Abstract
Studies of rDNA location in holocentric chromosomes of the Cyperaceae are scarce, but a few reports have indicated the occurrence of multiple 45S rDNA sites at terminal positions, and in the decondensed state of these regions in prometaphase/metaphase. To extend our knowledge of the number 45S and 5S rDNA sites and distribution in holocentric chromosomes of the Cyperaceae, 23 Brazilian species of Eleocharis were studied. FISH showed 45S rDNA signals always located in terminal regions, which varied from two (E. bonariensis with 2n = 20) to ten (E. flavescens with 2n = 10 and E. laeviglumis with 2n = 60). 5S rDNA showed less variation, with 16 species exhibiting two sites and 7 species four sites, preferentially at terminal positions, except for four species (E. subarticulata, E. flavescens, E. sellowiana and E. geniculata) that showed interstitial sites. The results are discussed in order to understand the predominance of terminal rDNA sites, the mechanisms involved in the interstitial positioning of 5S rDNA sites in some species, and the events of amplification and dispersion of 45S rDNA terminal sites.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
rDNA occurs as moderately repetitive DNA families in eukaryote genomes. 45S rDNA of plants, as well as in other organisms, appears as repeated and clustered segments at particular chromosomal positions. These segments predominantly occupy the chromosome ends (Furuta and Kondo 1999; Rego et al. 2009), varying in the number of sites among and within species (Vanzela et al. 1998; De Melo and Guerra 2003; Da Silva et al. 2008a; Raskina et al. 2008). 5S rDNA is also organized as repeated and clustered segments, occurring in most cases at chromosome positions different from those occupied by 45S rDNA (Moscone et al. 1999). However, they tend to occupy similar chromosomes and positions in closely related species, as in Aristolochia (Berjano et al. 2009).
The Cyperaceae are widely known as having holocentric chromosomes, and several studies involving chromosome counts and meiotic analysis were performed in order to understand the mechanisms of karyotype differentiation (Vanzela et al. 2000; Roalson 2008; Da Silva et al. 2010). However, few comparative studies have focused on the chromosome location of rDNA sites in species with holocentric chromosomes. Multiple 45S rDNA sites were detected in nine species of Rhynchospora (Vanzela et al. 1998, 2003) and seven species of Eleocharis (Da Silva et al. 2005, 2008a, b), both of the family Cyperaceae. Hoshi (1995) and Furuta and Kondo (1999) also reported multiple and terminal rDNA sites in Drosera. However, in Cuscuta approximata, one pair of 45S rDNA and three of 5S rDNA sites were reported (Guerra and García 2004). In spite of a general tendency towards a terminal location of 45S rDNA, 5S rDNA was reported to be preferentially located in interstitial regions in plants, but the only study of 5S rDNA location in the Cypercaeae showed interstitial hybridization sites associated with chromosome rearrangement (Da Silva et al. 2008b).
The family Cyperaceae possesses approximately 5,500 species (Govaerts et al. 2007), but less than 1% of the species have been studied by FISH using rDNA probes. These few reports do not discuss the location and amplification of rDNA based on models, as commonly done in organisms with monocentric chromosomes. The aim of this study was to localize the 45S and 5S rDNA sites using double-FISH in 23 Brazilian species of Eleocharis with different chromosome numbers and sizes. Data were discussed to assess the karyotypic diversity and to understand the possible mechanisms at work in rDNA amplification and distribution in these organisms that possess holocentric chromosomes.
Materials and methods
Three individuals of each of 23 species of Eleocharis were collected in different Brazilian states (Table 1). Samples were cultivated in a greenhouse at the Laboratório de Biodiversidade e Restauração de Ecossistemas (LABRE) at Universidade Estadual de Londrina, Paraná, Brazil. Vouchers were deposited at the ICN herbarium of the Universidade Federal do Rio Grande do Sul, Porto Alegre, Brazil (Table 1). Metaphase cells were obtained from root tips pre-treated with 2 mM 8-hydroxyquinoline for 24 h, fixed in ethanol:acetic acid (3:1, v:v) for 24 h, and stored at −20°C. Samples were softened in 4% cellulase plus 40% pectinase (w:v) at 37°C for 4 h and squashed in a drop of 60% acetic acid, and the cover slips were removed after freezing in liquid nitrogen.
Fluorescent in situ hybridization (FISH) was performed as described by Da Silva et al. (2008a). The pTa71 containing the 45S rDNA probe isolated from Triticum aestivum (Gerlach and Bedbrook 1979) was labeled with digoxigenin-11-dUTP, and the 5S rDNA probe isolated from Rhynchospora pubera (Angeles Cuadrado, unpublished) was labeled with biotin-14-dATP, both by nick translation. Probes were simultaneously utilized for FISH in a mixture of 34 μL containing 100% formamide (15 μL), 50% polyethylene glycol (6 μL), 20 × SSC (3 μL), 100 ng of calf thymus DNA (1 μL), 10% SDS (1 μL), and 200 ng of probes (4 μL each). The material was denatured at 90°C for 10 min, and hybridization was performed at 37°C overnight in a humid chamber. Post-hybridization washes were carried out with 70% stringency, using baths with SSC buffer.
The probes were simultaneously detected with avidin-FITC and anti-digoxigenin-rhodamine conjugates. The post-detection washes were performed in 4 × SSC/0.2% Tween 20, all at room temperature. Slides were mounted with 25 μL of DABCO solution, composed of glycerol (90%), 1,4-diaza-bicyclo(2,2,2)-octane (2.3%), 20 mM TrisHCl, pH 8.0 (2%), 2.5 mM MgCl2 (4%) and distilled water (1.7%), plus 4 μL of 2 μg/mL−1 DAPI.
All the chromosome images were separately acquired in grayscale with a Leica DM 4500 B microscope coupled with a DFC 300FX camera and overlapped with red color for DAPI, white for rhodamine (45S rDNA) and greenish–yellow (5S rDNA) for FITC, using the Leica IM50 4.0 software. The images were optimized for best contrast and brightness with Adobe Photoshop CS3 version 10.0 software.
Results
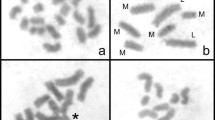
Twenty-three species were analyzed, of which 15 belonged to the subgenus Eleocharis, two to Scirpidium and six to Limnochloa (Table 1). The number of 45S rDNA sites varied from two to ten, independent of the subgenus to which the species belonged and the chromosome number. In contrast, the terminal location of 45S rDNA was constant in all species. The 5S rDNA probe showed a minor variation when compared to 45S rDNA, ranging from two to four chromosome sites which were terminal in 19 species and interstitial in four others (Table 1). The rDNA hybridization signals also varied in size between homologs, as observed in E. niederleinii (Fig. 1c), E. montana (Fig. 1g), E. debilis (Fig. 2f) and E. obtusetrigona (Fig. 3b). In all cases, the 45S rDNA hybridization signals appeared decondensed, unlike the 5S rDNA signals, even when they were located in the terminal region.
Metaphases of Eleocharis species double-probed with 45S rDNA and 5S rDNA (arrows) probes. a E. contracta (2n = 20) showing four hybridization signals of 45S rDNA and two of 5S. b E. nana (2n = 20) with two signals each of 45S and 5S rDNA. c E. niederleinii (2n = 20) with four 45S rDNA signals and two for 5S. d E. loefgreniana (2n = 20). e E. viridans (2n = 20). f E. subarticulata (2n = 6). g E. montana (2n = 40). h E. minima (2n = 20). i E. filiculmis (2n = 30). j E. capillacea (2n = 10). d, e, g, h, i, exhibited four 45S rDNA and two 5S rDNA signals at terminal positions. f E. subarticulata, exhibited 5S signals at interstitial positions. k E. flavescens (2n = 10) with ten 45S rDNA signals and two interstitial 5S signals. l E. maculosa (2n = 10) with four 45S rDNA and two of 5S. Bar represents 10 μm
Metaphases of Eleocharis species double-probed with 45S rDNA and 5S rDNA (arrows) probes. a E. sellowiana (2n = 10) with four 45S rDNA signals and two interstitial 5S signals. b E. sellowiana (2n = 20) with eight 45S rDNA signals and two interstitial 5S signals. c and d E. geniculata (2n = 10 and 20) both with four 45S rDNA signals and two interstitial 5S signals, respectively. e Eleocharis sp. (2n = 10) showing six hybridization signals of 45S rDNA and two of 5S. f E. debilis (2n = 30) with four signals of 45S rDNA and two of 5S. g E. bonariensis (2n = 20) with four 45S rDNA signals and four of 5S. Bar represents 10 μm
Metaphases of subgenus Limnochloa species double-probed with 45S rDNA and 5S rDNA (arrows) probes. a E. plicarhachis (2n = 54) showing ten hybridization signals of 45S rDNA and four of 5S. b E. obtusetrigona (2n = 52) with eight hybridization signals of 45S rDNA and four of 5S. c E. liesneri (2n = 50) with ten hybridization signals of 45S rDNA and four of 5S. d and e E. acutangula (2n = 54) and E. interstincta (2n = 40) both with eight hybridization signals of 45S rDNA and four of 5S. f E. laeviglumis (2n = 60) with ten signals of 45S rDNA and four of 5S. Bar represents 10 μm
The representatives of the subgenus Eleocharis showed no correspondence between polyploidy level and rDNA site number. Eleocharis flavescens, with 2n = 10, exhibited 45S rDNA hybridization signals in all chromosomes of the complement and only one pair with interstitial 5S rDNA signal (Fig. 1k), while E. nana with 2n = 20, showed two terminal signals of 45S and of 5S rDNA (Fig. 1b). Two interstitial signals of 5S rDNA were also found in E. subarticulata (2n = 6), E. sellowiana and E. geniculata, but, in the last two species, karyotypes with 2n = 10 and 20 were found. These species also showed variation in the number of terminal 45S rDNA sites: four in E. subarticulata (Fig. 1f), in the diploid and polyploid forms of E. geniculata (Fig. 2c, d), and in the diploid form of E. sellowiana; and eight in the polyploid form of E. sellowiana (Fig. 2a, b). Eleocharis contracta (Fig. 1a), E. niederleinii (Fig. 1c), E. loefgreniana (Fig. 1d), E. viridans (Fig. 1e), E. montana (Fig. 1g), E. minima (Fig. 1h), E. filiculmis (Fig. 1i), E. capillacea (Fig. 1j), E. maculosa (Fig. 1l) and E. debilis (Fig. 2f) exhibited different chromosome numbers but equal numbers of rDNA sites (four 45S rDNA and two 5S) always at the terminal positions. (Table 1).
The two species of the subgenus Scirpidium varied in number and position of rDNA sites. Eleocharis bonariensis with 2n = 20 showed four 45S rDNA sites and four signals of 5S rDNA, always at terminal positions (Fig. 2g), while Eleocharis sp., with 2n = 10 showed six chromosomes with 45S rDNA signals and another two with 5S rDNA, also always at terminal positions (Fig. 2e).
Six species of the subgenus Limnochloa were studied. These species showed different chromosome numbers (40, 50, 52, 54 and 60) and exhibited the smallest chromosomes of Eleocharis. Four 5S rDNA terminal sites were found in all species, but the number of terminal 45S rDNA sites varied. Eleocharis obtusetrigona, E. acutangula and E. interstincta showed eight hybridization signals (Fig. 3b, d and e), while E. plicarhachis, E. liesneri and E. laeviglumis showed 10 sites (Fig. 3a, c and f).
Discussion
The Cyperaceae exhibit a high variability in chromosome number (Roalson 2008), due mainly to events of symploidy (fusion), agmatoploidy (fission) and polyploidy (Håkanson 1958; Vanzela et al. 2000; Yano et al. 2004; Hipp et al. 2009). The basic chromosome number, x = 5, proposed by Löve et al. (1957) for the Cyperaceae is accepted by several authors (Vanzela et al. 1998; Yano et al. 2004; Da Silva et al. 2008a), even considering the high numeric variability. Therefore, we would expect the karyotypes with few chromosomes to also have few 45S rDNA sites. However, we found species with 2n = 10 with four signals (the majority of species), six signals (Eleocharis sp.) and signals in all chromosomes, such as in E. flavescens. Species with multiple 45S rDNA hybridization sites were previously reported, as in the genera Eleocharis (Da Silva et al. 2008a), Rhynchospora (Vanzela et al. 1998) and Drosera (Hoshi 1995).
The polyploids should have double the number of hybridization sites in relation to their diploid representatives, as reported in Rhynchospora tenuis with 2n = 4 and 2n = 8, possessing two and four 45S rDNA sites, respectively (Vanzela et al. 2003). This was also observed in E. sellowiana (with four and eight 45S rDNA signals). On the other hand, Eleocharis geniculata exhibited the same number of sites in both 2n = 10 and 2n = 20. The remaining polyploid species with 2n = 20, 30 and 40 showed two and four 45S rDNA signals (see Table 1); however, we did not find their representative diploids, making it impossible to establish the relationship between ploidy level and number of hybridization sites. The polyploids mentioned, namely E. filicumis and E. montana, possess regular meiosis (Da Silva et al. 2010), and the occurrence of few 45S rDNA sites number in these species could be explained by the loss of rDNA sites during the diploidization process, as proposed by Leitch and Bennett (1997). Similar events were reported in Passiflora (De Melo and Guerra 2003).
Species of the subgenus Limnochloa showed karyotypes with many and small chromosomes and little variation in size among them. Yano et al. (2004) and Da Silva et al. (2008a) have proposed that these karyotypes may have been generated by chromosome fission, but it was not confirmed in a later study by Da Silva et al. (2010). We found here that karyotypes of Limnochloa had multiple 45S rDNA sites (eight and ten). When we compared the FISH data in relation to karyotype organization, i.e., chromosome number and symmetry (see Da Silva et al. 2010), regardless of systematic arrangement, it was possible to detect that the species with a more symmetrical karyotype, namely E. interstincta (Limnochloa), Eleocharis sp. (Scirpidium), E. flavescens and E. sellowiana (the last two of the subgenus Eleocharis), exhibited a greater number of 45S rDNA sites. The karyotypes with a greater variation in chromosome size, i.e., more asymmetric, as in E. montana, E. nana and E. viridans (all of the subgenus Eleocharis), showed fewer 45S rDNA sites.
Cases of multiple 45S rDNA sites have been described in both plants with holocentric chromosomes (Vanzela et al. 1998) and in plants with monocentric ones (Hasterok and Maluszynska 2000; Lim et al. 2001; Kwon and Kim 2009). Some mechanisms of amplification/dispersion of repetitive DNA have been proposed or mentioned for the monocentric system: i) rDNA site mobility associated with co-location and activity of transposable elements (Shubert and Wobus 1985; Raskina et al. 2008) and ii) equilocal dispersion of heterochromatin and rDNA (Schweizer and Loidl 1987; Pederson and Linde-Laursen 1994, respectively). In relation to the first model, there are no reports in the literature involving the distribution of transposable elements in plant holocentric chromosomes. For the second model, however, Guerra (2000) proposed that the equidistribution of repetitive DNA may depend on some structural or functional similarity of each chromosomal region. In this sense, the polarization of rDNA sites observed in the interphase of some Rhynchospora species may be due to an association of active sites building a single nucleolus (Vanzela et al. 1998). Except for E. filicumis with 2n = 30 (Fig. 1i), all other species of Eleocharis exhibited 45S rDNA hybridization sites at the ends of chromosomes of similar size. Thus, it is possible that the grouping of 45S rDNA regions to form the nucleolus could contribute to the accumulation/dispersion of 45S rDNA segments at the ends of chromosomes that are similar in size and are near the nucleolus. This could explain the occurrence of multiple rDNA sites preferably in karyotypes whose chromosomes have the same size (symmetrical). Idiograms and chromosome measurements showing karyotype symmetry/asymmetry were previously detailed by Da Silva et al. (2010).
The 45S and 5S rDNA probes hybridized to different chromosome regions. The occurrence of 45S (terminal) and 5S rDNA (interstitial) sites in the same chromosome pair was found in E. flavescens, precisely because the 45S rDNA sites were distributed in the terminal regions of all chromosomes. The 5S rDNA hybridization signals were also detected at interstitial positions in E. sellowiana, E. geniculata, E. subarticulata and E. maculosa, but, in the last two species, 5S positioning was associated with chromosome rearrangements (Da Silva et al. 2005, 2008b, respectively). As E. sellowiana and E. geniculata exhibit regular meiosis (Da Silva et al. 2010), the interstitial positioning of 5S rDNA in these species could not be explained by agmatoploidy and symploidy, but possibly by inversion.
Here, we completed a comparative analysis of a large number of Eleocharis species: about 30% of Brazilian species in three subgenera. Our study showed that 45S rDNA is more variable in the number of sites than 5S, but it is unclear why the 45S rDNA sites never appear at interstitial positions, as observed with the 5S rDNA sites, since rearrangements are very common in the karyotype differentiation of the Cyperaceae (Vanzela et al. 2003, Da Silva et al. 2005, 2008b). Besides, it was unclear why the events that favor the multiplication and dispersion of 45S rDNA did not favor the same direction for 5S rDNA segments. In the future, efforts should be directed towards the investigation of the following aspects: (i) relationships between the location of 45S and 5S rDNA sites in other Cyperaceae groups, (ii) the functionality of 45S rDNA sites, and (iii) relationships between rDNA segments detected by FISH, pseudogenes and transposable elements.
References
Berjano R, Roa F, Talavera S, Guerra M (2009) Cytotaxonomy of diploid and polyploid Aristolochia (Aristolochiaceae) species based on the distribution of CMA/DAPI bands and 5S and 45S rDNA sites. Plant Syst Evol 280:219–227. doi:10.1007/s00606-009-0184-6
Da Silva CRM, González-Elizondo MS, Vanzela ALL (2005) Reduction of chromosome number in Eleocharis subarticulata (Cyperaceae) by multiple translocation. Bot J Linn Soc 149:457–464. doi:10.1111/j.1095-8339.2005.00449.x
Da Silva CRM, González-Elizondo MS, Rego LNAA, Torezan JMD, Vanzela ALL (2008a) Cytogenetical and cytotaxonomical analysis of some Brazilian species of Eleocharis. Aust J Bot 56:82–90. doi:10.1071/BT070170067-1924/08/010082
Da Silva CRM, González-Elizondo MS, Vanzela ALL (2008b) Chromosome reduction in Eleocharis maculosa (Cyperaceae). Cytogenetic and Genome Res 122:175–180. doi:10.1159/000163096
Da Silva CRM, Trevisan R, González-Elizondo MS, Ferreira JM, Vanzela ALL (2010) Karyotypic diversification and its contribution to the taxonomy of Eleocharis (Cyperaceae) from Brazil. Aust J Bot 58:1–12. doi:10.1071/BT091850067-1924/10/010001
De Melo NF, Guerra MS (2003) Variability of the 5S and 45S rDNA sites in Passiflora L. species with distinct base chromosome numbers. Ann Bot 92:309–316. doi:10.1093/aob/mcg138
Furuta T, Kondo K (1999) Sites of 18S–5.8S–26S rDNA sequences in diffused-centromeric chromosomes of Drosera falconerii. Chromosome Sci 3:69–73. doi:00A0408029
Gerlach WL, Bedbrook JR (1979) Cloning and characterization of ribosomal RNA genes from wheat and barley. Nucleic Acids Res 7:1869–1885. doi:10.1093/nar/7.7.1869
González-Elizondo MS, Peterson PM (1997) A classification of and key to the supraspecific taxa in Eleocharis (Cyperaceae). Taxon 46:433–449. doi:10.2307/1224386
Govaerts R, Simpson DA, Bruhl JJ, Egorova T, Goetghebeur P, Wilson K (2007) World checklist of Cyperaceae. Kew Publishing, Sedges. Kew
Guerra MS (2000) Patterns of heterochromatin distribution in plant chromosomes. Genet Mol Biol 23:1029–1041. doi:10.1590/S1415-47572000000400049
Guerra MS, García M (2004) Heterochromatin and rDNA sites distribution in the holocentric chromosomes of Cuscuta approximata Bab(Convolvulaceae). Genome 47:134–140. doi:10.1139/G03-098
Håkansson A (1958) Holocentric chromosomes in Eleocharis. Hereditas 44:531–540
Hasterok R, Maluszynska J (2000) Cytogenetic markers of Brassica napus L. chromosome. J Appl Genet 41:1–9
Hipp AL, Rothrock PE, Roalson EH (2009) The evolution of chromosome arrangements in Carex (Cyperaceae). Bot Rev 75:96–109. doi:10.1007/s12229-008-9022-8
Hoshi Y (1995) Chromosome studies in Drosera, the Droseraceae in connection with possible origin of the basic chromosome number of x = 10 well-differentiated in the North-hemisphere. Dissertation, Hiroshima University
Kwon J, Kim B (2009) Localization of 5S and 25S rRNA genes on somatic and meiotic chromosomes in Capsicum species of chili pepper. Mol Cells 27:205–209. doi:10.1007/s10059-009-0025-z
Leitch IJ, Bennett MD (1997) Polyploidy in angiosperms. Trend in Plant Science 2:470–476. doi:10.1016/S1360-1385(97)01154-0
Lim K, Wennekes J, Jong JH, Jacobsen E, Tuyl JM (2001) Karyotype analysis of Lilium longiflorum and Lilium rubellum by chromosome banding and fluorescence in situ hybridization. Genome 44:911–918. doi:10.1139/gen-44-5-911
Löve A, Löve D, Raymond M (1957) Cytotaxonomy of Carex section Cappilares. Can J Bot 35:715–761
Moscone EA, Klein F, Lambrou M, Fuchs J, Schweizer D (1999) Quantitative karyotyping and dual-color FISH mapping of 5S and 18S–25S rDNA probes in the cultivated Phaseolus species (Leguminosae). Genome 42:1224–1233. doi:10.1139/gen-42-6-1224
Pedersen C, Linde-Laursen I (1994) Chromosomal locations of four minor rDNA loci and a marker microsatellite sequence in barley. Chromosome Res 2:65–71. doi:10.1007/BF01539456
Raskina O, Barber JC, Nevo E, Belyayev A (2008) Repetitive DNA and chromosomal rearrangements: speciation-related events in plant genomes. Cytogenet Genome Res 120:351–357. doi:10.1159/000121084
Rego LNAA, Da Silva CRM, Torezan JMD, Gaeta ML, Vanzela ALL (2009) Cytotaxonomical study in Brazilian species of Solanum, Lycianthes and Vassobia (Solanaceae). Plant Syst Evol 279:93–102. doi:10.1007/s00606-009-0149-9
Roalson EH (2008) A synopsis of chromosome number variation in the Cyperaceae. Bot Rev 74:209–393. doi:10.1007/s12229-008-9011-y
Schubert I, Wobus U (1985) In situ hybridization confirms jumping nucleolus organizing regions in Allium. Chromosoma 92:143–148. doi:10.1007/BF00328466
Schweizer D, Loidl J (1987) A model for heterochromatin dispersion and the evolution of C-banded patterns. Chromosom today 9:61–74
Vanzela ALL, Cuadrado A, Jouve N, Luceño M, Guerra M (1998) Multiple locations of the rDNA sites in holocentric chromosomes of Rhynchospora (Cyperaceae). Chromosome Res 6:345–349. doi:10.1023/A:1009279912631
Vanzela ALL, Luceño M, Guerra M (2000) Karyotype evolution and cytotaxonomy in Brazilian species of Rhynchospora Vahl (Cyperaceae). Bot J Linn Soc 134:557–566. doi:10.1006/bojl.2000.0352
Vanzela ALL, Cuadrado A, Guerra M (2003) Localization of 45S rDNA and telomeric sites on holocentric chromosomes of Rhynchospora tenuis Link (Cyperaceae). Genet Mol Biol 26:199–201. doi:10.1590/S1415-47572003000200014
Yano O, Katsuyama T, Tsubota H, Hoshino T (2004) Molecular phylogeny of Japanese Eleocharis (Cyperaceae) based on ITS sequence data, and chromosomal evolution. J Plant Res 117:409–419. doi:10.1007/s10265-004-0173-3
Acknowledgments
The authors would like to thank the Brazilian agencies Fundação Araucária and CNPq for financial support. Dr. A. Leyva helped with English editing of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
da Silva, C.R.M., Quintas, C.C. & Vanzela, A.L.L. Distribution of 45S and 5S rDNA sites in 23 species of Eleocharis (Cyperaceae). Genetica 138, 951–957 (2010). https://doi.org/10.1007/s10709-010-9477-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10709-010-9477-5