Abstract
LEA (late embryogenesis abundant) proteins are firstly discovered in seeds and then identified in vegetative tissues of different plant species. They are mainly regulated under abiotic stress conditions. Although genome wide studies of different gene family members have been performed in cucumber, there is no such a study for LEA genes. We have identified 79 LEA genes in the cucumber genome. Based on phylogenetic analysis, CsLEA genes could be classified into seven groups in which structural motifs are relatively conserved. Tandem duplications play an important role in cucumber genome for LEA gene expansion. Orthologous and chromosomal relationships of CsLEA genes were observed based on comparative mapping analysis with other species. The in silico micro-RNA (miRNA) target analyses indicated that 37 CsLEA genes were targeted by different miRNAs, especially mir854 and mir414 are the most abundant identified ones. Public available RNA-seq data were analyzed for expression analysis of CsLEA genes in different tissues of cucumber. According to genome-wide expression analysis, nine CsLEA genes showed higher expression profiles in all tissues. The expression profiles of ten CsLEA genes in the root and leaf tissues of drought-stressed cucumber were examined using qRT-PCR. Among them, CsLEA-54 induced after stress application in leaf and root tissues and might provide adaptation to drought stress for cucumber. CsLEA-09, CsLEA-32 and CsLEA-57 genes responded to drought after 3 h later and might be considered as early response genes to water limitation. This research could help us to improve understanding of contribution of CsLEAs to drought tolerance in cucumber.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cucumber (Cucumis sativus L.) is an important crop and belongs to Cucurbitacae family which yields about 192 million tons of fruits, vegetables and seeds annually worldwide (http://faostat.fao.org). Besides, these family members are used as a model system for sex determination and plant vascular biology studies (Xoconostle Cázares et al. 1999; Tanurdzic and Banks 2004; Lough and Lucas 2006).
LEA (late embryogenesis abundant) proteins were firstly discovered from seeds (Dure and Galau 1981). Further studies revealed that LEA proteins were abundant in other plant tissues (Oztur et al. 2002; Dalal et al. 2009; George et al. 2009; Olvera-Carrillo et al. 2010). In addition, many LEA homologues have been identified in different organisms such as Caenorhabditis elegans, Artemia franciscana, Dictyostelium discoideum etc. (Gal et al. 2004; Eichinger et al. 2005; Kikawada et al. 2006; Boswell et al. 2009; Denekamp et al. 2010). LEA protein gene family members are very hydrophilic proteins with unstructured constitution which have repeating amino acid motifs and tendency to alpha helix structure (Rorat 2006; Tunnacliffe et al. 2010). Identified LEA proteins were classified into at least seven different groups according to sequence homologies and presence of distinctive motifs. These groups were LEA1, LEA2, LEA3, LEA4, LEA5, dehydrin and SMP (Seed Maturation Protein). However, no distinct universal classification criterion for LEA proteins has been established yet (Dure et al. 1989; Hunault and Jaspard 2010).
LEA proteins have significant roles in normal plant growth as well as in abiotic stress conditions to protect plants from the effects of inconvenient conditions. Their production can be stimulated by different abiotic stresses such as water deficiency, salinity, and cold. They also play important functions, including protection of cellular structures from the effects of water loss and desiccation (Serrano and Montesinos 2003), protection of proteins from stress-induced damage (Serrano and Montesinos 2003; Grelet et al. 2005; Shao et al. 2005) sequestration of ions (Grelet et al. 2005), and folding of denatured proteins (Bray 1993). LEA proteins can also act as chaperone proteins to resist cellular damage (Wise 2003; Umezawa et al. 2006). Studies revealed that LEA proteins have been related to abiotic stress tolerance especially drought, cold and salt stress conditions. For instance, a cDNA clone FsDhn1 (codes for type-II LEA proteins) upregulated by ABA from Fagus sylvatica L. seeds involved in drought responses in artificially dried seeds (Jimenez et al. 2008). Besides, a wheat LEA gene (TaLEA3) was cloned into Leymus chinensis, which is an perennial grass, by Agrobacterium mediated transformation and transgenic L. chinensis plants showed enhanced growth ability under water deficiency conditions (Wang et al. 2009). Transgenic sweet potato plants overexpressing sweet potato LEA 14 (IbLEA14) gene displayed enhanced tolerance to salt and drought stress conditions (Park et al. 2011). Moreover, over expression of AtLEA3-3 gene in Arabidopsis thaliana caused an increase tolerance to salt and osmatic stress (Zhao et al. 2011). Furthermore, Arabidopsis plants which are lack of LEA4 protein family members were more susceptible to water limitation than wild types (Olvera-Carrillo et al. 2010). Additionally, group 1 LEA proteins of Artemia franciscana embryos were cloned and expressed in Drosophila melanogaster cells. Analysis displayed the role of the LEA proteins on increment of viability resistance of Drosophila melanogaster cells during water deficiency and protection of mitochondrial function (Marunde et al. 2013).
Up to date, a limited number of studies have been conducted for determination of LEA gene family members in Arabidopsis, rice, soybean, tomato, potato, poplar etc. using whole genomic data. LEA gene characteristics, locations on chromosomes and gene duplications, phylogenetic analysis and determination of conserved motifs in LEA proteins, genome wide tissue specific expression profiles and response of this protein family under abiotic stress conditions were generally evaluated in these studies (Wang et al. 2007; Hundertmark and Hincha 2008; Li et al. 2011; Lan et al. 2013; Cao and Li 2015; Charfeddine et al. 2015).
Whole genome sequence of cucumber plant was published in 2009 (Huang et al. 2009). Whole genomic data enables to identify genes through genome wide using bioinformatics tools in cucumber and different plant species (Yu et al. 2007; Liu et al. 2011; Cai et al. 2014; Wen et al. 2014; Lv et al. 2015). However, gene identification and characterization studies are limited using cucumber genome up to now (Ling et al. 2011; Li et al. 2012; Baloglu et al. 2014; Baloglu 2014; Baloglu et al. 2015). The objective of this study is to identify LEA gene members and their conserved motifs and genomic disposition in cucumber genome as our study group interested in genome wide analysis in the Cucurbitacae family. In addition, expression profiles of these gene family members in cucumber were analyzed according to publicity open data and experimental analysis. This analysis is the first comprehensive study of the LEA gene family in cucumber and provides new opportunities for cloning and functional analyses, which may be used in further studies for improving stress tolerance in plants. This study also will enable further characterization of this important gene family in cucumber to explore their functions and evolution.
Materials and methods
Analysis and characterization of LEA genes
Three approaches were employed to determine LEA genes in cucumber genome. Firstly, LEA protein sequences which belong to 14 different plant species (Arabidopsis thaliana, Gossypium hirsitum, Oryza sativa, Glycine max, Sorghum bicolor, Triticum aestivum, Triticum durum, Pisum sativum, Zea mays, Hordeum vulgare, Brassica napus, Medicago truncatula, Nicotiana tabacum, Vitis vinifera) were derived from LEAP database which provides data about the LEA proteins for the analysis of their structure–function relationships (http://forge.info.univangers.fr/~gh/Leadb/index.php?action=0&mode=0) (Hunault and Jaspard 2010). These sequences were utilized for determination of homolog peptides with cucumber genome in PHYTOZOME v10.2 database by BLASTP search (Goodstein et al. 2012). This database is a plant comparative genomics portal and provides sequenced and annotated green plant genomes. Besides, the database searched for ‘LEA’ key word. Hidden Markov Model (HMM) forms of LEA proteins in Pfam database (http://pfam.sanger.ac.uk/) which includes a large collection of protein families and provides investigation of functional regions called as domains of proteins were also compared with Cucumis sativus sequences in PHYTOZOME database. In addition, to determine all members of LEA proteins in cucumber, searches were implemented by TBLASTN program at NCBI database against the EST sequences of cucumber genome. Collocated sequences with expected values <1.0 were included where redundant ones removed by ‘Decrease redundancy tool’ which enables to reduce the redundancy in a set of aligned or unaligned sequences (http://web.expasy.org/decrease_redundancy). Furthermore, conserved domains of every LEA protein was checked by using SMART (Simple Modular Architecture Research Tool) which allows the identification and annotation of genetically mobile domains and analysis of their architectures (http://smart.emblheidelberg.de/) (Letunic et al. 2012) and Pfam (http://pfam.sanger.ac.uk/) databases. Isoelectric points (pI), instability index and molecular weights were estimated by applying to ProtParam Tool which provides the computation of various physical and chemical parameters for a given protein (http://web.expasy.org/protparam).
Determination of chromosomal locations, estimation of gene structure and calculation of genomic distribution of LEA genes
Chromosomal locations of the LEA genes in cucumber were determined at PHYTOZOME database selecting Cucumis sativus via BLASTP scanning. Then, cucumber LEA protein sequences from PHYTOZOME database were used to search against Cucurbit Genomics Database which provides genomic search in genomes of Cucurbitacae family members (http://www.icugi.org/cgi-bin/ICuGI/index.cgi).
Tandem and segmental duplications were determined via a method based on Plant Genome Duplication Database-PGDD which enables to study homologous genes (that have maintained relative chromosomal positions) in sequenced genomes (http://chibba.pgml.uga.edu/duplication/index/blast) (Tang et al. 2008). In summary, BLASTP search was served against all predicted LEA proteins of Cucumis sativus and first five matches with ≤1e−05 was regarded as potential anchors. Collinear blocks were determined by MCScan, and alignments with ≤1e−10 were selected as important matches (Tang et al. 2008; Du et al. 2013). Tandem duplications were also defined as adjacent genes of same sub-family located within ten predicted genes apart or within 30 kbp of each other (Shiu and Bleecker 2003; Du et al. 2013). In addition, exon–intron organizations of LEA genes was conducted through ‘gene structure display server’ (http://gsds.cbi.pku.edu.cn/) (Guo et al. 2007a, b), which is based on comparing full complementary DNAs (cDNA) or estimated coding sequences’ relevant genomic sequences.
Sequence alignment, phylogenetic classification and identification of conserved motifs of CsLEAs
Amino acid sequences were loaded to MEGA6 (Tamura et al. 2013) program which allows researchers to conduct sequence alignment, to infer phylogenetic trees and to estimate divergence times and multiple sequence alignments was made by using ClustalW program with a gap open and gap extension penalties of 10 and 0.1, respectively (Thompson et al. 1997). Aligned sequence file was utilized to form (Saitou and Nei 1987) phylogenetic tree through neighbor joining method with bootstrap analysis for 1000 iterations. Jones–Taylor–Thornton (JTT) substitution model were used for phylogeny reconstruction and rates among sites were gamma distributed (G). Protein sequence domains were detected by using DNA domain search tool (MEME) (http://meme.nbcr.net/meme3/meme.html) which provides to discover novel motifs in collections of unaligned nucleotide or protein sequences (Bailey and Elkan 1994) and were used to identify motifs in candidate sequences. The parameters for the analysis were: number of repetitions-any; maximum number of motifs-20 and optimum width of motif ≥2 and ≤300. Detected MEME domains with ≤1e−30 were scanned at Interpro database through InterProScan which makes functional analysis of proteins by classifying them into families and predicting domains and important sites (Quevillon et al. 2005).
Gene ontology (GO) analysis
Functional analysis of LEA sequences were conducted by utilizing Blast2GO (http://www.blast2go.com) program which is a bioinformatics platform for high-quality functional annotation and analysis of genomic datasets. (Conesa and Götz 2008). Amino acid sequences of LEA proteins were loaded to Blast2GO program. Three categories of GO classification (determination of biological functions, cellular content and molecular functions) were achieved with this program.
Comparative physical mapping of cucumber LEA proteins and other species
BlastP program was used to discover orthologous relationships between cucumber chromosomes and other species. Arabidopsis, poplar and maize peptide sequences (www.phytozome.net), which were equal to cucumber LEA proteins amino acid sequences, were scanned. The ones whose e-value is ≤1e−5 and at least %80 identical ones were accepted as meaningful. Orthologous relationships between LEA genes in cucumber and Arabidopsis, poplar and maize chromosomes were monitored with MapChart program which is software for the graphical presentation of linkage maps and QTL (Voorrips 2002).
Estimating rates of synonymous and non-synonymous substitution
The amino-acid sequences belonging to duplicated protein-encoding LEA protein members and orthologous gene-pairs between cucumber and poplar, rice, Arabidopsis and maize were aligned by using a tool called CLUSTALW, which is an multiple sequence alignment tool. CODEML (http://www.bork.embl.de/pal2nal/) program, which converts a multiple sequence alignment of proteins and the corresponding DNA (or mRNA) sequences into a codon alignment (Suyama et al. 2006) was used to calculate synonymous (Ks) and non-synonymous (Ka) change ratio through a method in which LEA genes’ amino acid sequences and their original complementary DNA sequence alignment were made. Time (million years ago, Mya) of duplication and divergence of each LEA genes were calculated with the formula (T = Ks/2λ (λ = 6.5 × 10−9) and by using mutation ratio corresponding to every synonymous area and every year (Lynch and Conery 2000; Yang et al. 2008).
Determination of miRNAs CsLEA target genes
Determination of miRNA controlled gene targets is important for releasing of miRNA functions. Formerly known plant pre-miRNA sequences gathered from miRBase v20.0 (http://www.mirbase.org) and plant miRNA database (http://bioinformatics.cau.edu.cn/PMRD) were used for determination of miRNAs targeting LEA genes. psRNA Target Server (http://plantgrn.noble.org/psRNATarget/) was used with this aim; all known plant miRNAs and cucumber LEA gene transcripts were aligned and all the assumed plant cucumber miRNAs were detected. The parameters which had been used by Zhang (2005) were utilized and all the known plant miRNAs and their potential target or targets were assessed. miRNA targets, determined with computer method, were utilized to determine the assumed homologue with BLASTX scanning and analyzed.
Homology modeling of LEA proteins
All the cucumber LEA genes were scanned at Protein Data Bank (PDB) which provides information about the 3D structures of biological molecules, including proteins and nucleic acids (Berman et al. 2000) by using BLASTP to determine homologous sequence and known the best sample, having three-dimensional structure. For gathered information Phyre2 (Protein Homology/Analog/YRecognition Engine; http://www.sbg.bio.ic.ac.uk/phyre2) program which is a web-based service for protein structure prediction was used and protein structure was estimated with homology modeling (Kelley and Sternberg 2009).
Expression profiles of cucumber LEA genes by utilizing of transcriptome data
For RNA-Seq analyses, all the Illimuna HiSeq readings were obtained from an open data bank archive (SRA, Sequence Read Archive) which stores raw sequencing data and alignment information from high-throughput sequencing platforms with following accession numbers; SRR351476 (cucumber ovary tissue, unexpanded), SRR351489 (cucumber expanded ovary tissue, fertilized), SRR351495 (cucumber expanded ovary tissue, unfertilized), SRR351499 (cucumber root tissue), SRR351905 (cucumber 1 stem tissue), SRR351906 (cucumber leaf tissue), SRR351908 (cucumber male flower tissue), SRR351910 (cucumber tendril tissue) SRR351911 (cucumber tendril tissue basal), SRR351912 (cucumber female flower tissue). All readings were obtained as raw sequence data “.sra” and transformed to “fastq” format. After having eliminated low quality (Q) score <20), the rest of clean readings have been subjected to FastQC analysis in order to check sequence quality for per-base sequence qualities, per-sequence quality scores, per-base nucleotide content and sequence duplication levels. CLC Genomic Workbench version 7.5 was used for normalization of raw count data, after that hierarchical clustering heat map were generated by log2 RPKM values using PermutMatrix software (Caraux and Pinloche 2005) in this system.
Plant materials, growth conditions and treatments
Cucumber seeds were provided from Monsanto Gıda ve Tarım Tic. Ltd. Şti (Antalya). The pods of seeds were removed and seeds were washed three times with distillate water. After that, they were placed to pots and grown in 400 μmol m−2 s−1 light intensity and 24 ± 2 °C and 16-hour light and 8-hour dark photoperiod in a plant growing cabinet, a culture medium which was consisting of Hoagland solution (Hoagland and Arnon 1950), for 14 days.
Polyethylene glycol 6000 (PEG-6000) (10 %) has been introduced to Hoagland solution for drought stress. Stress treatment was started at 14th day of normal growing period. The plants having stress treatment and control plants were grown within the same growing conditions in plant growing cabinet. The plants were exposed to drought stress at different time courses including 0, 3, 12 and 24th hours. Root and leaf tissues from control and stressed ones were harvested for tissue specific expression analysis. Time point zero (0 h) was used as a control. Tissue samples, gathered by conducting biological sampling three by three, were used in tissue specific gene expression measurements by being frosted with liquid nitrogen.
RNA isolation and quantitative real-time PCR analysis
Trizol solution (Life Technologies Corporation, Grand Island, NY, USA) was utilized for total RNA isolation. Isolations were made as in Unver and Budak (2009). In short, 100 mg plant samples were taken to pieces in 2 ml sterile tubes in 1 ml Trizol reactive by using liquid nitrogen and homogenized. Homogenized samples were kept waiting in room temperature for 5 min and with this way full decomposition of nucleoprotein complex. For 1 ml Trizol reactive, 0.2 ml chloroform was added. The caps of tubes were closed properly and mixed by shaking strongly by hands for 15 s and it was kept waiting in room temperature for 2–3 min and centrifuged at 4 °C, 15.000 rpm for 20 min. The liquid phase on the top was transferred to a new tube and precipitation of RNA was provided by being mixed with isopropyl alcohol. Isopropyl alcohol was used as half of Trizol reactive used in the course of homogenization. The samples were kept waiting in room temperature for 10 min and then centrifuged at 15.000 rpm, 4 °C for 10 min. RNA precipitation was washed with 75 % ethanol for once and 1 ml ethanol was added for each used 1 ml Trizol reactive. The samples were mixed with vortex and centrifuged at 10.000 rpm 4 °C for 5 min. After all processes, RNA precipitation was put to dry for 5–10 min and RNA was dissolved in 30 ml sterile water and kept at −80 °C until it was used after being kept waiting at 55–60 °C for 10 min. Isolated RNAs were exposed to 1 U DNase I (Thermo, Lithuania) for 1 µg RNA to remove genomic DNA completely. Quality and durability of the isolated RNA was checked by using agarose-gel electrophoresis and Multi Scan Go device (Thermo Fisher Scientific, USA).
For real time PCR (qRT-PCR) analysis, 10 LEA genes were selected for each plant and specific primers were designed for these LEA genes by using Primer 5 software program (http://www.primer-e.com/index.htm). Primer list used in the qRT-PCR is presented in Table S6. Rotor-Gene qPCR detection system (Qiagen, Hilden, Germany) and Qiagen SYBR Green master mix (Qiagen, Hilden, Germany) were used for qPCR analysis. PCR reaction tube was containing 0.1 μl reverse and forward primers (100 pmol), 2 μl of cDNA, 10 μl SYBR Green master mix and nuclease-free water was added up to 20 μl. The 18S rRNA gene was determined as the internal control. To be used 18S rRNA gene primer sequence was as below (GenBank ID: X51542.1): 5′-GTGACGGGTGACGGAGAATT-3′ and 5′-GACACTAATGCGCCCGGTAT-3′ (Baloglu et al. 2014). The qRT-PCR was implemented as follows: 40 cycles of 95 °C for 10 s and 60 °C for 40 s. The melting curves were conformed to 95 °C for 5 s and 55 °C for 1 min and then cooled to 40 °C for 30 s. Before real time PCR analysis, the possibility of this control genes being reference gene was tried under drought stress conditions and it was observed that there was no change in expression analysis of this gene after relevant stress factors (Ling et al. 2011). Optimization of primers was made by determining the appropriate RT-PCR conditions for Tm values of primers. Sampling was made three times for every step and triple quantitative PCR was applied for any duplication.
ΔCT ve ΔΔCT values, ΔCT = CTsample − CTreference and ΔΔCT = ΔCTtreated sample − ΔCTcontrol (0 h) was calculated and the difference between expression levels were determined as 2−ΔΔCt (Livak and Schmittgen 2001). Student t test was utilized in the statistical analysis of the difference between the samples, having stress treatment and control samples. If p values <0.01, it was observed that the expression difference in LEA gene is meaningful.
Results and discussion
Identification of the CsLEA family member genes in cucumber genome
To identify LEA genes in cucumber genome, LEA protein sequences which belong to 14 different plant species including Arabidopsis, cotton, rice, soybean, sorghum, common wheat, durum wheat, pea, maize, barley, rape, barrel medic, tobacco and grape were derived from LEAP database (Hunault and Jaspard 2010). Multiple searches employed using these sequences and as a result, 79 LEA family members were determined in cucumber genome (Table S1). Identified LEA proteins named from CsLEA-01 to CsLEA-79 according to their appropriate location on cucumber chromosomes of 1–7. CsLEA-79 gene (Cucsa.328650) location couldn’t find on any chromosome.
Amino acid contents and molecular weights of CsLEA proteins varied from 65 to 600 amino acid in length and 6848.5 to 65404.2 Da, respectively. According to physiochemical analysis, only 21 of CsLEA proteins were considered to be acidic (pI < 7) whereas 58 of them were of a basic (pI > 7) character. These findings are consistent with the studies which found that most of LEA protein members in purple false brome and tomato were of a basic nature (Filiz et al. 2013; Cao and Li 2015). Detailed information about parameters of CsLEA protein sequences were given in Table S1.
Genome wide studies are very limited for LEA gene family members in plants. In Arabidopsis, a total of 51 LEA gene family members were defined (Hundertmark and Hincha 2008). Moreover, Wang et al. (2007) found 34 LEA genes in rice genome. In addition, different studies revealed that numbers of identified LEA protein family members in Chinese plum (Du et al. 2013), soybean (Li et al. 2011), tomato (Cao and Li 2015), potato (Charfeddine et al. 2015), purple false brome (Filiz et al. 2013) and poplar (Lan et al. 2013) were 30, 36, 27, 29, 36 and 53 respectively. Our findings showed that cucumber has the highest number of LEA genes compared to LEA genes found in other plants. This may be arisen from detail bioinformatics analysis of many different plants and using many different identification tools.
Chromosomal location and structure of CsLEA Genes
CsLEA genes were spread to all cucumber chromosomes and the exact positions of CsLEA genes on seven cucumber chromosomes were given in Table S1. Sixteen CsLEA genes located on Chromosome 6 which includes the highest number of CsLEA gene and this chromosome followed by Chromosome 3 and Chromosome 7 with the number of 15 and 14 CsLEA genes, respectively. Some transcription factor genes (TF) like basic leucine zipper TF family (bZip) and growth regulating factor TF family members were mostly located on chromosome 3 in cucumber (Baloglu et al. 2014; Baloglu 2014) as LEA gene family members. Besides, only 5 CsLEA genes found in chromosome 2 which includes the lowest number of CsLEA gene.
Gene duplication events which are an explanation of ortholog genes are commonly observed in organisms (Mehan et al. 2004). Tandem and segmental duplications of LEA genes were searched in cucumber genome. Fifty-six LEA genes showed tandem duplication which accounting for the total of 70 % of all CsLEA genes. In addition, tandem duplication rates were the highest in the 3rd chromosome (Fig. 1). Tandem duplications also repeated on chromosome 4 (CsLEA-29, CsLEA-30, CsLEA-33, CsLEA-34, CsLEA-35 and CsLEA-36), chromosome 6 (CsLEA-55, CsLEA-56, CsLEA-58, CsLEA-59, CsLEA-60 and CsLEA-61) and chromosome 7 (CsLEA-65, CsLEA-67, CsLEA-69, CsLEA-70, CsLEA-72 and CsLEA-74). Observed tandem duplication rates in Arabidopsis, rice, tomato, potato, Chinese plum and poplar were 33, 17, 29, 7, 40, and 30 %, respectively (Wang et al. 2007; Hundertmark and Hincha 2008; Du et al. 2013; Lan et al. 2013; Cao and Li 2015; Charfeddine et al. 2015). Tandem duplication has a key role in gene duplications. Tandem duplication rates were higher in cucumber LEA genes than in Arabidopsis, rice, tomato, potato and Chinese plum. LEA gene duplication events may be more common in cucumber when compared with LEA genes in other studied plants. These findings may explain the fact that among the other plant species, the highest number of LEA gene members was observed in cucumber genome. These data can be suggested that tandem duplication events may be the main reason for the mechanisms which are responsible for CsLEA gene number expansion. On the other hand, segmental duplication was not found in cucumber while this event was ranging between 14 and 51 % in Arabidopsis, rice, Chinese plum, tomato and potato (Wang et al. 2007; Hundertmark and Hincha 2008; Du et al. 2013; Cao and Li 2015; Charfeddine et al. 2015). This can be attributed to the no function of segmental duplication events on LEA gene family expansion in cucumber genome.
Distributions of CsLEA genes on cucumber chromosomes. Physical locations of CsLEA genes are showed on cucumber chromosomes (numbered 1–7) using MapChart programme and chromosomal distances are given as Mbp. Only CsLEA-79 gene can’t locate on any chromosome and scaffold number was given. Tandem duplicated genes are indicated in the boxes
Also exon–intron structure was analyzed in CsLEA gene family members. We found that 44 CsLEA genes had no introns, which represented 55 % of these gene family members in cucumber. The great majority of intronless CsLEA genes were classified in Cluster IV and Cluster VII (Fig. S1). Intron containing genes had mostly one or two intron regions and this situation was similar in tomato (Cao and Li 2015), potato (Charfeddine et al. 2015) and Chinese plum (Du et al. 2013) which can be attributed to evolutionary preservation According to our results, exon–intron organizations of each CsLEA gene clusters were well preserved especially in Cluster VII. This may show the functional preservation of CsLEA proteins in different clusters. In addition, a few intron contents of CsLEA genes are consistent with the opinion that the stress response genes especially have lower intron number (Lan et al. 2013).
Phylogenetic classification and identification of conserved motifs of CsLEAs
A phylogenetic tree was constructed to evaluate relationships between CsLEA genes using Neighbour-Joining method with 1000 bootstrap replicates by MEGA v.6 software (Molecular Evolutionary Genetics Analysis) (Tamura et al. 2013). According to phylogenetic tree, CsLEA genes can be categorized into seven main clusters (Fig. 2). Using Pfam family term, LEA2 group member proteins were dominant between CsLEA proteins and found especially in 2nd, 4th and 7th clusters of the phylogenetic tree. LEA3 group proteins featured in the 1st cluster with the four members (CsLEA-29, CsLEA 30, CsLEA-73, and CsLEA-75) while LEA6 group proteins were represented by two members in the 3rd cluster (CsLEA-63, CsLEA-64). All LEA5 (CsLEA-35, CsLEA-36) and Seed maturation protein (SMP) group member (CsLEA-7, CsLEA-70, CsLEA-72) proteins accumulated in the 5th cluster but dehydrin group proteins were observed in different clusters depending on their different protein motif contents (CsLEA-10, CsLEA-11, CsLEA-31, CsLEA-54). Predominant LEA protein groups in tomato, maize, Chinese plum, poplar and Arabidopsis were dehydrin, LEA3, dehydrin and LEA2, LEA4 and LEA4 groups, respectively based on Pfam domain analysis (Hundertmark and Hincha 2008; Du et al. 2013; Lan et al. 2013; Cao and Li 2015, Li and Cao 2016). However, LEA2 group proteins in cucumber were predominant and included in all clusters of cucumber LEA proteins. These differences may be arisen from their response mechanisms to different stress conditions.
Phylogenetic classification of CsLEA proteins. The protein sequences are loaded to MEGA 6 program and aligned by CLUSTALW and phylogenetic tree is constructed by neighbor-joining method with bootstrap analysis for 1000 iterations. The proteins are classified into seven distinct clusters and each cluster is distributed by different color. (Color figure online)
Moreover, motif composition of CsLEA proteins was surveyed to test the reliability of the phylogenetic tree. Sixty-eight of CsLEA proteins contained motifs and 15 different motifs were observed using MEME software (Fig. S2). Conserved amino acid sequences of identified motifs were shown in Table 1. According to the data, LEA5, LEA6 and dehydrin group proteins in CsLEA proteins had no conserved motifs. Conversely, two distinct sequence motifs (K and Y segments) were observed in dehydrin proteins of potato (Charfeddine et al. 2015). In addition, K segment which is a lysine rich motif was reported in Chinese plum dehydrins (Du et al. 2013). Motif 1, 2, 3, 4 and 8 were predominantly present in LEA2 group proteins and also motif 5, 10 and 15 were observed in some LEA2 group proteins which were in different clusters according to phylogenetic tree. This may be an explanation for location of cucumber LEA2 group genes in different clusters. All LEA4 group proteins (CsLEA-15, CsLEA-26, CsLEA-53 and CsLEA-76) included only motif 6. Besides, motif 6 was present in LEA1 group proteins (CsLEA-28 and CsLEA-38), too. LEA4 and LEA1 group proteins were similar because of the presence of the same motif. Some members of LEA3 group proteins have no motifs but only CsLEA-29 contained motif 8 in this group. Motif 13 was shared by all members of SMP group proteins (CsLEA-7, CsLEA-70, CsLEA-72). Motif analysis of LEA proteins indicated the fact that CsLEA proteins in the same LEA group shared same motifs. It can be concluded that the presence of special motifs in LEA proteins may determine LEA group formation. Motif conservation in clusters may be suggested that LEA proteins have different functions which are cluster specific. Besides, LEA genes sharing same motifs in the same cluster may be raised by gene expansion. Variability between different clades according to motif analysis may be attributed to evolution of clades from different ancestors.
Determination of LEA gene orthologs between cucumber and other species
Physically mapped CsLEA genes on cucumber chromosomes were matched with LEA genes on chromosomes of poplar, Arabidopsis and maize (Fig. S3). An average of 32 % orthologous relationships of 79 CsLEA genes were observed with these species. Orthologous relationships of CsLEA genes with poplar, Arabidopsis, maize and rice have a range of 81, 33, 10 and 4 %, respectively. Maximum orthologous relationship of CsLEA genes were considered with poplar. These results were consistent with the study of Baloglu et al. (2014) who found that cucumber bZIP transcription factors showed maximum orthology with poplar genome. In addition, these observations may show the role of chromosomal rearrangements on organization of cucumber, poplar, Arabidopsis, maize and rice genomes. Moreover, evolution of LEA genes can be revealed by genetic comparison maps between cucumber and other related organisms. Besides, this can be helpful for isolation of orthologous LEA genes from cucumber by using this genetic map information of other related plants for genetic expansion.
Gene ontology analysis
Cellular localization, biological process and molecular function of CsLEA genes were determined by the GO slim analysis using Blast2GO software. Analysis revealed that response to stimulus, stress or abiotic stress were the most abundant functions as biological processes which were consistent with the roles of LEA proteins in a cell. In addition, prediction of CsLEA proteins’ function indicated that they had mainly binding capacity and this can be attributed to their roles such as enzyme protector activity (Goyal et al. 2005; Reyes et al. 2008) in water deficiency conditions or membrane stabilization activity to protect membranes in freezing temperatures or drought conditions (Kosová et al. 2007; Tolleter et al. 2010). CsLEA proteins commonly localized in the cell or on the plasma membrane according to cellular roles for membrane maintenance (Wolkers et al. 2001; Liu et al. 2010) (Fig. 3; Table S3). Besides, they can accumulate in the subcellular parts of the cell such as cytoplasm, organelles and nucleus in cucumber cells. LEA protein localization was showed especially in cytoplasmic or nuclear regions in tomato (Cao and Li 2015) whereas; localization of this protein family was distributed in diverse subcellular regions including mitochondria and chloroplast in purple false brome (Filiz et al. 2013). In addition, previous analysis revealed that LEA2 group proteins localized in cytoplasm, nucleus or other cell regions like mitochondria (Battaliga et al. 2008). These studies are consistent with our findings and this can be suggested that LEA proteins locate in different subcellular regions.
Gene ontology analysis of CsLEA proteins in three categories (Biological processes, molecular functions and cellular components) using Blast2Go program. Different colors which are indicated near the graphics show different molecular functions, biological roles and cellular localizations of CsLEA proteins. (Color figure online)
Gene duplications and divergence rates of the CsLEA proteins
Gene duplication events may provide evolution of new functions for proteins (Kondrashov et al. 2002). In the present study, non-synonymous (Ka) versus synonymous (Ks) substitution rates (Ka/Ks) were calculated for 56 tandem duplicated CsLEA genes to evaluate evolutionary radiation of this gene family. Also these substitution rates estimated for orthologous genes of CsLEA from Arabidopsis (26- pairs), poplar (64-pairs), maize (8-pairs) and rice (3-pairs) (Fig. 4). Ka/Ks ratios were between 0.008 and 0.436 and the average was 0.14 for tandem duplication. The Ka/Ks ratios of CsLEA genes were <1 and this can be attributed to the strong purifying pressure occurred for these family members in cucumber. These duplication events can be suggested that recent time of divergence ranged between approximately 4 and 398 million years ago (MYA). Moreover, mean average of Ka/Ks ratios between cucumber and poplar, Arabidopsis, maize and rice were 0.08, 0.06, 0.03 and 0.01, respectively when surveyed orthologous gene pairs. Considering divergence times between CsLEA genes and these of other plant species, the earliest diverged genes from CsLEA genes were with maize genes with the average of 245 MYA. This rate was followed by rice, Arabidopsis and poplar with the average divergence rates of 204, 176 and 78 MYA, respectively (Table S2, Table S5). From these results, cucumber and poplar may be exposed to a largely purifying selection. In addition, cucumber and poplar LEA genes may be closer evolutionary than other studied plants. Segmental and tandem duplication events enable gene family expansions (Cao et al. 2012; Chen and Cao 2014). Exploration of genome wide studies about LEA genes revealed that a total of 22 genes in tomato (Cao and Li, 2015), eight genes in potato (Charfeddine et al. 2015), eight genes in purple false brome (Filiz et al. 2013), 17 genes in Chinese plum (Du et al. 2013) and 42 genes in Arabidopsis (Hundertmark and Hincha 2008) showed tandem and segmental duplication events. This can be attributed to the LEA gene family expansion between different plant species.
Determination of miRNAs CsLEA target genes
miRNAs are one of the methods used to inhibit target gene expression in abiotic and biotic stress conditions in plants through cleavage of the target transcripts (Bartel 2004; Jones-Rhoades et al. 2006; Ambros and Chen 2007).
psRNA Target database was employed to determine targeting CsLEA transcripts using default parameters according to the scoring schema of miRU (Zhang 2005) to score the complementarity between small RNA and their target transcript. The maximum expectation which is the threshold of the score was setted as 3.0. Also the accessibility of mRNA target site to small RNA has been identified as one of the important factors. They are involved in target recognition and the database enables to calculate target accessibility, which is represented by the energy required to open secondary structures around target site of target mRNA. The less energy means the more possibility that small RNA is able to contact target mRNA. The UPE parameter represents the energy that is required to open secondary structures around target site.
In the present study, 50 different miRNAs were determined for target genes by using psRNATarget: A Plant Small RNA Target Analysis Server. A total of 37 CsLEA genes (CsLEA01-03-04-06-09-12-14-15-16-18-21-22-23-25-26-27-32-33-34-37-44-49-50-51-52-53-54-55-57-58-62-65-67-71-72-77-79) targeted by miRNA’s were summarized in Table S4. Among cucumber LEA genes, CsLEA-06 and CsLEA-37 genes were the most targeted genes by miRNAs. Besides, mir854 and mir414 are most abundant miRNAs. According to Go analysis in Arabidopsis, targets of mir414 were especially transcriptional regulators including bZIP, WRKY, MYB, B3, heat shock proteins and TCP which are important in plant growth, development and defense mechanisms (Eulgem et al. 2000; Gurley 2000; Jakoby et al. 2002; Suo et al. 2003; Guo et al. 2007a, 2007b; Romanel et al. 2009; Guleria and Yadav 2011). Furthermore, studies revealed that up-regulation of mir854 in drought stressed rice but mechanism of this role is unclear (Zhou et al. 2010). mir854 has target regions in the 3’-untranslated region of oligouridylate-binding protein1b (UBP1b) which is coding a nuclear RNA-binding protein contributed to the regulation of pre-mRNA maturation (Lambermon et al. 2000). mir854 family which is a regulator element of transcriptional mechanisms is included by both animals and plants (Arteaga-Va´ zquez et al. 2006). Identification of CsLEA genes targeted by these miRNAs may facilitate the understanding of their roles in cucumber. On the other hand, this study is the first study determining LEA proteins targeted by miRNAs. Discovery of miRNA structure may allow determination of miRNAs which are specific to each tissue.
Homology modeling of cucumber LEA proteins
BLASTP search was implemented for homology modeling in Plant Data Bank (PDB). Hidden Markov Model (HMM) search was selected using detection rate for determination of homology model in Phyre2 database which is available on the web to predict and analyze protein structure, function and mutations (Söding 2005). A total of 6 CsLEA proteins [CsLEA-9 (PDB ID: 1yyc), CsLEA-14 (PDB ID: 1xo8), CsLEA-18 (PDB ID: 1yyc), CsLEA-24 (PDB ID: 1xo8), CsLEA-52 (PDB ID: 1yyc), CsLEA-76 (PDB ID: 1xq8)] showed high homology and 75–90 % of residues modelled at >90 % confidence interval under intensive mode (Fig. 5). According to secondary structures of 6 CsLEA protein models, β sheets were predominant except CsLEA76. CsLEA76 protein had only α helix structures. A previous study reported that LEA2 group proteins had a structure including especially one α-helix and two antiparallel β-sheets. This is consistent with our results with the predominant β sheets and one or two α-helixes in predicted CsLEA protein structures. This structure may have a role in fluid loss to reduce the effects of stress or injuries on plant tissues (Singh et al. 2005; Li and Cao 2016). These predicted protein structures may facilitate understanding of molecular functions of LEA proteins.
Predicated three dimensional configurations of CsLEA proteins. The configuration of 6 CsLEA proteins [CsLEA-9 (PDB ID: 1yyc), CsLEA-14 (PDB ID: 1xo8), CsLEA-18 (PDB ID: 1yyc), CsLEA-24 (PDB ID: 1xo8), CsLEA-52 (PDB ID: 1yyc), CsLEA-76 (PDB ID: 1xq8)] which are modelled at >90 % confidence interval are distributed
Tissue specific expression profiles and expression analysis of LEA genes under drought stress
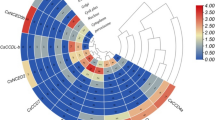
A RNA seq approach was implemented using Sequence Read Archive (SRA) data to explore tissue specific gene expression profiles of LEA genes in cucumber. Hierarchically ranked heat map, which includes expression patterns of CsLEA genes in ovary (unexpanded, fertilized and unfertilized), leaf, root, tendril, tendril base parts, stem, male flower and female flower tissues, was shown in Fig. S4. According to this map, CsLEA-09, CsLEA-27, CsLEA-40, CsLEA-54, CsLEA-61, CsLEA-71, CsLEA-73, CsLEA-77 and CsLEA-79 genes exhibited very high expression profiles in all studied tissues. Expression of approximately all LEA genes were observed in root tissues, in contrast, most of the CsLEA genes had no expression in leaf tissues. In addition, CsLEA-17 and CsLEA-56 gene expressions were not seen in any tissues. CsLEA-18, CsLEA-20, CsLEA-30, CsLEA-35, CsLEA-36, CsLEA-46, CsLEA-47, CsLEA-48 and CsLEA-52 genes indicated an expression profile in one or two tissues.
Additionally, qRT-PCR was implemented to explore drought responsive LEA gene family members in cucumber. For this purpose, expression profiles of ten CsLEA genes (CsLEA-09, CsLEA-32, CsLEA-37, CsLEA-49, CsLEA-54, CsLEA-57, CsLEA-71, CsLEA-73, CsLEA-76 and CsLEA-77) were evaluated using leaf and root tissues of cucumber under drought stress conditions. Transcriptome data leaded to selection of these genes. According to this data, LEA genes which have high expression levels in all tissues were selected. Moreover, LEA genes were determined between genes which are highly expressed in only leaf or in root tissues and their expression levels were studied (Fig. 6). Among these genes, expression level of CsLEA-54 gene increased in all studied hours (3rd h, 12th h and 24th h) after drought stress application in leaf tissues. This gene may be highly expressed under drought stress and further studies required to evaluate its role under water limitation conditions. In addition, CsLEA-09, CsLEA-32 and CsLEA-57 genes responded to drought by increase in expression level at the 3rd h similar to expression pattern of CsLEA-54. These genes may be early response genes to water limitation among cucumber LEA genes.
Although CsLEA-37, CsLEA-49, CsLEA-71, CsLEA-76 and CsLEA-77 genes showed an increase pattern at 3rd h, their gene expression levels were maximum at 12th h. These genes may be response genes to drought stress after the 3rd h. CsLEA-09, CsLEA-73, CsLEA-76, CsLEA-77 genes were first response genes to drought in root tissues. Besides, gene expression levels of CsLEA-32, CsLEA-37, CsLEA-49, CsLEA-54, CsLEA-57 and CsLEA-71 genes increased at 24th h in root tissue. These genes may be considered as a secondary response genes to water deficiency.
Expression levels of all selected genes raised at 3rd h or 24th h in root or leaf tissue. These genes can be thought to have a role in drought response together with other transcription factors and stress genes. Between these genes, CsLEA-09, CsLEA-32, CsLEA-54, CsLEA-57 genes showed a similar pattern in leaf and root tissues and upregulated in leaf than in root tissues. When compared expression levels of these genes in leaf and root tissues, CsLEA-37, CsLEA-49, CsLEA-76 and CsLEA-77 genes were late response genes.
Transcriptome data and real time PCR results were convenient together. However, CsLEA-32, CsLEA-37 and CsLEA-57 genes showed expression only in root tissue in transcriptome data while their expression were observed both in root and leaf tissues in real time PCR results. Application of drought stress condition and the cultivar of the cucumber plant may be the reason of these differences.
According to orthologous gene analysis from Arabidopsis named as #4, #14 and #10, LEA genes had orthologous relations with CsLEA54 gene whose expression level upregulated in leaf tissues under drought conditions. Also #4 (AT1G20440.1) and #10 (AT1G76180.1) Arabidopsis LEA genes were induced under different stress conditions especially, under cold stress. Besides, CsLEA73 gene expression augmented by water limitation in cucumber and Arabidopsis LEA #41 gene expression, which is the orthologous of CsLEA73 gene, was upregulated by drought stress, too. In addition, expression level of SAG gene (senescence-associated gene) 21 (#38) from Arabidopsis, which is another gene ortholog of CsLEA73 gene, raised under cold, salt and drought stress conditions (Hundertmark and Hincha 2008; Weaver et al. 1998). These findings are relevant with our results and suggest that these proteins may have essential roles under drought stress conditions in these plants. Furthermore, five members of LEA genes among studied eight ones in tomato were upregulated when LEA gene expression patterns explored under salt, water deficiency or cold conditions (Cao and Li 2015). In addition, gene expression levels of some LEA proteins in potato (StDHN2a, TAS14 and StLEA27) were expressed only in water deficiency and/or salt conditions while StDHN1 (YSK2) and StDHN25 (SK3) genes were expressed in all tissues which may explain the roles of these proteins in the normal plant development stages (Charfeddine et al. 2015). Our findings are consistent with previous studies revealing functional roles of LEA gene family under abiotic stress conditions. This protein family may provide a protection for embryonal structures under drought stress conditions (Hanin et al. 2011; Du et al. 2013; Lan et al. 2013)
This study reports a detailed genome wide identification of LEA genes in cucumber genome. Physiological characteristics of CsLEA proteins and their systematic positions were evaluated in this study. Our findings suggest that LEA genes may be potential response genes under the water limitation conditions. Further detailed studies should be done for understanding of this important gene family function in cucumber.
References
Ambros V, Chen X (2007) The regulation of genes and genomes by small RNAs. Development 134(9):1635–1641
Arteaga-Va´zquez M, Caballero-Pe´rez J, Vielle-Calzada JP (2006) A family of microRNAs present in plants and animals. Plant Cell 18:3355–3369
Bailey TL, Elkan C (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. In: Proceedings of the second international conference on intelligent systems for molecular biology, AAAI Press, Menlo Park, California, pp 28–36
Baloglu MC (2014) Genome-wide in silico identification and comparison of Growth Regulating Factor (GRF) genes in Cucurbitaceae family. Plant Omics J 7:260–270
Baloglu MC, Eldem V, Hajyzadeh M, Unver T (2014) Genome-wide analysis of the bZIP transcription factors in cucumber. PLoS One 9(4):e96014. doi:10.1371/journal.pone.0096014
Baloglu MC, Ulu F, Altunoglu YC, Pekol S, Alagoz G, Ese O (2015) Identification, molecular characterization and expression analysis of RPL24 genes in three Cucurbitaceae family members: cucumber, melon and watermelon. Biotechnol Biotechnol Equip 26(6):1024–1034
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116(2):281–297
Battaliga M, Olvera-Carrillo Y, Garciarrubio A, Campos F, Covarrubias AA (2008) The enigmatic LEA proteins and other hydrophilins. Plant Physiol 148:6–24
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The protein data bank. Nucleic Acids Res 28:235–242
Boswell LC, Menze MA, Hand SC (2009) Multiple isoforms of late embryogenesis abundant proteins in Artemia franciscana embryos. Integr Comp Biol 49:E203
Bray EA (1993) Molecular responses to water deficit. Plant Physiol 103:1035–1040
Cai X, Ye J, Hu T, Zhang Y, Ye Z, Li H (2014) Genome-wide classification and expression analysis of nucleobase–ascorbate transporter (NAT) gene family in tomato. Plant Growth Regul 73(1):19–30
Cao J, Li X (2015) Identification and phylogenetic analysis of late embryogenesis abundant proteins family in tomato (Solanum lycopersicum). Planta 241(3):757–772
Cao J, Shi F, Liu X, Huang G, Zhou M (2012) Phylogenetic analysis and evolution of aromatic amino acid hydroxylase. FEBS Lett 584:4775–4782
Caraux G, Pinloche S (2005) PermutMatrix: a graphical environment to arrange gene expression profiles in optimal linear order. Bioinform Appl Note 21(7):1280–1281
Charfeddine S, Saïdi MN, Charfeddine M, Gargouri-Bouzid R (2015) Genome-wide identification and expression profiling of the late embryogenesis abundant genes in potato with emphasis on dehydrins. Mol Biol Rep 42(7):1163–1174
Chen Y, Cao J (2014) Comparative genomic analysis of the Sm gene family in rice and maize. Gene 539:238–249
Conesa A, Götz S (2008) Blast2GO: a comprehensive suite for functional analysis in plant genomics. Int J Plant Genom. doi:10.1155/2008/619832
Dalal M, Tayal D, Chinnusamy V, Bansal KC (2009) Abiotic stress and ABA-inducible group 4 LEA from Brassica napus plays a key role in salt and drought tolerance. J Biotechnol 139(2):137–145. doi:10.1016/j.jbiotec.2008.09.014
Denekamp NY, Reinhardt R, Kube M, Lubzens E (2010) Late Embryogenesis Abundant (LEA) proteins in nondesiccated, encysted, and diapausing embryos of rotifers. Biol Reprod 82(4):714–724. doi:10.1095/biolreprod.109.081091
Du D, Zhang Q, Cheng T et al (2013) Genome-wide identification and analysis of late embryogenesis abundant (LEA) genes in Prunus mume. Mol Biol Rep 40(2):1937–1946. doi:10.1007/s11033-012-2250-3
Dure LI, Galau GA (1981) Developmental biochemistry of cottonseed embryogenesis and germination XIII. Regulation of biosynthesis of principal storage proteins. Plant Physiol 68(1):187–194
Dure L, Crouch M, Harada J, Ho THD, Mundy J, Quatrano R, Thomas T, Sung ZR (1989) Common amino acid sequence domains among the LEA proteins of higher plants. Plant Mol Biol 12(5):475–486
Eichinger L, Pachebat JA, Glockner G, Rajandream MA, Sucgang R, Berriman M (2005) The genome of the social amoeba Dictyostelium discoideum. Nature 435:43–57
Eulgem T, Rushton PJ, Robatzek S, Somssich IE (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5(5):199–206
Filiz E, Ozyigit II, Tombuloglu H, Koc I (2013) In silico comparative analysis of LEA (Late Embryogenesis Abundant) proteins in Brachypodium distachyon L. Plant Omics 6(6):433
Gal TZ, Glazer I, Koltai H (2004) An LEA group 3 family member is involved in survival of C. elegans during exposure to stress. FEBS Lett 577:21–26
George S, Usha B, Parida A (2009) Isolation and characterization of an atypical LEA protein coding cDNA and its promoter from drought-tolerant plant Prosopis juliflora. Appl Biochem Biotechnol 157(2):244–253. doi:10.1007/s12010-008-8398-6
Goodstein DM, Shu S, Howson R et al (2012) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40:D1178–D1186. doi:10.1093/nar/gkr944
Goyal K, Walton LJ, Tunnacliffe A (2005) LEA proteins prevent protein aggregation due to water stress. Biochem J 388:151–157
Grelet J, Benamar A, Teyssier E et al (2005) Identification in pea seed mitochondria of a late-embryogenesis abundant protein able to protect enzymes from drying. Plant Physiol 137:157–167
Guleria P, Yadav SK (2011) Identification of miR414 and expression analysis of conserved miRNAs from Stevia rebaudiana. Genom Proteom Bioinform 9(6):211–217
Guo AY, Zhu QH, Chen X, Luo JC (2007a) GSDS: a gene structure display server. Yi chuan Hereditas/Zhongguo yi chuan xue hui bian ji 29(8):1023–1026
Guo Q, Xiang A, Yang Q, Yang Z (2007b) Bioinformatic identification of microRNAs and their target genes from Solanum tuberosum expressed sequence tags. Chin Sci Bull 52(17):2380–2389
Gurley WB (2000) HSP101: a key component for the acquisition of thermotolerance in plants. Plant Cell 12(4):457–460
Hanin M, Brini F, Ebel C, Toda Y, Takeda S, Masmoudi K (2011) Plant dehydrins and stress tolerance. Plant Signal Behav 6:1503–1509
Hoagland DR, Arnon DI (1950) The water-culture method for growing plants without soil. Calif Agric Exp Stn Circ 347:1–32
Huang S, Li R, Zhang Z et al (2009) The genome of the cucumber Cucumis sativus L. Nat Genet 41(12):1275–1281
Hunault G, Jaspard E (2010) LEAPdb: a database for the late embryogenesis abundant proteins. BMC Genom 11:221. doi:10.1186/1471-2164-11-221
Hundertmark M, Hincha DK (2008) LEA (Late Embryogenesis Abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genom. doi:10.1186/1471-2164-9-118
Jakoby M, Weisshaar B, Dröge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F (2002) bZIP transcription factors in Arabidopsis. Trends Plant Sci 7(3):106–111
Jimenez AJ, Alonso-Ramirez A, Nicolas C (2008) Two cDNA clones (FsDhn1 and FsClo1) up-regulated by ABA are involved in drought responses in Fasus sylvatica L seeds. J Plant Physiol 165(17):1798–1807. doi:10.1016/j.jplph.2007.11.013
Jones-Rhoades MW, Bartel DP, Bartel B (2006) MicroRNAs and their regulatory roles in plants. Annu Rev Plant Biol 57:19–53. doi:10.1146/annurev.arplant.57.032905.105218
Kelley LA, Sternberg MJE (2009) Protein structure prediction on the web: a case study using the Phyre server. Nat Protoc 4:363–371
Kikawada T, Nakahara H, Kanamori Y, Iwata K, Watanabe M, McGee B, Tunnacli A, Okuda T (2006) Dehydration induced expression of LEA proteins in an anhydrobiotic chironomid. Biochem Biophys Res Commun 348:56–61
Kondrashov FA, Rogozin IB, Wolf YI, Koonin EV (2002) Selection in the evolution of gene duplications. Genome Biol 3(2):research0008.1–research0008.9
Kosová K, Vítámvás P, Prášil IT (2007) The role of dehydrins in plant response to cold. Biol Plant 51(4):601–617
Lambermon MH, Simpson GG, Wieczorek Kirk DA, Hemmings- Mieszczak M, Klahre U, Filipowicz W (2000) UBP1, a novel hnRNP-like protein that functions at multiple steps of higher plant nuclear pre-mRNA maturation. EMBO J 19:1638–1649
Lan T, Gao J, Zeng QY (2013) Genome-wide analysis of the LEA (late embryogenesis abundant) protein gene family in Populus trichocarpa. Tree Genet Genomes 9(1):253–264
Letunic I, Doerks T, Bork P (2012) SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res 40(D1):D302–D305. doi:10.1093/nar/gkr931
Li X, Cao J (2016) Late Embryogenesis Abundant (LEA) Gene Family in Maize: identification, evolution, and expression profiles. Plant Mol Biol Report 34(1):15–28
Li L, Xu HL, Yang XL, Li YX, Hu YK (2011) Genome-wide identification, classification and expression analysis of LEA gene family in soybean. Sci Agric Sin 44(19):3945–3954
Li Q, Zhang C, Li J, Wang L, Ren Z (2012) Genome-wide identification and characterization of R2R3MYB family in Cucumis sativus. Plos One 7(10):e47576
Ling J, Jiang W, Zhang Y et al (2011) Genome-wide analysis of WRKY gene family in Cucumis sativus. BMC Genom 12(1):471
Liu Y, Zheng Y, Zhang Y, Wang W, Li R (2010) Soybean PM2 protein (LEA3) confers the tolerance of Escherichia coli and stabilization of enzyme activity under diverse stresses. Curr Microbiol 60:373–378
Liu Y, Jiang H, Chen W, Qian Y, Ma Q, Cheng B, Zhu S (2011) Genome-wide analysis of the auxin response factor (ARF) gene family in maize (Zea mays). Plant Growth Regul 63(3):225–234
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25(4):402–408
Lough TJ, Lucas WJ (2006) Integrative plant biology: role of phloem long-distance macromolecular trafficking. Annu Rev Plant Biol 57:203–232
Lv J, Rao J, Johnson F, Shin S, Zhu Y (2015) Genome-wide identification of jasmonate biosynthetic genes and characterization of their expression profiles during apple (Malus × domestica) fruit maturation. Plant Growth Regul 75(1):355–364
Lynch M, Conery JS (2000) The evolutionary fate and consequences of duplicate genes. Science 290:1151–1155. doi:10.1126/science.290.5494.1151
Marunde MR, Samarajeewa DA, Anderson J, Li S, Hand SC, Menze MA (2013) Improved tolerance to salt and water stress in Drosophila melanogaster cells conferred by late embryogenesis abundant protein. J Insect Physiol 59(4):377–386
Mehan MR, Freimer NB, Ophoff RA (2004) A genome-wide survey of segmental duplications that mediate common human genetic variation of chromosomal architecture. Hum Genom 1(5):335–344
Olvera-Carrillo Y, Campos F, Luis Reyes J, Garciarrubio A, Covarrubias AA (2010) Functional analysis of the group 4 late embryogenesis abundant proteins reveals their relevance in the adaptive response during water deficit in Arabidopsis. Plant Physiol 154(1):373–390. doi:10.1104/pp.110.158964
Oztur ZN, Talame V, Deyholos M, Michalowski CB, Galbraith DW, Gozukirmizi N, Tuberosa R, Bohnert HJ (2002) Monitoring large-scale changes in transcript abundance in drought- and salt stressed barley. Plant Mol Biol 48(5–6):551–573. doi:10.1023/a:1014875215580
Park SC, Kim YH, Jeong JC, Kim CY, Lee HS, Bang JW, Kwak SS (2011) Sweetpotato late embryogenesis abundant 14 (IbLEA14) gene influences lignification and increases osmotic and salt stress-tolerance of transgenic calli. Planta 233(3):621–634. doi:10.1007/s00425-010-1326-3
Quevillon E, Silventoinen V, Pillai S et al (2005) InterProScan: protein domains identifier. Nucleic Acids Res 33:W116–W120. doi:10.1093/nar/gki442
Reyes JL, Campos F, Wei H, Arora R, Yang Y, Karlson DT, Covarrubias AA (2008) Functional dissection of hydrophilins during in vitro freeze protection. Plant Cell Environ 31:1781–1790
Romanel EA, Schrago CG, Couñago RM, Russo CA, Alves-Ferreira M (2009) Evolution of the B3 DNA binding superfamily: new insights into REM family gene diversification. PLoS One 4(6):e5791
Rorat T (2006) Plant dehydrins—tissue location, structure and function. Cell Mol Biol Lett 11:536–556
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Serrano R, Montesinos C (2003) Molecular bases of desiccation tolerance in plant cells and potential applications in food dehydration. Food Sci Technol Int 9:157–161
Shao HB, Liang ZS, Shao MA (2005) LEA proteins in higher plants: structure, function, gene expression and regulation. Colloid Surf B 45:131–135
Shiu SH, Bleecker AB (2003) Expansion of the receptor-like kinase/pelle gene family and receptor-like proteins in Arabidopsis. Plant Physiol 132:530–543. doi:10.1104/pp.103.021964
Singh S, Cornilescu CC, Tyler RC, Cornilescu G, Tonelli M, Lee MS, Markley JL (2005) Solution structure of a late embryogenesis abundant protein (LEA14) from Arabidopsis thaliana, a cellular stressrelated protein. Protein Sci 14(10):2601–2609
Söding J (2005) Protein homology detection by HMM-HMM comparison. Bioinformatics 21:951–960
Suo J, Liang X, Pu L, Zhang Y, Xue Y (2003) Identification of GhMYB109 encoding a R2R3 MYB transcription factor that expressed specifically in fiber initials and elongating fibers of cotton (Gossypium hirsutum L.). Biochim Biophys Acta Gene Struct Expr 1630(1):25–34
Suyama M, Torrents D, Bork P (2006) PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res 34:W609–W612. doi:10.1093/nar/gkl315
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30(12):2725–2729. doi:10.1093/molbev/mst197
Tang H, Bowers JE, Wang X et al (2008) Synteny and collinearity in plant genomes. Science 320:486–488. doi:10.1126/science.1153917
Tanurdzic M, Banks JA (2004) Sex-determining mechanisms in land plants. Plant Cell 16:S61–S71
Thompson JD, Gibson TJ, Plewniak F et al (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882. doi:10.1093/nar/25.24.4876
Tolleter D, Hincha DK, Macherel D (2010) A mitochondrial late embryogenesis abundant protein stabilizes model membranes in the dry state. Biochim Biophys Acta 1798:1926–1933
Tunnacliffe A, Hincha DK, Leprince O, Macherel D (2010) LEA Proteins: versatility of form and function. In: Lubzens E, Cerda J, Clark M (eds) Sleeping beauties—dormancy and resistance in harsh environments. Springer, Berlin, pp 91–108
Umezawa T, Fujita M, Fujita Y, Yamaguchi-Shinozaki K, Shinozaki K (2006) Engineering drought tolerance in plants: discovering and tailoring genes unlock the future. Curr Opin Biotechnol 17:113–122
Unver T, Budak H (2009) Conserved microRNAs and their targets in model grass species Brachypodium distachyon. Planta (ISI) 230(4):659–669
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78
Wang XS, Zhu HB, Jin GL, Liu HL, Wu WR, Zhu J (2007) Genome-scale identification and analysis of LEA genes in rice (Oryza sativa L.). Plant Sci 172(2):414–420
Wang L, Li X, Chen S, Liu G (2009) Enhanced drought tolerance in transgenic Leymus chinensis plants with constitutively expressed wheat TaLEA (3). Biotechnol Lett 31(2):313–319. doi:10.1007/s10529-008-9864-5
Weaver LM, Gan S, Quirino B, Amasino RM (1998) A comparison of the expression patterns of several senescence-associated genes in response to stress and hormone treatment. Plant Mol Biol 37:455–469
Wen X, Niu T, Kong X (2014) In silico analysis of PHB gene family in maize. Plant Growth Regul 73(2):181–191
Wise MJ (2003) LEA ping to conclusions: a computational reanalysis of late embryogenesis abundant proteins and their possible roles. BMC Bioinform 4:52
Wolkers WF, McCready S, Brandt WF, Lindsey GG, Hoekstra FA (2001) Isolation and characterization of a D-7 LEA protein from pollen that stabilizes glasses in vitro. Biochim Biophys Acta 1544:196–206
Xoconostle Cázares B, Xiang Y, Ruiz-Medrano R, Wang HL, Monzer J, Yoo BC, McFarland KC, Lucas WJ (1999) Plant paralog to viral movement protein that potentiates transport of mRNA into the phloem. Science 283:94–98
Yang Z, Gu S, Wang X et al (2008) Molecular evolution of the cpp-like gene family in plants: insights from comparative genomics of Arabidopsis and rice. J Mol Evol 67:266–277. doi:10.1007/s00239-008-9143-z
Yu Y, Xia X, Yin W, Zhang H (2007) Comparative genomic analysis of CIPK gene family in Arabidopsis and Populus. Plant Growth Regul 52(2):101–110
Zhang Y (2005) miRU: an automated plant miRNA target prediction server. Nucleic Acids Res 33:W701–W704
Zhao P, Liu F, Ma M, Gong J, Wang Q, Jia P, Zheng G, Liu H (2011) Overexpression of AtLEA3-3 confers resistance to cold stress in Escherichia coli and provides enhanced osmotic stress tolerance and ABA sensitivity in Arabidopsis thaliana. Mol Biol 45(5):785–796. doi:10.1134/s0026893311050165
Zhou L, Liu Y, Liu Z, Kong D, Duan M, Luo L (2010) Genome-wide identification and analysis of drought-responsive microRNAs in Oryza sativa. J Exp Bot 61(15):4157–4168
Acknowledgments
This work was financially supported by The Scientific and Technological Research Council of Turkey (TUBITAK) with grant number 114O761.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1
Exon-intron organization of cucumber CsLEA genes. LEA gene family members in cucumber are classified according to Fig. 2 Clusters. Exons are showed as yellow boxes and introns are grey lines (JPEG 215 kb)
Fig. S2
Presentation of motif clades for the CsLEA proteins. Fifteen different MEME motifs are distributed as different colors (JPEG 54 kb)
Fig. S3
Comparative physical mapping indicated high degree of orthologous relationships of LEA genes located on cucumber chromosomes with poplar, Arabidopsis and maize. Orthologous genes between cucumber and poplar, Arabidopsis and maize are presented by red lines (JPEG 322 kb)
Fig. S4
Heatmap distribution of CsLEA genes, which showing expression patterns of these gene family members in the 10 different tissues and organ systems of cucumber using transcriptome data. Expression values indicated as color gradient from no expression (green) to high expression (red) levels (TIFF 3108 kb)
Table S4
miRNA targets identified by psRNATarget (XLSX 14 kb)
Rights and permissions
About this article
Cite this article
Celik Altunoglu, Y., Baloglu, P., Yer, E.N. et al. Identification and expression analysis of LEA gene family members in cucumber genome. Plant Growth Regul 80, 225–241 (2016). https://doi.org/10.1007/s10725-016-0160-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10725-016-0160-4