Abstract
Recent studies have found that the relative importance of predictors of metacommunity structure is dependent on different factors. Low explanatory power of multivariate models is a frequent result. To increase this power, ecologists have suggested different strategies, including the use of functional approaches. Using a phytoplankton dataset from 17 reservoirs in Southern Brazil, sampled seasonally over eight years, we tested the hypothesis that the explanatory power of multivariate models would be higher when the analyses were based on functional groups than when based on a taxonomic approach. We also modeled the temporal variation in the strength of species sorting (as given by the adjusted coefficient of determination derived from environmental variables). We found high temporal variability in the strength of species sorting, indicating that results from snapshot surveys should be interpreted cautiously. When compared to the taxonomic approach, we did not find an increase in the explanatory power of multivariate models when the analyses were based on a functional approach. The main correlates of the temporal variation in the strength of species sorting were insolation, water temperature, and environmental heterogeneity, suggesting that conditions related to productivity and heterogeneity are important in determining the role of species sorting in phytoplankton communities.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The metacommunity concept proposes a framework to understand the roles of niche and dispersal-based processes in generating different species compositions. Originally, four paradigms were proposed—neutral, patch dynamics, species sorting, and mass effects—to emphasize different sets of metacommunity dynamics and provided a useful strategy for discussing metacommunity scenarios (Leibold et al., 2004). Patterns should, however, fit within the broader framework of metacommunity theory as a continuous and multidimensional space defined according to species equivalence, dispersal abilities, and environmental heterogeneity (Logue et al., 2011).
Any study seeking the causes of community variation may consider that the structure of local communities varies through time (Bengtsson et al., 1997), as well as the predictors of community variation (Bellier et al., 2014). Nonetheless, very few local communities are sampled over long time periods and are available to test temporal variation in the predictors of community variation. For instance, one can expect that correlation with environmental variables may be higher or lower depending on temporal fluctuations of key environmental factors. Thus, a comprehensive understanding of how metacommunities are organized should consider the temporal variation in correlates of community structure. In addition, one can also evaluate the role of earlier metacommunity data to explain current metacommunity structure (Castillo-Escrivà et al., 2017). This represents the potential effect of earlier occupation on current community composition of the set of local communities that composes the metacommunity.
Predictability of community structure may also vary depending on species traits. Small-sized aquatic organisms can show efficient dispersal and they can thus reach most sites (Finlay & Fenchel, 2004). In this perspective, differences between local microalgal communities, for example, may be explained by species responses to local environmental features (Martiny et al., 2006; see also Leibold et al., 2004 for the definition of species sorting mechanisms). Even so, previous studies have reported spatial structure in microalgal metacommunities; thus, in addition to the role of local environmental factors, spatial variables have also been found to be influential, suggesting some level of dispersal limitation (Soininen et al., 2007; Vyverman et al., 2007; Vanormelingen et al., 2008; Heino et al., 2010).
Differences in the responses of different groups of microalgae to environmental conditions suggest that taxonomic classification of species may not be sufficient to disentangle community assembly processes. For instance, planktonic algae responses may be different from those of attached algae because they are probably more susceptible to hydrological variations, resulting in high (passive) dispersal rates (Wetzel et al., 2012) and possibly lower effects of earlier occupation. In this sense, analysis considering traits should also be used as a powerful and complementary approach to understand community assembly (McGill et al., 2006; Litchman et al., 2010). Several classifications of phytoplankton into functional groups have been proposed (e.g., Reynolds et al., 2002; Salmaso & Padisák, 2007; Padisák et al., 2009; Kruk et al., 2010), and such groupings can be used to understand community assembly processes. In short, functional groups are based on between-species similarity of relevant traits that respond similarly to community assembly.
Phytoplankton is a group that is particularly appropriate for investigating determinants of metacommunity organization because it consists of a diverse group of species that typically respond in a predictable way to the environment (Reynolds, 1984; 1989). Reservoirs also provide good model systems to study spatial patterns due to their relative isolation in the landscape. We explored if such variation has a seasonal pattern using data sampled over 32 time periods (i.e., spring, summer, autumn, and winter seasons from 2005 to 2013). In this study, we used phytoplankton and environmental data from a set of 17 reservoirs in Southern Brazil, sampled seasonally over eight years. For each period, we quantified the relative contributions of local environmental factors, spatial variables, and earlier community data to the structure of phytoplankton metacommunities and functional groups. Mechanisms related to both environmental and spatial predictors are now accepted as simultaneously important for community assembly (Soininen et al., 2007; Hájek et al., 2011; Chust et al., 2013; Gallego et al., 2014), as well as the earlier community structure (Castillo-Escrivà et al., 2017). We expected a high temporal variation in the relative contribution of these predictors (i.e., environmental, spatial, and earlier community) for both species composition and functional groups.
We also tested for correlates of the strength of species sorting (SSS hereafter), estimated by the variation in community structure that is (uniquely) explained by environmental variables. We hypothesized that SSS would be higher in trait-based analyses than in analyses considering species composition. We anticipated that the SSS would be relatively stronger in periods with high environmental heterogeneity given that a wide environmental gradient may promote species sorting (Leibold et al., 2004; McCreadie & Bedwell, 2013). Also, we expected that SSS may be lower in periods with a high dominance of Cyanobacteria. In such periods, dominance of Cyanobacteria may decrease the community turnover that can be explained, as a consequence of the well-known negative effects of Cyanobacteria on other algae (Suikkanen et al., 2005). SSS may also be low during periods with high total species richness given the high amount of information in biological matrices to be explained (see also Low-Décarie et al., 2014). Bini et al. (2014) found a negative correlation between SSS and nutrient concentrations. Also, Chase et al. (2010) showed that beta diversity was positively correlated with productivity due to a strong role of stochastic assembly processes. Based on these findings, we predicted a negative relationship between SSS and productivity.
Materials and methods
Study area and survey program
We used a dataset obtained by a water quality monitoring program run by “COPEL” (The Energy Company of the State of Paraná, Brazil), consisting of phytoplankton densities and environmental variables sampled in 17 reservoirs (Fig. 1) during four austral seasons (spring, summer, fall, and winter), from 2005 to 2013 (see also Wojciechowski et al., 2017). All the reservoirs are in the State of Paraná, Southern Brazil. Paraná State covers an area of 199,727 km2 corresponding to 2.34% of the Brazilian territory. Although reservoirs vary in level of degradation and uses (including drinking water supply, recreation, and hydroelectricity generation), Cyanobacterial blooms are frequent in most of them (Fernandes et al., 2005; 2014b; IAP, 2009).
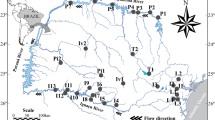
State of Paraná with the geographic location of the reservoirs. Samples were taken seasonally in each reservoir from 2005 to 2013. Main rivers of each drainage basin are also shown. AP Apucaraninha (n = 32 samplings), CP Capivari (n = 32), CV Cavernoso (n = 33), CXI Caxias I (n = 34), CXII Caxias II (n = 34), CH Chopim (n = 32), GU Guaricana (n = 34), JO Jordão (n = 33), ME Melissa (n = 31), MO Mourão (n = 33), PI Pitangui (n = 33), RP Rio dos Patos (n = 33), SV Salto do Vaú (n = 33), SJ São Jorge (n = 33), SEI Segredo I (n = 33), SEII Segredo II (n = 33), VO Vossoroca (n = 34)
Subsurface samples (at ca. 15 cm water depth) for both phytoplankton density (cell ml−1) and environmental variables were taken simultaneously from the lacustrine zone of each reservoir. Summer, fall, winter, and spring samples were taken in January, April, July, and October, respectively, totalizing 32 sampling periods (=8 years × 4 seasons). Phytoplankton samples were fixed with Lugol’s solution and stored in amber flasks. Phytoplankton taxa were identified to the species level, whenever possible. The estimation of the phytoplankton density was the same during the period of study. Cellular density (cells.ml−1) was estimated according to the Utermöhl (1958) sedimentation technique and by counting random fields under inverted microscope Olympus IX70 (600x). All the species present in at least 20 fields or 100 cells of the most abundant species were counted (Lund et al., 1958). The following environmental variables were obtained according to APHA (2005): water temperature (°C), dissolved oxygen (DO, mg l−1), pH, and conductivity (μS cm−1) were measured in situ; water samples were taken to the laboratory for analysis of total phosphorus (TP, mg l−1), total nitrogen (TN, mg l−1), ammonium (NH4 +, mg l−1), nitrate (NO3 −, mg l−1), and nitrite (NO2 −, mg l−1) concentration. Water transparency (m) was obtained using a Secchi disk at the same sampling sites. These environmental variables varied widely over time (Table S2; see also Wojciechowski et al., 2017).
Biological data
Details of phytoplankton community in the reservoirs of Paraná have already been described in Wojciechowski et al. (2017). The dataset included 606 phytoplankton taxa in all periods and reservoirs studied (see also Table S1 in Online Resource for the most common species recorded). Chlorophyceae was the predominant class in terms of number of taxa (273 taxa), followed by Bacillariophyceae (104 taxa), Cyanophyceae (82 taxa), Zygnemaphyceae (56), and Chrysophyceae (52). Other phytoplankton classes were represented by a lower number of taxa, ranging from 36 to 11 taxa. The most abundant algae were mainly Cyanobacteria, e.g., Cylindrospermopsis raciborskii, some species of the genera Aphanizomenon, Dolichospermum, and Microcystis. Most of the Cyanobacteria (67% of the species) were considered among the most abundant in at least one sampling period. Only 51 species were considered common (i.e., appearing in at least 10% of samples; Table S1 in Online Resource).
The phytoplankton density (cell.ml−1) data were classified taxonomically (based on species identities) and functionally (using functional group classifications). For functional groups, we used a classification based exclusively on morphology, the “Morphologically Based Functional Groups” (MBFG) proposed by Kruk et al. (2010). This decision was made based on the simplicity of the classification that divides phytoplankton species into seven groups. This classification is based on the following traits: volume (µm3), surface area (µm2), surface/volume ratio (µm−1), maximum linear dimension (µm), and presence/absence of mucilage, flagella, gas vesicles, heterocysts, and siliceous structures (Kruk et al., 2010). The complete list of species composing each functional group is available in Table S1 (Online Resource 1). The seven groups are the following: (I) Small organisms (<2 µm) with high surface/volume ratio, without siliceous structures (e.g., small species of Chlorococcales); (II) Flagellated organisms of small size with siliceous exoskeletons, such as species of the genus Chromulina; (III) Large filaments containing aerotopes, mainly represented by species of Nostocales in our study; (IV) Individuals of medium size lacking mucilage, flagella, or exoskeletons (e.g., species of the genera Monoraphidium and Scenedesmus); (V) Unicellular individuals of medium to large size (>2 µm), with flagella, such as species of Cryptophyceae and Dinophyceae; (VI) Non-flagellated organisms with siliceous exoskeletons (Bacillariophyceae); (VII) Large mucilaginous colonies, such as species of the genera Eutetramorus and Microcystis (Kruk et al., 2010).
According to our goals, we generated the following response matrices: reservoirs (rows) × density of species (columns) for each period; reservoirs × density of the seven functional groups for each period; and reservoirs × density of species for each functional group for each period. Given that functional groups have different biological traits, we anticipated that correlates of SSS may differ among them, and some predictions can be specified. Similar to what is expected considering the entire community, SSS of groups with more species may be less predicted than the SSS of groups with few species. Groups I and III were those with the lowest number of species, and groups IV, V, and VI were those with the highest number of species. The groups I and II, composed of small algae, may show high dispersal rates and therefore may be more explained by SSS than groups III–VII, composed of medium to large organisms. The SSS for group III, composed of Cyanobacteria, may be mainly correlated to productivity, given that species of this group respond quickly to an increase in productivity, usually resulting in summer blooms over multiple reservoirs (Paerl, 2014).
Data analysis
Our main goal was to explain temporal variation in SSS, differently from Wojciechowski et al. (2017) which aimed to explain temporal variation in beta diversity. Two main steps were undertaken to accomplish this task. First, SSS (our response variable) was estimated, for each sampling period, using a Partial Redundancy Analysis (pRDA; Borcard et al., 1992; Legendre & Legendre, 2012) and variation partitioning (Peres-Neto et al., 2006). Community structure may be influenced by local conditions (e.g., abiotic variables), spatial variables (used as proxy of dispersal), and earlier community structure. Therefore, we partitioned the variation in phytoplankton community structure in each sampling period in three pure fractions: (a) proportion of variance explained exclusively by the environmental variables; (b) proportion of variance explained by the spatial variables; and (c) proportion of variance explained by earlier community structure. Fiver other fractions were also estimated: (d) the variation in biological data explained by spatially structured environmental variables; (e) the variation in biological data explained by the common structure of environmental variables and earlier community structure; (f) the variation explained by the earlier community structure that was spatially structured; (g) the variation in biological data explained by the common structure of environmental variables, spatial variables, and earlier community structure; (h) unexplained variation in biological data.
To increase comparability between sampling periods, we decided to use the first four PCA axes to summarize the environmental variables, the first four Moran’s Eigenvector Maps to represent spatial variables (Dray et al., 2006), and the first four PCoA vectors to represent the earlier community structure in pRDA and variation partitioning, as explained subsequently. The first four axes of PCA and PCoA usually explained a high proportion of the variance in the original data (from 89.9 to 96.9% for PCA, and from 50.7 to 66.5% for PCoA). Also, the first four Moran’s Eigenvector Maps (MEMs) were associated with the highest (and always positive) eigenvalues, describing positive autocorrelation at global scales (Dray et al., 2006). PCA axes were calculated using all the environmental variables described above. Environmental variables were log + 1 transformed, except pH, before analysis. The PCA axes were estimated using the function “princomp” in the package vegan (Oksanen et al., 2015) for the software R 3.1.3 (R Core Team, 2015). Moran’s Eigenvector Maps (MEMs; Dray et al., 2006) were created using the geographic coordinates of the 17 reservoirs and the function “pcnm” in the R package PCNM (Legendre et al., 2013). PCoA scores (using Bray–Curtis dissimilarity) were calculated using all the species present in the previous sampling period using the function “pcoa” in the R package ape (Paradis et al., 2004). The sum of all fractions [a + b + c + d + e + f + g] corresponds to the total variance explained by environmental, spatial, and earlier community occupation. Fractions [a], [b], and [c] were tested for statistical significance using randomization tests with 999 runs. As the earlier community structure was measured using the community data from the previous period, we showed fractions estimated for 31 periods. Thus, there are no results for the first sampling period (fall 2005). For each response matrix (species, functional, and species separated into different functional groups), we estimated the fractions [a], [b], and [c] for each of the 31 periods separately. Fraction [a] was our measure of SSS. Phytoplankton density data and environmental variables were, respectively, Hellinger-transformed (Legendre & Gallagher, 2001) and standardized before the analyses. Our pRDA models were estimated using the function “varpart” in the R vegan package (Oksanen et al., 2015). We performed a paired Wilcoxon test to compare the adjusted coefficients of determination (adjusted R 2), estimated by RDA models, for functional groups matrices and phytoplankton species matrix, using the function “wilcox.test” in the stats package. We compared the global adjusted R 2 and the adjusted R 2 considering the SSS (fraction [a]; see above).
We applied a General Least-Square Model (GLS, Pinheiro & Bates, 2000) to explain the temporal variation in SSS. We used the following predictors: environmental heterogeneity (EH), total species richness of the period (SR), Cyanobacterial dominance (CYANO), and an environmental variable indicating productivity. To estimate EH, we applied an analysis of homogeneity of multivariate dispersions to the abiotic matrix in each period, as described by Anderson et al. (2006, 2011). All the environmental variables described above were previously standardized and used to estimate EH. As a proxy of productivity, we added one of the following climate variables in alternative models: total insolation (INS, kWh m−2), precipitation (PRE, mm), or water temperature (WT, °C). It is well-known that all climatic variables above are positively correlated with productivity, and also that they vary seasonally in Subtropics (Ball, 2015). Total insolation and precipitation were obtained from INMET (National Institute of Meteorology) in BDMEP (Database of Meteorological Data for Education and Research) from a meteorological station located in the center of the Paraná State (−25°46′W, −50°63′S). We also used the mean density (cells mL−1) of all the Cyanobacterial taxa occurring in each period as a predictor of SSS. However, we did not use this predictor for Group III, given that this group is composed entirely of Cyanobacteria species. For our GLS model, we assumed an autoregressive model of order 1 (Pinheiro & Bates, 2000). This model was estimated using the nlme package in R (Pinheiro et al., 2015). The best approximating model for our data was selected using the Akaike Information Criterion (AIC), following Burnham & Anderson (2004). In this case, we considered plausible models those with ΔAIC lower than 5.
Results
Variation partitioning of community data
The relative importance of environmental, spatial, and historical predictors (i.e., pure fractions [a], [b], and [c]) varied widely over time (Table S3 in Online Resource), and significant predictors were found in some sampling periods (Table S3 in Online Resource).
The average total variance (all fractions summed) explained by our predictor matrices ranged from 21% (Group IV) to 54% (Group I and III). Data on earlier community structure were, in average, the best predictor matrix of phytoplankton community structure, except for two cases (groups I and II). The environmental and spatial matrices were the main predictors of groups I and II, respectively. These matrices were unrelated to group IV, and no significant relationships were found between matrix group VI and earlier community structure (Table S3 in Online Resource).
In general, the total variation in phytoplankton community structure was better explained when the response matrix was organized according to a functional approach instead of a traditional (taxonomic) approach (Table S4 in Online Resource, Fig. 2). However, we found a higher total adjusted coefficient of determination among the different functional groups when compared to the species response matrix (Table S4 in Online Resource, Fig. 2). Groups I and III showed significant higher median values of SSS when compared to species response matrix (Table S4 in Online Resource, Fig. 2).
Median of the total adjusted coefficient of determination (Total Adjusted R 2) from RDA models considering the response matrices of phytoplankton species density (Species), functional groups density (Groups), and deconstructed into different phytoplankton group densities (Group I—VII). SSS = Strength of species sorting. “+” significantly higher than species data; and “−“ significantly lower than species data according to Wilcoxon test
The highest SSS values were found, in general, when the response matrix was organized by the density vectors of the functional groups (Fig. 3). Similar results were found for Group I and III (Fig. 3).
Time series of the adjusted coefficient of determination (Adjusted R 2) from RDA models considering environmental variables as explanatory variables and phytoplankton data as response matrices: species densities, functional groups density, and species density in each functional group (Group I—VII). SSS = Strength of species sorting
GLS models
There were no significant relationships between our predictor variables and our measure of the SSS for phytoplankton group and for most functional group (I, II, III, VI, and VII) composition (Table S5 in Online Resource). SSS estimated for species composition was positively related to INS (Table 1). Another predictor variable representing productivity (WT) was positively related to SSS for group IV, whereas the SSS for group V was positively related to EH. The next two best models (with delta AIC < 5; Table 1) provided similar results.
Discussion
A simultaneous role of environmental and spatial drivers has been recorded for several phytoplankton communities (Soininen et al., 2007; Hájek et al., 2011; Chust et al., 2013; Gallego et al., 2014). Yet, debate on the most important community assembly mechanism for a certain metacommunity is highly criticized and unproductive when not related to clear expectations (e.g., relative roles depend on organisms’ dispersal ability, spatial scale, and environmental gradients studied; Heino et al., 2015; see also Brown et al., 2017). Indeed, it is hard to conclude that environmental or spatial drivers are the most important predictors of a certain metacommunity given that considering all relevant variables in predictor matrices is highly difficult if not impossible (see also Vellend et al., 2014). Even so, we found that the earlier community composition was an important predictor set of the present community composition. Such ‘historical’ influences are rarely analyzed in studies on the composition of phytoplankton metacommunities. Thus, we suggest earlier occupation is included in future studies on correlates of phytoplankton community structure.
Our results also corroborated the view that determinants of community structure are highly variable over time (Heino & Mykrä, 2008; Erős et al., 2014; Fernandes et al., 2014a). Studies have demonstrated that explaining spatial patterns in microalgal metacommunities based on snapshot samplings may be biased (Vyverman et al., 2007; Verleyen et al., 2009; Heino et al., 2010; Hájek et al., 2011). Such assessments of metacommunity structure, despite being conducted over large spatial extents, do not capture the temporally dynamic nature of phytoplankton metacommunities (Beisner et al., 2006). Complementarily, we have already demonstrated in a previous study that beta diversity of phytoplankton in the same reservoirs also varies temporally (Wojciechowski et al., 2017). The present study adds to this knowledge because it shows that not only beta diversity, but also the determinants of metacommunity organization vary temporally.
We hypothesized that, using the same sets of predictors, phytoplankton composition would be better explained in trait-based analyses than in taxonomic ones. This hypothesis was based on the assumption that species within a functional group would have similar responses to environmental variation. However, we found limited support for this hypothesis. One explanation for this result may simply be that, in terms of community structure predictability, the deconstructive approach (based on functional groups; Marquet et al., 2004) is not so superior to the traditional, taxonomic-based, approach. Also, it is important to emphasize that we used literature data to create our functional groups. Thus, higher coefficients of determination could be obtained for the deconstructive approach, in comparison to the taxonomic approach, with the use of local data for the formation of the functional groups. The functional group classification used here uses basically morphology to classify species and were proposed using phytoplankton data from temperate sites very different from those studied here (e.g., Reynolds et al., 2002; Salmaso & Padisák, 2007; Padisák et al., 2009). Also, a better elucidation of how environmental variables related to functional traits could be obtained if ‘true’ functional composition was analyzed, e.g., either assessing traits responses to environmental gradients in a combination of RLQ and forth-corner analysis (Dray et al., 2014) or using community-weighted mean (CWM) response matrices (Lavorel et al., 2007) in pRDA instead of classifying taxa into functional groups proposed by experts. In this case, meaningful phytoplankton traits must be measured for all species in the dataset (Kleyer et al., 2012). However, we still do not have a set of meaningful traits for the phytoplankton species recorded here that allows us to use RLQ and forth-corner analysis or CWM-RDA.
We found, however, that SSS values over the time period were frequently higher when we used the density vectors of the functional groups instead of the total species composition. Furthermore, groups I (small organisms with high surface/volume ratio) and III (filamentous Cyanobacteria, mainly Nostocales) showed higher environment-related variation. These results suggest that, even with the limited support for our general hypothesis, trait-based analyses are central to understanding the likely causes underlying community assembly. Indeed, while species compositions were mainly affected by productivity, species composition in certain groups was affected by productivity or environmental heterogeneity.
Functional group classification was also useful with regard to our expectation that communities with high species richness would be less well explained by environmental predictors (Low-Décarie et al., 2014). Although we did not find a significant correlation between SSS and total species richness, species-rich groups (e.g., groups IV and V) exhibited higher SSS than species-poor groups (e.g., groups I and III). This observation also highlights that community responses to environmental variation are better elucidated by investigating groups of species that respond similarly to environmental gradients (i.e., the functional groups).
We also observed that variation in SSS among the different functional groups is commonly unpredictable (Soininen, 2014), and explanatory power for certain functional groups is higher than for the others (Fig. 2). Therefore, it seems that community assembly mechanisms may be more evident for some functional groups than for others. This was the case of the relationship with productivity, as proxied by temperature. A likely positive relationship between productivity and SSS was observed for group IV (composed of organisms of medium size lacking specialized traits, i.e., very common Chlorophytes, as Monoraphidium and Scenedesmus). Although we did not use a proxy for eutrophication as a predictor, this is in line with a previous study suggesting that eutrophication determines community assembly (Donohue et al., 2009) if more productive periods coincide with occurrences of eutrophication.
Phytoplankton communities can be structured by several factors, such as water chemical characteristics (especially nutrients; Reynolds, 1984, 2006), morphological, physical and hydrological variables (i.e., water column mixing dynamics and water retention time; Domitrovic, Domitrovic 2003; Jones & Elliott, 2007), climate (Paerl & Huisman, 2008), and biological interactions (i.e., top-down control and viral infection; Lazzaro et al., 2003; Brussaard, 2004). This multiplicity of factors may contribute to low explanatory power of multivariate models. In addition, although we do not have data to test, we speculate that specific management strategies (e.g., flushing and dredging), which differ among the reservoirs, may also be of paramount importance to predict phytoplankton community structure (see Rangel et al., 2012). Also, phytoplankton metacommunity in the Paraná State reservoirs was predominantly composed of rare species (the few common species are shown in Table S1). Distributions of rare species are difficult to model (Heino & Soininen, 2010; Siqueira et al., 2012), also contributing to the low variation explained in our analysis. Finally, the possibility of phytoplankton metacommunities being regulated by stochastic processes (De Meester et al., 2005; Chase, 2007; Vellend et al., 2014) should not be ignored.
To conclude, we showed that the strength of species sorting of phytoplankton metacommunities was highly variable in time. This finding prevailed irrespective of the dataset used (i.e., all species, species in each functional group, or aggregate functional groups), and suggest that the effects of species sorting in structuring metacommunities may be dependent on the time of sampling (Erős et al., 2014; Fernandes et al., 2014a). Hence, we strongly encourage researchers to go beyond snapshot sampling, as the mechanisms controlling local communities may be temporally variable and to some degree unpredictable. Finally, we have also demonstrated that SSS and its likely determinants depend on the functional group, highlighting the importance of trait-based analyses for studies aiming to understand the causes of community assembly in metacommunities.
References
Anderson, M. J., K. E. Ellingsen & B. H. McArdle, 2006. Multivariate dispersion as a measure of beta diversity. Ecology Letters 9: 683–693.
Anderson, M. J., T. O. Crist, J. M. Chase, M. Vellend, B. D. Inouye, A. L. Freestone, N. J. Sanders, H. V. Cornell, L. S. Comita, K. F. Davies, S. P. Harrison, N. J. B. Kraft, J. C. Stegen & N. G. Swenson, 2011. Navigating the multiple meanings of β diversity: a roadmap for the practicing ecologist. Ecology Letters 14: 19–28.
Apha, A. P. H. A., 2005. Standard methods for the examination of water & wastewater, 21st ed. American Public Health Association, Washington.
Ball, R., 2015. Trends in seasonal climate variance in the subtropical reservoir Joe Pool lake Texas: implications dissolved oxygen and nutrient distribution. The University of Texas at Arlington, Arlington: 1–97.
Beisner, B. E., P. R. Peres-Neto, E. S. Lindström, A. Barnett & M. L. Longhi, 2006. The role of environmental and spatial processes in structuring lake communities from bacteria to fish. Ecology 87: 2985–2991.
Bellier, E., V. Grøtan, S. Engen, A. K. Schartau, I. Herfindal & A. G. Finstad, 2014. Distance decay of similarity, effects of environmental noise and ecological heterogeneity among species in the spatio-temporal dynamics of a dispersal-limited community. Ecography 37: 172–182.
Bengtsson, J., S. R. Baillie & J. Lawton, 1997. Community variability increases with time. Oikos 78: 249–256.
Bini, L. M., V. L. Landeiro, A. A. Padial, T. Siqueira & J. Heino, 2014. Nutrient enrichment is related to two facets of beta diversity for stream invertebrates across the United States. Ecology 95: 1569–1578.
Borcard, D., P. Legendre & P. Drapeau, 1992. Partialling out the spatial component of ecological variation. Ecology 73: 1045–1055.
Brown, B. L., E. R. Sokol, J. Skelton & B. Tornwall, 2017. Making sense of metacommunities: dispelling the mythology of a metacommunity typology. Oecologia 183: 643–652.
Brussaard, C. P. D., 2004. Viral control of phytoplankton populations—a review. The Journal of Eukaryotic Microbiology 51: 125–138.
Burnham, K. P. & D. R. Anderson, 2004. Model selection and multimodel inference. Springer, New York.
Castillo-Escrivà, A., J. A. Aguilar-Alberola & F. Mesquita-Joanes, 2017. Spatial and environmental effects on a rock-pool metacommunity depend on landscape setting and dispersal mode. Freshwater Biology 62: 1004–1011.
Chase, J. M., 2007. Drought mediates the importance of stochastic community assembly. Proceedings of the National Academy of Sciences 104: 17430–17434.
Chase, J. M., 2010. Stochastic community assembly causes higher biodiversity in more productive environments. Science 328: 1388–1391.
Chust, G., X. Irigoien, J. Chave & R. P. Harris, 2013. Latitudinal phytoplankton distribution and the neutral theory of biodiversity. Global Ecology and Biogeography 22: 531–543.
De Meester, L., S. Declerck, R. Stoks, G. Louette, F. Van De Meutter, T. De Bie, E. Michels & L. Brendonck, 2005. Ponds and pools as model systems in conservation biology, ecology and evolutionary biology. Aquatic Conservation: Marine and Freshwater Ecosystems 15: 715–725.
de Domitrovic, Y. Z., 2003. Effect of fluctuations in water level on phytoplankton development in three lakes of the Paraná river floodplain (Argentina). Hydrobiologia 510: 175–193.
Donohue, I., A. L. Jackson, M. T. Pusch & K. Irvine, 2009. Nutrient enrichment homogenizes lake benthic assemblages at local and regional scales. Ecology 90: 3470–3477.
Dray, S., P. Legendre & P. R. Peres-Neto, 2006. Spatial modelling: a comprehensive framework for principal coordinate analysis of neighbour matrices (PCNM). Ecological Modelling 196: 483–493.
Dray, S., P. Choler, S. Dolédec, P. R. Peres-Neto, W. Thuiller, S. Pavoine & C. J. F. ter Braak, 2014. Combining the fourth-corner and the RLQ methods for assessing trait responses to environmental variation. Ecology 95: 14–21.
Erős, T., P. Sály, P. Takács, C. L. Higgins, P. Bíró & D. Schmera, 2014. Quantifying temporal variability in the metacommunity structure of stream fishes: the influence of non-native species and environmental drivers. Hydrobiologia 722: 31–43.
Fernandes, I. M., R. Henriques-Silva, J. Penha, J. Zuanon & P. R. Peres-Neto, 2014a. Spatiotemporal dynamics in a seasonal metacommunity structure is predictable: the case of floodplain-fish communities. Ecography 37: 464–475.
Fernandes, L. F., K. C. Gutseit, J. Wojciechowski, P. E. D. Lagos & C. F. Xavier, 2014b. Phytoplankton ecology in the Rio Verde reservoir. In Carneiro, C., C. V. Andreoli, C. L. N. Cunha & E. F. Gobbi (eds), Reservoir eutrophication: preventive management: an applied example of integrated basin management interdisciplinary research. IWA Publishing, London: 255–276.
Fernandes, L. F., P. E. D. Lagos, A. C. Wosiack, C. V. Pacheco, L. Domingues, L. Zenhder-Alves & V. Coquemala, 2005. Comunidades fitoplanctônicas em ambientes lênticos. In Andreoli, C. V. & C. Carneiro (eds), Gestão integrada de mananciais de abastecimento eutrofizados. Sanepar-Finep, Curitiba: 303–366.
Finlay, B. J. & T. Fenchel, 2004. Cosmopolitan metapopulations of free-living microbial eukaryotes. Protist 155: 237–244.
Gallego, I., T. A. Davidson, E. Jeppesen, C. Pérez-Martínez, F. Fuentes-Rodríguez, M. Juan & J. J. Casas, 2014. Disturbance from pond management obscures local and regional drivers of assemblages of primary producers. Freshwater Biology 59: 1406–1422.
Hájek, M., J. Roleček, K. Cottenie, K. Kintrová, M. Horsák, A. Poulíčková, P. Hájková, M. Fránková & D. Dítě, 2011. Environmental and spatial controls of biotic assemblages in a discrete semi-terrestrial habitat: comparison of organisms with different dispersal abilities sampled in the same plots. Journal of Biogeography 38: 1683–1693.
Heino, J., L. M. Bini, S. M. Karjalainen, H. Mykrä, J. Soininen, L. C. G. Vieira & J. A. F. Diniz-Filho, 2010. Geographical patterns of micro-organismal community structure: are diatoms ubiquitously distributed across boreal streams? Oikos 119: 129–137.
Heino, J., A. S. Melo, L. M. Bini, F. Altermatt, S. A. Al-Shami, D. G. Angeler, N. Bonada, C. Brand, M. Callisto, K. Cottenie, O. Dangles & D. Dudgeon, 2015. A comparative analysis reveals weak relationships between ecological factors and beta diversity of stream insect metacommunities at two spatial levels. Ecology and Evolution 5: 1235–1248.
Heino, J. & H. Mykrä, 2008. Control of stream insect assemblages: roles of spatial configuration and local environmental factors. Ecological Entomology 33: 614–622.
Heino, J. & J. Soininen, 2010. Are common species sufficient in describing turnover in aquatic metacommunities along environmental and spatial gradients? Limnology and Oceanography 55: 2397–2402.
IAP, 2009. Monitoramento da qualidade das águas dos reservatórios do Estado do Paraná. Curitiba. Available on: http://www.iap.pr.gov.br/modules/conteudo/conteudo.php?conteudo=809
Jones, I. D. & J. A. Elliott, 2007. Modelling the effects of changing retention time on abundance and composition of phytoplankton species in a small lake. Freshwater Biology 52: 988–997.
Kleyer, M., S. Dray, F. de Bello, J. Lepš, R. J. Pakeman, B. Strauss, W. Thuiller & S. Lavorel, 2012. Assessing species and community functional responses to environmental gradients: which multivariate methods? Journal of Vegetation Science 23: 805–821.
Kruk, C., V. L. M. Huszar, E. T. H. M. Peeters, S. Bonilla, L. Costa, M. Lürling, C. S. Reynolds & M. Scheffer, 2010. A morphological classification capturing functional variation in phytoplankton. Freshwater Biology 55: 614–627.
Lavorel, S., K. Grigulis, S. McIntyre, N. S. G. Williams, D. Garden, J. Dorrough, S. Berman, F. Quétier, A. Thébault & A. Bonis, 2007. Assessing functional diversity in the field—methodology matters! Functional Ecology 22: 134–147.
Lazzaro, X., M. Bouvy, R. A. Ribeiro-Filho, V. S. Oliviera, L. T. Sales, A. R. M. Vasconcelos & M. R. Mata, 2003. Do fish regulate phytoplankton in shallow eutrophic Northeast Brazilian reservoirs? Freshwater Biology 48: 649–668.
Legendre, P. & E. D. Gallagher, 2001. Ecologically meaningful transformations for ordination of species data. Oecologia 129: 271–280.
Legendre, P. & L. F. J. Legendre, 2012. Numerical ecology. Elsevier, Québec.
Legendre, P., D. Borcard, F.G. Blanchet, & S. Dray, 2013. PCNM: MEM spatial eigenfunction and principal coordinate analyses. R Package Version 2.1-2/r109
Leibold, M. A., M. Holyoak, N. Mouquet, P. Amarasekare, J. M. Chase, M. F. Hoopes, R. D. Holt, J. B. Shurin, R. Law, D. Tilman, M. Loreau & A. Gonzalez, 2004. The metacommunity concept: a framework for multi-scale community ecology. Ecology Letters 7: 601–613.
Litchman, E., P. T. de Pinto, C. A. Klausmeier, M. K. Thomas & K. Yoshiyama, 2010. Linking traits to species diversity and community structure in phytoplankton. Hydrobiologia 653: 15–28.
Logue, J. B., N. Mouquet, H. Peter & H. Hillebrand, 2011. Empirical approaches to metacommunities: a review and comparison with theory. Trends in Ecology & Evolution 26: 482–491.
Low-Décarie, E., C. Chivers & M. Granados, 2014. Rising complexity and falling explanatory power in ecology. Frontiers in Ecology and the Environment 12: 412–418.
Lund, J. W. G., G. Kipling & E. D. Le Creen, 1958. The inverted microscope method of estimating algae numbers and the statistical basis of estimation by counting. Hydrobiologia 11: 143–170.
Marquet, P. A., M. Fernández, S. A. Navarrete & C. Valdovinos, 2004. Species richness emerging: toward a deconstruction of species richness patterns. In Lomolino, M. & R. Lawrence (eds), Frontiers in biogeography: new directions in the geography of nature. Sinauer Associates Inc, Sunderland: 191–209.
Martiny, J. B., B. J. Bohannan, J. H. Brown, R. K. Colwell, J. A. Fuhrman, J. L. Green, M. C. Horner-Devine, M. Kane, J. A. Krumins, C. R. Kuske, P. J. Morin, S. Naeem, L. Ovreås, A. L. Reysenbach, V. H. Smith & J. T. Staley, 2006. Microbial biogeography: putting microorganisms on the map. Nature Reviews Microbiology 4: 102–112.
McCreadie, J. W. & C. R. Bedwell, 2013. Patterns of co-occurrence of stream insects and an examination of a causal mechanism: ecological checkerboard or habitat checkerboard? Insect Conservation and Diversity 6: 105–113.
McGill, B. J., B. J. Enquist, E. Weiher & M. Westoby, 2006. Rebuilding community ecology from functional traits. Trends in ecology & evolution (Personal edition) 21: 178–185.
Oksanen, J., F. G. Blanchet, R. Kindt, P. Legendre, P. R. Minchin, R. B. O’Hara, G. L. Simpson, P. Solymos, M. H. H. Stevens, & H. Wagner, 2015. Community Ecology Package, 2015.
Padisák, J., L. O. Crossetti & L. Naselli-Flores, 2009. Use and misuse in the application of the phytoplankton functional classification: a critical review with updates. Hydrobiologia 621: 1–19.
Paerl, H. W., 2014. Mitigating harmful cyanobacterial blooms in a human- and climatically-impacted world. Life 4: 988–1012.
Paerl, H. W. & J. Huisman, 2008. Climate. Blooms like it hot. Science 320: 57–58.
Paradis, E., J. Claude & K. Strimmer, 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20: 289–290.
Peres-Neto, P. R., P. Legendre, S. Dray & D. Borcard, 2006. Variation partitioning of species data matrices: estimation and comparison of fractions. Ecology 87: 2614–2625.
Pinheiro, J., D. M. Bates, S. DebRoy, D. Sarkar, & R Core Team, 2015. nlme: Linear and Nonlinear Mixed Effects Models.
Pinheiro, J. C. & D. M. Bates, 2000. Mixed-effects models in S and S-PLUS. Springer-Verlag, New York.
R Core Team, 2015. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/
Rangel, L. M., L. H. S. Silva, P. Rosa, F. Roland & V. L. M. Huszar, 2012. Phytoplankton biomass is mainly controlled by hydrology and phosphorus concentrations in tropical hydroelectric reservoirs. Hydrobiologia 693: 13–28.
Reynolds, C. S., 1984. Phytoplankton periodicity: the interaction of form, function and environmental variability. Freshwater Biology 14: 111–142.
Reynolds, C. S., 1989. Physical determinants of phytoplankton succession plankton ecology: succession in plankton communities. Springer, Berlin: 9–56.
Reynolds, C. S., 2006. Ecology of phytoplankton. Cambridge University Press, Cambridge.
Reynolds, C. S., V. L. M. Huszar, C. Kruk, L. Naselli-Flores & S. Melo, 2002. Towards a functional classification of the freshwater phytoplankton. Journal of Plankton Research 24: 417–428.
Salmaso, N. & J. Padisák, 2007. Morpho-functional groups and phytoplankton development in two deep lakes (Lake Garda, Italy and Lake Stechlin, Germany). Hydrobiologia 578: 97–112.
Siqueira, T., L. M. Bini, F. O. Roque, S. R. Marques Couceiro, S. Trivinho-Strixino & K. Cottenie, 2012. Common and rare species respond to similar niche processes in macroinvertebrate metacommunities. Ecography 35: 183–192.
Soininen, J., 2014. A quantitative analysis of species sorting across organisms and ecosystems. Ecology 95: 3284–3292.
Soininen, J., M. Kokociński, S. Estlander, J. Kotanen & J. Heino, 2007. Neutrality, niches, and determinants of plankton metacommunity structure across boreal wetland ponds. Ecoscience 14: 146–154.
Suikkanen, S., G. O. Fistarol & E. Granéli, 2005. Effects of cyanobacterial allelochemicals on a natural plankton community. Marine Ecology Progress Series 287: 1–9.
Utermöhl, H., 1958. Zur Vervollkommnung der quantitativen Phytoplankton-Methodik. Mitt. Int. Verein. Theor. Angew. Limnol 26: 11–20.
Vanormelingen, P., K. Cottenie, E. Michels, K. Muylaert, W. Vyverman & L. De Meester, 2008. The relative importance of dispersal and local processes in structuring phytoplankton communities in a set of highly interconnected ponds. Freshwater Biology 53: 2170–2183.
Vellend, M., D. S. Srivastava, K. M. Anderson, C. D. Brown, J. E. Jankowski, E. J. Kleynhans, N. J. B. Kraft, A. D. Letaw, A. A. M. Macdonald, J. E. Maclean, I. H. Myers-Smith, A. R. Norris & X. Xue, 2014. Assessing the relative importance of neutral stochasticity in ecological communities. Oikos 123: 1420–1430.
Verleyen, E., W. Vyverman, M. Sterken, D. A. Hodgson, A. De Wever, S. Juggins, B. Van de Vijver, V. J. Jones, P. Vanormelingen, D. Roberts, R. Flower, C. Kilroy, C. Souffreau & K. Sabbe, 2009. The importance of dispersal related and local factors in shaping the taxonomic structure of diatom metacommunities. Oikos 118: 1239–1249.
Vyverman, W., E. Verleyen, K. Sabbe, K. Vanhoutte, M. Sterken, D. A. Hodgson, D. G. Mann, S. Juggins, B. Van de Vijver, V. Jones, R. Flower, D. Roberts, V. A. Chepurnov, C. Kilroy, P. Vanormelingen & A. De Wever, 2007. Historical processes constrain patterns in global diatom diversity. Ecology 88: 1924–1931.
Wetzel, C. E., D. C. Bicudo, L. Ector, E. A. Lobo, J. Soininen, V. L. Landeiro & L. M. Bini, 2012. Distance decay of similarity in neotropical diatom communities. PLoS ONE 7: e45071.
Wojciechowski, J., J. Heino, L. M. Bini & A. A. Padial, 2017. Temporal variation in phytoplankton beta diversity patterns and metacommunity structures across subtropical reservoirs. Freshwater Biology 62: 751–766.
Acknowledgements
We warmly thank T. Siqueira and an anonymous reviewer for providing numerous and useful insights in previous drafts of this manuscript. We also acknowledge “Companhia Paranaense de Energia (COPEL)” for providing the data, and “Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES)” and “Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq)” for continuous financial support. J.W. received a PhD scholarship from CAPES.
Author information
Authors and Affiliations
Corresponding author
Additional information
Guest editors: Koen Martens, Sidinei M. Thomaz, Diego Fontaneto & Luigi Naselli-Flores / Emerging Trends in Aquatic Ecology II
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wojciechowski, J., Heino, J., Bini, L.M. et al. The strength of species sorting of phytoplankton communities is temporally variable in subtropical reservoirs. Hydrobiologia 800, 31–43 (2017). https://doi.org/10.1007/s10750-017-3245-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10750-017-3245-9