Modica, Y. Zhang, L. Sirovich, M.-C. Boisselier, C. Cruaud, M. Holford & S. Samadi, 2012. Large-scale species delimitation method for hyperdiverse groups. Molecular Ecology 21: 2671–2691.
Rix, M. G., D. L. Edwards, M. Byrne, M. S. Harvey, L. Joseph & J. D. Roberts, 2014. Biogeogrpahy and speciation of terrestrial fauna in the south-western Australian biodiversity hotspot. Biological Reviews of the Cambridge Philosophical Society 90: 762–793.
Ronquist, F., M. Teslenko, P. van der Mark, D. L. Ayres, A. Darling, S. Höhna, B. Larget, L. Liu, M. A. Suchard & J. P. Huelsenbeck, 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542.
Rambaut, A., A. J. Drummond, D. Xie, G. Baele & M. A. Suchard, 2018. Posterior summarization in Bayesian phylogenetics using tracer 1.7. Systematic Biology 67: 901–904.
Ratnasingham, S. & P. D. N. Hebert, 2013. A DNA-based registry for all animal species: the barcode index number (BIN) system. PLoS ONE 8: e66213.
Rozas, J., A. Ferrer-Mata, J. C. Sánchez-Del Barrio, S. Guirao-Rico, P. Librado, S. E. Ramos-Onsins & A. Sánchez-Gracia, 2017. DnaSP 6: DNA sequence polymorphism analysis of large datasets. Molecular Biology and Evolution 34: 3299–3302.
Sambrook, J., E. F. Fritsch & T. Maniatis, 1989. Molecular Cloning: A Laboratory Manual. Cold Spring Harbour Press, New York.
Sela, I., H. Ashkenazy, K. Katoh & T. Pupko, 2015. GUIDANCE2: accurate detection of unreliable alignment regions accounting for the uncertainty of multiple parameters. Nucleic Acids Research 43: W7–W14.
Sheldon, F., 2017. Variable plasticity in shell morphology of some Australian freshwater mussels (Unionoida, Hyriidae). Transactions of the Royal Society of South Australia 141: 193–208.
Sousa, R., A. Teixeira, A. Santos, H. Benaissa, S. Varandas, M. Ghamizi, V. Prié, E. Froufe & M. Lopes-Lima, 2018. Oued Bouhlou: a new hope for the Moroccan pearl mussel. Aquatic Conservation Marine and Freshwater Ecosystems 28: 247–251.
Strayer, D. L., 2008. Freshwater Mussel Ecology: A Multifactor Approach to Distribution and Abundance. University of California Press, Berkley.
Unmack, P. J., 2001. Biogeography of Australian freshwater fishes. Journal of Biogeography 28: 1053–1089.
Unmack, P. J., M. P. Hammer, M. Adams & T. E. Dowling, 2011. A phylogenetic analysis of pygmy perches (Teleostei: Percichthyidae) with an assessment of the major historical influences on aquatic biogeography in southern Australia. Systematic Biology 60: 797–812.
Vaughn, C. C., 2018. Ecosystem services provided by freshwater mussels. Hydrobiologia 810: 15–27.
Walker, J. M., J. P. Curole, D. E. Wade, E. G. Chapman, A. E. Bogan, G. T. Watters & W. R. Hoeh, 2006. Taxonomic distribution and phylogenetic utility of gender-associated mitochondrial genomes in the Unionoida (Bivalvia). Malacologia 48: 265–282.
Walker, J. M., A. E. Bogan, E. A. Bonfiglio, D. C. Campbell, A. D. Christian, J. P. Curole, J. L. Harris, R. J. Wojtecki & W. R. Hoeh, 2007. Primers for amplifying the hypervariable, male-transmitted COII-COI junction region in amblemine freshwater mussels (Bivalvia: Unionoidea: Ambleminae). Molecular Ecology Notes 7: 489–491.
Walker, K. F., H. A. Jones & M. W. Klunzinger, 2014a. Bivalves in a bottleneck: taxonomy, phylogeography and conservation of freshwater mussels (Bivalvia: Unionoida) in Australasia. Hydrobiologia 735: 61–79.
Walker, K. F., H. A. Jones & M. Klunzinger, 2014b. Hyridella glenelgensis. The IUCN Red List of Threatened Species 2014: e.T58609631A58628791.
Whelan, N. V., A. J. Geneva & D. L. Graf, 2011. Molecular phylogenetic analysis of tropical freshwater mussels (Mollusca: Bivalvia: Unionoida) resolves the position of Coelatura and supports a monophyletic Unionidae. Molecular Phylogenetics and Evolution 61: 504–514.
Whiting, M. F., 2002. Mecoptera is paraphyletic: multiple genes and phylogeny of Mecoptera and Siphonaptera. Zoologica Scripta 31: 93–104.
Wilson, B. E., 1995. A (not-so) radical solution to the species problem. Biology and Philosophy 10: 339–356.
Zhang, J., P. Kapli, P. Pavlidis & A. Stamatakis, 2013. A general species delimitation method with applications to phylogenetic placements. Bioinformatics 29: 2869–2876.
Acknowledgements
Genetic analyses were funded by FCT – Portuguese Foundation for Science and Technology COMPETE 2020, Portugal 2020 and the European Union through the ERDF under Project ConBiomics: the missing approach for the Conservation of freshwater Bivalves No NORTE-01-0145-FEDER-030286 and, in part, through a grant from the Holsworth Wildlife Research Endowment. AGS and MLL were also funded by FCT under grants SFRH/BD/137935/2018 and SFRH/BD/115728/2016, respectively. This research was supported by national funds through FCT - Foundation for Science and Technology within the scope of UIDB/04423/2020 and UIDP/04423/2020. Specimen field collection was funded by a Murdoch University PhD Scholarship and a grant from the Holsworth Wildlife Research Endowment. Wildlife collection permits were obtained from the Western Australian Department of Environment and Conservation (SF007049) and the Western Australian Department of Fisheries (1724-2010-06). We thank James Keleher for his assistance during field collection and Corey Whisson and the WA Museum for facilitating specimen collection management. We especially thank the editor and anonymous reviewers for their comments which significantly improved upon earlier versions of this paper.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Guest editors: Manuel P. M. Lopes-Lima, Nicoletta Riccardi, Maria Urbanska & Ronaldo G. Sousa / Biology and Conservation of Freshwater Molluscs
Rights and permissions
About this article
Cite this article
Klunzinger, M.W., Lopes-Lima, M., Gomes-dos-Santos, A. et al. Phylogeographic study of the West Australian freshwater mussel, Westralunio carteri, uncovers evolutionarily significant units that raise new conservation concerns. Hydrobiologia 848, 2951–2964 (2021). https://doi.org/10.1007/s10750-020-04200-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10750-020-04200-6