Abstract
Ascorbic acid or vitamin C is a wide spectrum antioxidant and plays a crucial role in a many metal-containing enzymes essential for humans, which are unable to synthesize the vitamin C and must obtain it from dietary sources. Ascorbic acid is transported by sodium-coupled ascorbic acid transporters or SVCTs in humans. However, little information is available about the nucleobase–ascorbate transporters (NATs) in tomato (Solanum lycopersicum). In the current study, we identified 12 NAT genes by screening SGN genome databases in tomato. A complete overview of this gene family in tomato is presented, including gene structures, chromosome distribution and localization, phylogenies, motif analysis and expression profiles. The SlNAT genes contained 14 exons, mostly, and dispersed on all the chromosomes except chromosome 8 and 9. All the SlNATs were located to plasma membrane, chloroplast thylakoid membrane, Golgi body, and endoplasmic reticulum (membrane). The phylogenetic tree showed that the plant NATs were divided into 4 clades, well-supported by the distribution of conserved motifs, and the SlNAT proteins shared higher similarity and clustered more closely with AtNAT proteins. Furthermore, the expression profiles of SlNAT genes in various organs showed 9 out of 12 SlNAT genes were constituently expression with differential expression levels under normal growth conditions. Our systematic analysis will provide a useful platform for molecular clone and functional identification of NAT genes in tomato and probably other Solanaceae plants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
l-Ascorbic acid (AsA), the reduced form of vitamin C, serves as an important essential antioxidant for many biological systems, is involved in the synthesis of collagen and carnitine, iron utilization, and immune cell development. AsA must be obtained via the diet by humans, primates and a few other animals due to the unable to synthesize AsA endogenously. Fresh fruits and vegetables, found in especially high amounts of AsA levels, are the main dietary source for humans. AsA is also an essential to plants as it is to animals. In plants, AsA functions as a major redox buffer play a critical role in responses to abiotic stresses, and as a cofactor for enzymes involved in regulating photosynthesis, hormone biosynthesis, and regenerating other antioxidants. AsA regulates cell cycle, through cell expansion and senescence, and is involved in signal transduction (Gallie 2013; Szarka et al. 2012).
In contrast to the single pathway synthesized AsA in animals, as many as four AsA biosynthetic pathways are present in plants (Wheeler et al. 1998; Agius et al. 2003; Wolucka and Van Montagu 2003; Lorence et al. 2004). Among those, the d-mannose/l-galactose pathway, or the Smirnoff-Wheeler pathway, is responsible for the accumulation of AsA synthesis (Wheeler et al. 1998). Although the initial steps of the pathway are located in the cytosol, AsA is synthesized on the inner membrane of the mitochondria (Bartoli et al. 2010), from where it is distributed to all the intracellular compartments and to the apoplast. AsA was detected in the nuclei and the cytosol of Arabidopsis thaliana and tobacco with highest concentration by using high-resolution immuno electron microscopy in leaf cells, whereas mitochondria, plastids and vacuoles were displayed the lowest ascorbate levels among the organelles. Dictyosomes, the cell walls or intercellular space of mesophyll cells was not detected AsA presence (Zechmann et al. 2011).
Long-distance phloem-mediated transport of AsA was demonstrated with feeding analysis. Feeding source leaves with the precursor to AsA resulted in an increase in AsA content in the treated leaf and in sink tissues in Arabidopsis, potato and tomato (Tedone et al. 2004; Franceschi and Tarlyn 2002; Badejo et al. 2012). Using radio-labeled AsA to leaves also demonstrated the AsA was accumulated in the phloem and transported to root tips, shoots, and floral organs, but not to mature leaves (Franceschi and Tarlyn 2002). All of those strongly suggested that ascorbate must be transported through the cytosol into other cell compartments and through source to sink. AsA is generally not considered to diffuse through lipid bilayers because of its negatively charged form at physiological pH values (Gallie 2013; Asard et al. 1992).
In humans, Na+-dependent vitamin C transporters SVCT1 (SLC23A1) and SVCT2 (SLC23A2) are well characterized and specificly transported for AsA. The SLC23 family belongs to the nucleobase–ascorbate transporter (NAT) family, also called the Nucleobase–Cation Symporter-2 (NCS2) family, which involved in uptake of nucleobases and identified in bacteria, archea, diatoms, fungi, plants, and animals (Burzle et al. 2013; de Koning and Diallinas 2000). Nevertheless, in plants, only one NAT protein, ZmLPE1 (leaf permease1 from maize), has been characterized by functional complementation of a purine transport-deficient A. nidulans strain (Argyrou et al. 2001), and 12 putative NAT genes are screened in Arabidopsis thaliana genome (Maurino et al. 2006).
Tomato fruits are considered a major dietary source of vitamin C in many countries, because it is consumed regularly and in large quantities. Tomato also offers a good model for other crops species whose fruit is also a fleshy berry. Previous reports have mainly focus on biosynthesis and metabolism pathway to enhance AsA contents in tomato, but the transportation of AsA from sours to sink or from the cytosol into other cell compartments remain vague.
In this study, we performed a genome-wide identification of NAT proteins in tomato and revealed that the tomato genome contained a total of 12 NAT members, named as SlNAT 1–12. The gene structure, chromosome distribution, conserved motif composition, localization predicted and expression pattern of tomato NATs were performed. In addition, the comparison phylogenetic tree was constructed to evaluate the evolutionary relationships of NAT proteins in tomato and other species. Our systematic analysis in model species provides a foundation for further functional dissection of NAT genes in tomato, and could also help to elucidate the NAT gene function in others species.
Materials and methods
Database searches for the identification of NAT family members in tomato
The NAT family genes in tomato (Solanum lycopersicum) were collected by searching against SOL Genomics Network (SGN) Unigene database (http://solgenomics.net/search/loci) using key word of NAT. The Arabidopsis AtNAT12 protein was also subjected to BLASTN searches in SGN to confirm omission in database. The gene loci for each NAT proteins in organism of tomato were recovered, including locus name, gene description, chromosome arm, and SGN Unigenes. The putative open reading frames (ORFs) and proteins sequences were predicted by GENSCAN (http://genscanw.biosino.org/). The NAT domain of deduced tomato NAT amino acid sequences were confirmed by ScanProsite (http://www.expasy.ch/tools/scanprosite/) and InterProScan (http://www.ebi.ac.uk/Tools/InterProScan/). Pseudogenes were determined according to their gene annotation or when their coding sequences were obviously terminated by premature stop codons. If there was more than one allele, the longest allele was chosen as representative. The localization of tomato NATs was predicted by PROST (http://psort.hgc.jp/form.html and http://wolfpsort.org/).
NAT sequences of Arabidopsis, rice, and other species
The Arabidopsis (Arabidopsis thaliana) NAT gene name list and the corresponding coding and protein sequences were obtained from Arabidopsis transcription factor databases (http://datf.cbi.pku.edu.cn). The rice (Oryza sativa) NAT gene name list and the corresponding coding and protein sequences were downloaded from rice MPSS database (http://mpss.udel.edu/rice/mpss_index.php), the rice NAT genes were renamed according to the chromosome and their position, because of lacking of detail information (Maurino et al. 2006). Sequences of NAT proteins from other species were also retrieved from the NCBI database.
Sequence structure analysis and its distribution on chromosome
The exon/intron structure of SlNAT genes was generated using GSDS (http://gsds.cbi.pku.edu.cn/) by aligning the cDNA sequences with the corresponding genomic sequences. To determine the location of SlNAT genes on tomato chromosomes, the BLASTN search with SGN genomes (chromosome) database was processed. The resulting position of SlNAT genes on tomato chromosome were manually marked on bar.
Phylogenetic tree
A multiple alignment analysis was performed with multiple sequence alignment (Corpet 1988), the phylogenetic tree were created by MEGA4 program (Tamura et al. 2007). Phylogenetic tree was constructed using the neighbor-joining (NJ) method (Saitou and Nei 1987) with the Poisson correction, random seed of phylogeny test, and the pairwise deletion option parameters engaged. The reliability of the trees obtained was tested using bootstrap test with 1,000 replicates, clades with the test value higher than 50 and compared with other’s result were selected for the consensus tree. Images of the phylogenetic trees were also drawn using MEGA4.
Identification of conserved motifs
Protein motifs of the NAT protein sequences were identified statistically using MEME program (http://meme.nbcr.net/meme/) (Bailey and Elkan 1994) with motif length set as 6–100, motif sites 2–120, maximum number of motifs to find was set at 25, searching given strand only and the distribution of one single motif was any number of repetitions. The functional annotation of the identified motifs was implemented by ScanProsite and InterProScan. The phosphorylation sites were identified by Motifscan (http://myhits.isb-sib.ch/cgi-bin/motif_scan).
Analysis of expression profile of SlNAT genes in tomato various tissues
The expression profile was determined through analyzing the RNA-seq data based on locus gene name. The RNA-seq datas were downloaded from Tomato Functional Genomics Database (http://ted.bti.cornell.edu/cgi-bin/TFGD/digital/home.cgi), including the sequenced data of various tissues in tomato cultivar Heinz and the wild species S. pimpinellifolium, LA1589. Seeds for Heinz and LA1589 were greenhouse grown for 3 weeks in flats that were then transferred to growth chambers with no light for 72 h to promote starch degradation. Fresh meristematic expanding leaves were harvested and frozen in liquid nitrogen and stored at −80 °C. Only genes with at least one average RPKM value from all 11 tissues ≥2 in this study were considered to be expressed.
Results
The NAT gene family in tomato
A systematic analysis was performed to identify NAT genes in the tomato genome according to the SGN database, a total of 12 non-redundant SlNAT genes (Table 1) were identified and manually verified their uniqueness by removing redundant sequences from the databases and different transcripts of the same gene. These numbers were similar to the number of NAT genes present in the rice (11 NAT genes) and Arabidopsis (12 NAT genes) genomes (Maurino et al. 2006). Since there was no standard annotation assigned to these newly identified genes, we named these SlNAT genes as SlNAT1 to SlNAT12 based on the order of their location on the chromosomes. The identified nucleotide and amino acid sequences are presented in Supplementary S1.
The SlNAT genes name, the locus gene name, the chromosome, exon numbers, ORF length, isoelectric point (pI), molecular weight (Mw) and predicted localization was showed in Table 1. All of the 12 SlNAT genes belonged to the Xanthine/uracil/vitamin C permease family confirmed by ScanProsite. The identified nucleotide and amino acid sequences were presented in Supplemental S1. All the deduced SlNATs shared the similar amino acid sequences length with 530 AA, and a predicted molecular mass of approximately 60 kDa, excepted for SlNAT10 and SlNAT11, which the amino acid residue and the molecular mass was 713 AA and 77 kDa, respectively; but their predicted pIs were very divergent, ranging from 8.67 to 9.63. All the SlNATs shared the similar localization to plasma membrane, chloroplast thylakoid membrane, Golgi body, and endoplasmic reticulum (membrane) (Table 1).
The structural analyses and genome distribution of SlNAT genes
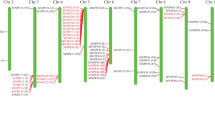
Structural analyses could provide valuable information involved in duplication events and evolution pattern when processing phylogenetic relationships within gene families. Thus, the exon/intron structure of each member of the SlNAT family was analyzed. Like rice and Arabidopsis NAT genes structures, the number of exons determined for the SlNAT genes were 14 expected for SlNAT4, SlNAT10 and SlNAT11, which had 13, 10 and 10 exons, respectively (Table 1; Fig. 1). These suggested a conservative evolution pattern of NAT genes.
Structure analysis of tomato SlNAT genes. Gene structures were generated from GSDS (http://gsds.cbi.pku.edu.cn/chinese.php)
To determine the genomic distribution of the SlNAT genes, we identified the position based on SGN genome database. The result showed the SlNAT genes were dispersed on all the chromosomes except chromosome 8 and 9. Chromosome 4 and 11 were found to possess two SlNAT genes each, while only one SlNAT gene was found on the rest of chromosome. The most of SlNAT were present at the telomeric ends on a single chromosome (Fig. 2; Table 1).
Positions of SlNAT gene family members on the tomato chromosomes. Scale represents a 10 Mb chromosomal distance. Chromosomal mapping was based on the physical position (Mb) in 12 tomato chromosomes. The chromosome number is indicated at the top of each chromosome. Chromosomal positions of the tomato NAT genes are indicated by gene name (assigned in Table 1)
Evolutionary relationships between the NAT family in tomato and other species
It is possible to analyze the same gene family among different species through the comparative genomics. To investigate the molecular evolution and phylogenetic relationships among NATs in tomato and other species, a multiple sequence alignment was conducted with 38 NAT protein sequences from plants, and used for the construction of a phylogenetic tree (Fig. 3). The information of NAT proteins in Arabidopsis, rice and other species were presented in Supplementary Table S1. We constructed a phylogenetic tree by the NJ method with poisson correction and bootstrap analysis (1,000 replicates), based on the alignment of all the NAT amino acid sequences (Supplementary Fig. 1). All tomato NAT proteins shared 27–89 % identical amino acid residues with each other and 18–89 % similarity with NAT proteins from other plants (Supplementary Table S2).
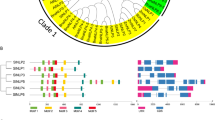
The NJ phylogenetic tree of the NAT members. The unrooted tree, constructed with MEGA4.0, has been generated using full-length amino acid sequences from tomato, Arabidopsis, rice and other species NATs. The tree shows the four phylogenetic subfamilies (a, b, c, d) with high predictive values (bootstrap support of 50 or greater)
The NJ phylogenetic tree (Fig. 3) showed that all the NAT genes were divided into 4 well-supported clades, similarly to that of other’s report (Maurino et al. 2006). All SlNAT proteins shared higher similarity with AtNAT proteins and clustered more closely together in the phylogenetic tree than they did with those from rice and other species, suggesting that two dicot plants had a closer evolutionary relationship than with the monocot plants and consistent with the fact that both tomato and Arabidopsis are diverged more recently from a common ancestor.
Accordingly, the tree clades of NAT genes were classified into four groups, A, B, C, and D (Fig. 3). Among these, the group C constituted the largest clade containing 12 members, each species of tomato, Arabidopsis and rice had four members; the group A formed the second largest clade containing 11 members, 4 belonged to rice NATs, 3 to tomato NATs, 3 to Arabidopsis NATs and one to cotton NAT gene; the B group were made up of 9 members, tomato and Arabidopsis each had 3 NATs, and rice, maize and ice plant had one NAT gene each; group D consisted of 6 NAT genes; tomato, Arabidopsis and rice each had 2 members. Interestingly, there were specific-species subgroups of rice, tomato and Arabidopsis NAT genes in group A and C, indicating that there was a presumed gene loss/gain event between the dicot-monocot split.
Within each class, six pairs of orthologous proteins were found at the terminus of the phylogenetic tree with 1,000 replicates: ZmLpe and OsNAT4, SlNAT5 and AtNAT3, SlNAT8 and AtNAT1, SlNAT2 and GhNAT1, SlNAT10 and AtNAT11 and SlNAT11 and AtNAT12, and 7 pairs of homologous/paralogous proteins were identified according to the phylogenetic tree; 3 pairs in rice and Arabidopsis each, and one pairs in tomato (e.g. SlNAT1 and SlNAT9; AtNAT5 and AtNAT6; OsNAT3 and OsNAT5), suggested higher identities and similarities. The identities were ranged from 78 to 89 % (Supplementary Table 2). This result suggested that some members of the SlNAT, OsNAT and AtNAT gene families might originate from the same ancestral genes before divergence of monocots and dicots.
Motifs were identified with the MEME software using the complete amino acid sequences of NAT genes. Multilevel consensus sequences for the MEME defined motifs are listed in Table 3.
Domains and Motifs analyses in NATs family in plant
To further reveal the diversifications of NATs in tomato, putative motifs were predicted by the program MEME and 25 distinct motifs were identified (Table 2). The schematic distribution of the 25 motifs among the different gene groups was showed in Table 2; these motifs were represented in their relative location within the protein. The indentified multilevel consensus sequence for the motifs was showed in Table 3. The 25 motifs identified by MEME were annotated by Motif Scan and Sbase; among the 25 motifs, motifs 1, 2, 3, 6, 8, and 9 were annotated to Xanthine/uracil/vitamin C permease, together comprised the named NAT domain (Table 3), were shared by all of the members expected for OsNAT3, AtNAT9 and OsNAT7, which had partial deletion in the NAT domain. The motif 1 uniformly observed in all NAT proteins was the so-called ‘NAT signature motif’, which was present in a domain critical for substrate recognition in Aspergillus UapA and UapC purine transporters (Meintanis et al. 2000).
Numbers correspond to the motifs described in Table 2. Sequences obtained from the analysis of all the NAT complete proteins with the MEME tools. The highly conserved QH motif (bold and box) and signature motif (underline) are highlighted in motif 2 and 1, respectively. cAMP- and cGMP-dependent protein kinase phosphorylation site (green), N-glycosylation sites (blue) and N-myristoylation sites (italic and bold) are indicated. Protein kinase C (PKC) phosphorylation sites are indicated with bold and red, and Casein kinase II (CK2) phosphorylation sites are showed with box.
As expected, most of the closely related members in the phylogenetic tree shared common motif compositions, suggesting functional similarities among the NAT proteins within the same subfamily, but the unique motifs were shared by different groups (Table 1; Fig. 3). The group C showed the same motif compositions excepted for OsNAT3, the distribution structure of group C was [11]_[2]_[16]_[3]_[10]_[13]_[7]_[8]_[6]_[9]_[1]_[4]_[5], while in group B, interestingly, shared similar motifs compositions with group C, but found a motif (16) deletion (for the most of NATs in this group), suggested that those NATs shared common ancestral genes. Accordingly, those NATs could form a big clade, supporting by bootstrap values (100 %) in the phylogenetic tree (Fig. 3). A unique motif (11) was also found in these two groups (Table 1). The group A, B and C were further clustered forming a large clade (Fig. 3), and similar motifs compositions were found (Table 1). In group A, there was a replacement of motif 11 to motif 19/10 compared to group B, the group A could further clustered four clades, the same as displayed in phylogenetic tree (Fig. 3). The motif 19 and 24 were unique motif in subgroup, containing SlNAT5, AtNAT3, OsNAT11, SlNAT8 and AtNAT1, while in another subgroup, two motifs 10 was found in the NATs. In the group D, much more unique motifs (15, 20, 22, 23, 17, 14, 18 and 12) were detected and were common inner the group (Table 2), suggesting a functional divergence. Moreover, motif 10, 4 and 7 were dispersed in all the NATs, motif 13 and 5 were the common motif shared by group A, B and C, implying that they were likely to be necessary for NAT function. Motif scan analysis revealed that many of these motifs possess phosphorylation sites such as protein kinase C, N-myristoylation and casein kinase II (Table 3).
Expression patterns of SlNAT genes in cultivar and wild tomato
The expression patterns of the 12 tomato NAT genes in the different organs were analyzed through searching the RNA-seq data. The tomato NAT gene family was expressed in distinct patterns (Tables 4, 5). The expression profiles reveal that 3 genes, SlNAT1, SlNAT9 and SlNAT4, from group B showed no expression in cultivar Heinz and had a low expression level in wild tomato LA1589. In cultivar Heinz, the remained SlNAT genes displayed spatial variations in different tomato organs. Some of them were constitutively expressed in every organ investigated and the expression levels were high, especially in young immature fruit, and descend with fruit ripening, such as SlNAT3 and SlNAT7 from group C, whereas SlNAT12 and SlNAT5 shared similar expression pattern with high expressed in mature fruit. Some SlNAT genes were constitutively expressed with low levels in every organ tested, such as those of SlNAT6, SlNAT2, SlNAT8, SlNAT10 and SlNAT11, among them, SlNAT6, SlNAT2 and SlNAT8, SlNAT10 and SlNAT11 displayed similar expression pattern (Table 4). In wild tomato LA1589, the similar expression pattern was also observed in SlNAT7, SlNAT2, SlNAT8 and SlNAT10 with constitutively low expression in every organ investigated. SlNAT3 and SlNAT5 were expressed in every organ tested and the expression levels were relatively higher in 10DPA, 20 DPA, hypocotyl and cotyledon. SlNAT6, SlNAT11 and SlNAT12 showed similar expression pattern with high expression in young flower bud, young leaf, vegetative meristem, hypocotyl, 10DPA and 20 DPA, the expression levels was low at the later ripening stage. These expression profiles suggest a divergence in the biological functions of SlNAT genes during plant development (Table 5).
Discussion
The NAT family is one of the five known families of transporters that use nucleobases as their principal substrates and the only one that is evolutionarily conserved and widespread in all major taxa of organisms. In humans the AsA was transported mainly through SVCT1 and SVCT2 (Burzle et al. 2013). Whereas in plant, only one nucleobase transporters, ZmLPE1 from maize, has been characterized by functional complementation of fungal purine transport mutants. The ZmLPE1 specific transport for uric acid, xanthine and can also bind, but do not transport ascorbate. Its function is necessary for proper chloroplast development (Argyrou et al. 2001). Recently, 12 NAT genes sharing high similarity with known NATs from other species were classified through analysis of the Arabidopsis thaliana genome (Maurino et al. 2006). However, virtually nothing is known about this family in tomato. Therefore, Genome-wide survey and characterization of NAT genes in tomato would facilitate a better understanding this gene superfamily and provide potential candidate NATs for further gene function analysis.
The NAT proteins in tomato genomes
This study has identified 12 SlNAT genes in tomato from available tomato genomic sequences (SGN), classified by the presence of a highly conserved NAT domain. The numbers of NAT genes are similar to previous studies in Arabidopsis and rice (Maurino et al. 2006). Like others NAT genes in rice and Arabidopsis, the tomato NAT genes also shared the similar exons numbers, sequences length, pI and Mw (Table 1 and Supplementary Table S1). The chromosomal location analyses (Fig. 2) of the SlNAT genes showed the SlNAT genes distributed on all the chromosomes except chromosome 8 and 9. Each two genes were located on each of Chromosome 4 and 11, while the remaining SlNAT genes appeared randomly scattered throughout the chromosomes. In addition, the SlNAT genes seemed to present at the termination on the chromosome (Fig. 2).
The deduced SlNATs shared the similar amino acid sequences length with 530 AA, and a predicted molecular mass of approximately 60 kDa, excepted for SlNAT10 and SlNAT11, which the amino acid residue and the molecular mass was 713 AA and 77 kDa, respectively; but their predicted pIs were very divergent, ranging from 8.67 to 9.63. Those results are similar with NATs in Arabidopsis, ten AtNATs showed similar amino acid residue with 530 AA, but AtNAT11 and AtNAT12 shared 711 AA (Maurino et al. 2006), suggesting the functional similarity and evolutional conservation.
All the SlNATs shared the similar localization to plasma membrane, chloroplast thylakoid membrane, Golgi body, and endoplasmic reticulum (membrane) (Table 1). Due to the fact that the last step in the AsA biosynthetic pathway takes place on the inner membrane of mitochondria, AsA must be transported to other cell across the membranes. It was described previously the ascorbate was transported across plasma and plastid membranes (Horemans et al. 1998; 2000). Consequently, membranes location of SlNATs means that they would be transporters for AsA in tomato.
Structural divergences have been demonstrated playing a more important role during the evolution of multiple gene family, mainly through three types of mechanisms, exon/intron gain/loss, exonization/pseudoexonization, and insertion/deletion, each of which contributed differently to structural divergence (Xu et al. 2012). Our structural analyses showed that the SlNAT genes contain 14 exons expected for SlNAT4, SlNAT10 and SlNAT11, which had 13, 10 and 10 exons, respectively, suggesting a conservative evolution pattern, but occurrence the generation of functionally divergences of NAT genes (Table 1; Fig. 1).
Motifs analyses in NATs family in tomato
Identification and characterization of the conserved motifs are increasingly significant, so we further analyzed the conserved motifs in tomato NAT family and other NATs by MEME. The majority of SlNAT proteins in the same group or subgroup shared similar motifs, suggesting that these conserved motifs play crucial roles in group or subgroup-specific functions. Multiple alignments of NAT sequences reveal the presence of several highly conserved sequence motifs. Of particular importance is the motif (Q/E/P)NXGXXXXT(R/K/G), as shown in motif 1 in this study, which is located downstream from transmembrane domains 8 (Burzle et al. 2013), which was called the ‘NAT signature motif’ and critical for the function and specificity of Aspergillus UapA and UapC purine transporters (Meintanis et al. 2000). Besides, 5 other recognizable conserved motifs, 2, 3, 6, 8, and 9 are together responsibility for the named NAT domain, which were broadly distributed in the all the NAT protein sequences, expected for OsNAT3, AtNAT9 and OsNAT7 with partial deletion in the NAT domain (Table 2).
There also motifs, such as motif 10, 4 and 7, were wide dispersed in all the NATs, or were the common motif shared by group A, B and C, like motif 13 and 5, implying that they were likely to be necessary for NAT function. In addition, some other motifs were also identified uniquely between different clades. The motif distribution analysis confirmed the conservative and functional divergence with NAT proteins over evolutionary history and also correlated well with the phylogenetic analysis.
Comparative genomic analysis of the tomato and other NAT proteins
The NJ phylogenetic tree (Fig. 3) showed that all the NATs were divided into 4 well-supported clades, similarly to that of other’s report (Maurino et al. 2006). Accordingly, the tree clades of NATs were classified into four groups, A, B, C, and D (Fig. 3). The plant NAT proteins shared 18–89 % homogeneous amino acid sequences with or within NAT proteins from others. The similarity of the plant NAT proteins to the vitamin C transporter from mammals and to the nucleobase transporters from A. nidulans is about 14–35 and <17 %, respectively (Supplementary Fig. 1). All SlNAT proteins shared higher similarity with AtNAT proteins and clustered more closely together in the phylogenetic tree than they did with those from rice and other species, and each group contained the same members from tomato and Arabidopsis (Fig. 3 and Supplementary Fig. 1), suggesting that two dicot plants had a closer evolutionary relationship than with the monocot plants and consistent with the fact that both tomato and Arabidopsis are diverged more recently from a common ancestor.
Interestingly, there were specific-species subgroups of tomato and rice NAT genes in group A and C, such as SlNA3, SlNAT7 and SlNAT12; OsNAT3, OsNAT5, OsNAT8 and OsNAT10 in group C; OsNAT1, OsNAT7 and OsNAT9 in group A, indicating that there was a presumed gene loss/gain event between the dicot-monocot split.
Organ- and tissue-specific expression of tomato NAT genes
The expression patterns of the 12 tomato NAT genes in the different organs were analyzed through searching the RNA-seq data. Four different expression patterns were obtained (Tables 4, 5). Three genes, SlNAT1, SlNAT9 and SlNAT4, from group B showed no expression in cultivar Heinz or had a low expression level in wild tomato LA1589. In Arabidopsis, the expression of two genes, AtNAT9 and AtNAT10, could not be detected in tested tissues. The GUS activity analysis of transgenic A. thaliana plants driven by AtNAT promoters showed that the AtNAT9 and AtNAT10 promoter was barely active and inactivity, respectively (Maurino et al. 2006). Other SlNAT genes were constitutively expressed with spatial variations in every organ investigated. The second expression pattern were expressed with high levels, especially in young immature fruit, and descend with fruit ripening, such as SlNAT3 and SlNAT7 from group C, whereas SlNAT12 and SlNAT5 representative for the third expression pattern shared similar expression pattern with high expressed in mature fruit. The last expression pattern were constitutively expressed but with low levels in every organ tested, such as SlNAT6, SlNAT2, SlNAT8, SlNAT10 and SlNAT11. AtNAT7 and AtNAT8, coming from the same group with SlNAT3, SlNAT7, SlNAT6 and SlNAT12, were localized to the plasma membrane and involved in the transport of signal molecules or substrates needed during the fast growth of undifferentiated tissues, whereas the ascorbate and GSH content and the ratio of reduced/oxidized were not significant differences in the nat7 and nat8 mutants as compared with the wild type (Maurino et al. 2006).
In wild tomato LA1589, the similar expression pattern was observed compared with cultivar Heinz but with a different expression levels (Table 5). SlNAT7, SlNAT2, SlNAT8 and SlNAT10 with constitutively low expression in every organ investigated. SlNAT3 and SlNAT5 were expressed with relatively higher in 10DPA, 20 DPA, hypocotyl and cotyledon. SlNAT6, SlNAT11 and SlNAT12 showed high expression in young flower bud, young leaf, vegetative meristem, hypocotyl, 10DPA and 20 DPA, the low expression levels at the later ripening stage. These expression profiles suggest a divergence in the biological functions of SlNAT genes during plant development.
Conclusions
Recently, significant progress has been made toward the identification and characterization of NAT genes in Arabidopsis and rice; however, little is known concerning this gene family in tomato with berry fruit. In the present study we identified 12 NAT genes in tomato, the characteristics of SlNATs, including locus gene name, the chromosome, exon numbers, number of nucleotide and amino acids (length), pI, Mw and predicted localization was investigated. The separation of the tomato NAT proteins into 4 groups was mutually supported by their exon/intron structure, phylogeny, and the distribution of conserved motifs. The expression profiles of SlNAT genes in various organs showed 9 out of 12 SlNAT genes were constituently expression with differential expression levels under normal growth conditions. Our systematic analysis furthers the understanding of NAT genes in plants and provides a framework for future functional studies of the NAT family in tomato.
References
Agius F, Gonzalez-Lamothe R, Caballero JL, Munoz-Blanco J, Botella MA, Valpuesta V (2003) Engineering increased vitamin C levels in plants by overexpression of a d-galacturonic acid reductase. Nat Biotechnol 21(2):177–181
Argyrou E, Sophianopoulou V, Schultes N, Diallinas G (2001) Functional characterization of a maize purine transporter by expression in Aspergillus nidulans. Plant Cell 13(4):953–964
Asard H, Horemans N, Caubergs RJ (1992) Transmembrane electron transport in ascorbate-loaded plasma membrane vesicles from higher plants involves a b-type cytochrome. FEBS Lett 306(2):143–146
Badejo AA, Wada K, Gao Y, Maruta T, Sawa Y, Shigeoka S, Ishikawa T (2012) Translocation and the alternative d-galacturonate pathway contribute to increasing the ascorbate level in ripening tomato fruits together with the d-mannose/l-galactose pathway. J Exp Bot 63(1):229–239
Bailey TL, Elkan C (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2:28–36
Bartoli CG, Pastori GM, Foyer CH (2010) Ascorbate biosynthesis in mitochondria is linked to the electron transport chain between complex III and IV. Plant Physiol 123:335–343
Burzle M, Suzuki Y, Ackermann D, Miyazaki H, Maeda N, Clemencon B, Burrier R, Hediger MA (2013) The sodium-dependent ascorbic acid transporter family SLC23. Mol Aspects Med 34(2–3):436–454
Corpet F (1988) Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res 16(22):10881–10890
de Koning H, Diallinas G (2000) Nucleobase transporters (review). Mol Membr Biol 17(2):75–94
Franceschi VR, Tarlyn NN (2002) l-Ascorbic acid is accumulated in source leaf phloem and transported to sink tissues in plants. Plant Physiol 130:649–656
Gallie DR (2013) l-ascorbic acid: a multifunctional molecule supporting plant growth and development. Scientifica. doi:10.1155/2013/795964
Horemans N, Potters G, Caubergs R, Asard H (1998) Transport of ascorbate into protoplasts of Nicotiana tabacum Bright Yellow-2 cell line. Protoplasma 205(1):114–121
Horemans N, Foyer CH, Asard H (2000) Transport and action of ascorbate at the plant plasma membrane. Trends Plant Sci 5(6):263–267
Lorence A, Chevone BI, Mendes P, Nessler CL (2004) Myo-inositol oxygenase offers a possible entry point into plant ascorbate biosynthesis. Plant Physiol 134(3):1200–1205
Maurino VG, Grube E, Zielinski J, Schild A, Fischer K, Flugge UI (2006) Identification and expression analysis of twelve members of the nucleobase–ascorbate transporter (NAT) gene family in Arabidopsis thaliana. Plant Cell Physiol 47(10):1381–1393
Meintanis C, Karagouni AD, Diallinas G (2000) Amino acid residues N450 and Q449 are critical for the uptake capacity and specificity of UapA, a prototype of a nucleobase–ascorbate transporter family. Mol Membr Biol 17(1):47–57
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4(4):406–425
Szarka A, Banhegyi G, Asard H (2012) The inter-relationship of ascorbate transport, metabolism and mitochondrial, plastidic respiration. Antioxid Redox Signal. doi:10.1089/ars.2012.5059
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24(8):1596–1599
Tedone L, Hancock RD, Alberino S, Haupt S, Viola R (2004) Long-distance transport of l-ascorbic acid in potato. BMC Plant Biol 4:16. doi:10.1186/1471-2229-4-16
Wheeler GL, Jones MA, Smirnoff N (1998) The biosynthetic pathway of vitamin C in higher plants. Nature 393(6683):365–369
Wolucka BA, Van Montagu M (2003) GDP-mannose 3′,5′-epimerase forms GDP-l-gulose, a putative intermediate for the de novo biosynthesis of vitamin C in plants. J Biol Chem 278(48):47483–47490
Xu G, Guo C, Shan H, Kong H (2012) Divergence of duplicate genes in exon-intron structure. Proc Natl Acad Sci USA 109(4):1187–1192
Zechmann B, Stumpe M, Mauch F (2011) Immunocytochemical determination of the subcellular distribution of ascorbate in plants. Planta 233(1):1–12
Acknowledgments
This study was supported by grants from the State Major Basic Research Development Program (No. 2011CB100600), and National Natural Science Foundation of China (No. 31171974).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Cai, X., Ye, J., Hu, T. et al. Genome-wide classification and expression analysis of nucleobase–ascorbate transporter (NAT) gene family in tomato. Plant Growth Regul 73, 19–30 (2014). https://doi.org/10.1007/s10725-013-9864-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10725-013-9864-x