Abstract
Aristolochia is the largest genus of the family Aristolochiaceae and the only one with large chromosome number variation. A combination of fluorochrome banding and in situ hybridization of 5S and 45S rDNA probes was used to evaluate the structural karyotype variability of representatives of two subgenera: Siphisia, which seems to have a single chromosome number (2n = 32), probably derived from an old polyploidization event, and Aristolochia, including the Old World section Diplolobus and the New World Gymnolobus. Based on chromosome morphology and on the degree of diploidization of rDNA sites, A. serpentaria (Siphisia) was identified as an old hexaploid, whereas A. paucinervis (Diplolobus) seemed to be a recent hexaploid (2n = 34). The karyotypes of the five analyzed species of section Gymnolobus were structurally more stable than those from Diplolobus, which varied considerably in the type of heterochromatin, chromosome number, and morphology. These data indicate that fluorochrome banding and rDNA localization may substantially improve the cytotaxonomical analysis of this genus.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Aristolochia L. “sensu lato” (Aristolochiaceae, Aristolochioideae) is the largest and most diverse genus of the family, consisting of about 500 species characterized by their peculiar, zygomorphic tubular flowers that exhibit a particular fly-trap mechanism of pollination (Sakai 2002). Species of the genus are generally perennial herbs, shrubs, or lianas, and are distributed mainly in tropical regions, although they are also found in subtropical or temperate habitats (De Groot et al. 2006). However, the circumscription of this genus is unclear and several authors have proposed up to 15 segregates from Aristolochia s.l. (Wanke et al. 2006). Among the different taxonomic frameworks suggested for the genus Aristolochia s.l., it is noteworthy the recognition of three subgenera: Siphisia, Pararistolochia, and Aristolochia (González and Stevenson 2000). The latter is the largest and comprehends the section Diplolobus, composed of ca. 120 species from Europe, N. and E. Africa, Asia, and N. Australia, and section Gymnolobus, with ca. 210 species from the New World. More recent studies propose that the genus Aristolochia s.l. consists of two major lineages, divided into four genera: Endodeca and Isotrema within the subtribe Isotrematinae (previously treated as subgenus Siphisia) and Pararistolochia and Aristolochia “sensu stricto” within Aristolochiinae (Kelly and González 2003; Neinhuis et al. 2005). Despite the different taxonomic ranks, these systems are highly similar.
Studies on chromosome numbers have helped to characterize clades in Aristolochia s.l., with most of them characterized by a single chromosome number (Sugawara et al. 2001). Thus, within the subgenus Aristolochia, 2n = 12 is predominant in the subsection Podanthemum, 2n = 16 in the series Thyrsicae, and 2n = 14 in the subsections Aristolochia and Pentandrae. The other two subgenera, Siphisia and Pararistolochia, have rather invariable chromosome numbers of 2n = 32 and 2n = 12, respectively (Ohi-Toma et al. 2006). According to Grant (1982), the basic number in Aristolochia should be x = 7, but this assumption is now barely sustainable. The variation in chromosome number currently known is much higher than before (2n = 6, 8, 10, 12, 14, 24, 26, 28, 32, and 36) and the distribution of these numbers in the phylogenetic tree is rather complex (Ohi-Toma et al. 2006). Furthermore, the number 2n = 28, which supported Grant’s assumption of x = 7 and which was reported earlier for several Aristolochia species, is probably a miscount (Sugawara et al. 2001). The base number of a genus should parsimoniously explain the chromosome variability of that group and have a clear relationship with the closest related groups, as reviewed by Guerra (2008). However, Thottea, the sister group of the genus Aristolochia, has x = 13 (Morawetz 1985), a number found only in a single species of the section Diplolobus.
According to Sugawara et al. (2001), species of Aristolochia have, in general, symmetrical karyotypes with small (0.5–2 μm) meta or submetacentric chromosomes, so that karyotype differentiation would be practically restricted to chromosome number variation. Techniques such as chromosome banding or fluorescent in situ hybridization (FISH) have proved to be useful tools for additional karyotype differentiation, helping to clarify evolutionary and phylogenetic relationships among species (Vaio et al. 2005; Almeida et al. 2007; de Moraes et al. 2007), but, till now, they have never been applied to Aristolochia species.
Base-specific fluorochromes are generally used to identify heterochromatic regions, which, in general, consist of non-coding tandemly repetitive DNA sequences. The most used fluorochromes in plant cytogenetics are chromomycin A3 (CMA), which binds preferentially to GC-rich DNA, and 4′,6-diamidino-2-phenylindole (DAPI), which preferentially stains AT-rich regions (Schweizer 1976; Guerra 2000). FISH with rDNA probes reveals valuable additional markers for chromosome and genome characterization. Tandemly repetitive 45S rRNA genes are usually located on the secondary constriction and adjacent heterochromatin, both of which constitute the nucleolar organizer region (NOR). These sites are commonly detected with fluorochromes such as CMA+/DAPI− bands (Cabral et al. 2006; Fregonezi et al. 2006), but FISH with 45S rDNA probes provides a more reliable recognition of the nucleolar organizer regions (NORs), especially the minor and inactive sites (Brasileiro-Vidal et al. 2007). On the other hand, 5S rDNA sites are exclusively detected by FISH, as they do not generate chromosome constrictions and are rarely visible as heterochromatin blocks (Cabral et al. 2006).
In the present work, a combination of CMA/DAPI fluorochrome staining and FISH with 5S and 45S rDNA probes was used to characterize the karyotypes of ten species of Aristolochia s.l. belonging to the subgenera Siphisia and Aristolochia, aiming to understand the karyotype evolution and the taxonomic relationships among some species of this complex group.
Materials and methods
Plant material
Ten species of Aristolochia, one of them belonging to subgenus Siphisia (A. serpentaria) and nine from the subgenus Aristolochia, were analyzed. Tubers or seeds of Aristolochia species were collected and maintained in cultivation at the greenhouses of the University of Seville, Spain. Most vouchers were deposited at the SEV herbarium, University of Seville, Spain, with the exception of A. birostris, which is at the EAN herbarium, Federal University of Paraíba, Areia, Brazil. For collection localities and voucher numbers, see Table 1.
The root tips of A. brasiliensis were obtained from seeds germinating in Petri dishes, whereas for the other species, they were obtained from plants growing in pots. They were pretreated with 2 mM 8-hydroxyquinoline for 3.5 h at room temperature, fixed overnight in 3:1 ethanol–acetic acid (v/v) at room temperature, and stored at −20°C.
Fluorochrome staining
For chromosome preparations, fixed root tips were washed in distilled water, digested in 2% (w/v) cellulase Onozuka R-10 (Serva) and 20% (v/v) pectinase (Sigma), and squashed in 45% acetic acid. Coverslips were removed by freezing in liquid nitrogen and the slides were air-dried. The best slides were selected after a brief staining with a DAPI (2 mg/ml):glycerol mixture (1:1, v/v), destained in 3:1 ethanol–acetic acid for 30 min, and dehydrated in 100% ethanol for at least 2 h.
For CMA/DAPI staining, the slides were aged for 3 days and stained with 0.5 mg/ml CMA (Sigma) for 1 h and with 2 μg/ml DAPI (Sigma) for 30 min. The slides were mounted in McIlvaine’s pH 7 buffer:glycerol (1:1, v/v). Images were acquired with a Leica DM LB microscope equipped with a Cohu CCD camera and Leica QFISH software. The images were optimized for best contrast and brightness with Adobe Photoshop CS3 (Adobe Systems, Inc.). The slides were destained again and stored at −20°C until use for FISH.
Fluorescent in situ hybridization
The FISH procedure was based on Moscone et al. (1996) with minor modifications. Two rDNA probes, R2 and D2, were used to locate the 45S and 5S rDNA sites, respectively. R2 is a 6.5-kb fragment containing an 18S–5.8S–25S rDNA repeat unit from Arabidopsis thaliana, and D2 is a 500-bp fragment of the 5S rDNA repeat unit from Lotus japonicus (see Almeida et al. 2007). They were labeled using a nick translation kit (Gibco) with digoxigenin-11-dUTP (Roche) and biotin-11-dUTP (Sigma), respectively. The 5S rDNA was detected with mouse anti-biotin (Roche) and the signals were amplified with rabbit anti-mouse TRITC conjugate (Dako). The 45S rDNA was detected with sheep anti-digoxigenin FITC conjugate (Roche) and amplified with rabbit anti-sheep FITC conjugate (Dako). All preparations were counterstained with DAPI (2 μg/mL) and mounted in Vectashield (Vector). The cell images were acquired as above. Ideograms were constructed based on the analysis of at least five well-spread metaphases using Adobe Photoshop CS3 software. Chromosomes were ordered from the largest one to the shortest.
Results
Most species (A. baetica, A. brasiliensis, A. loefgrenii, A. cymbifera, A. clematitis, A. birostris, and A. gigantea) had 2n = 14 and exhibited more or less symmetrical karyotypes. Nevertheless, chromosome number 1 was conspicuously larger than the others in A. clematitis, A. birostris, and A. gigantea (Fig. 1). Among the remaining species, A. serpentaria (2n = 32) and A. paucinervis (2n = 34) were polyploids with symmetrical karyotypes (Figs. 1 and 2a, g), while A. rotunda had a reduced chromosome number (2n = 12) and a rather asymmetrical karyotype (Figs. 1 and 3a), with remarkable chromosome size variation. The smallest chromosome on each karyotype was about 1.5–2.2 μm in all of the analyzed species, whereas the largest reached 2.5–3.9 μm. Chromosome morphology as well as the positions of 5S and 45S rDNA sites and band patterns obtained with CMA and DAPI fluorochromes were summarized in the ideograms of Fig. 1. Known phylogenetic relationships among these species are shown by the adapted cladograms on the right side of this figure. Only in A. serpentaria and A. clematitis was the chromosome morphology unclear, so that the centromere position was not indicated. Likewise, in A. rotunda, the number and position of 5S rDNA sites were not reliably identified.
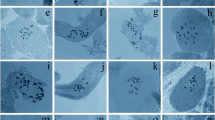
Distribution of heterochromatin and rDNA sites in the chromosome complements of species of subgenus Siphisia (a, b) and subgenus Aristolochia, section Diplolobus (c–h). a, b A. serpentaria. c, d A. baetica. e, f A. clematitis. g, h A. paucinervis. CMA (yellow), DAPI (blue), 5S rDNA (red), 45S rDNA (green). The grey color in the inserts of d–f shows DAPI pseudocolor merged with green or red images from FISH. Observe in a and b the distended secondary constriction between two chromosome parts (arrows). The arrows in c point to secondary constrictions. The arrows in b, h and the inserts of d–f show 5S or 45S rDNA sites. The bar in h represents 10 μm
Distribution of heterochromatin and rDNA sites in the chromosome complements of subgenus Aristolochia sections Diplolobus (a, b) and Gymnolobus (c–l). a, b A. rotunda. c A. birostris. d, e A. brasiliensis. f A. gigantea. g–i A. cymbifera. j–l A. loefgrenii. CMA (yellow), DAPI (blue), 5S rDNA (red), 45S rDNA (green). The grey color in the inserts of c, f show DAPI contrast merged with green or red images from FISH. Compare the centromeric heterochromatin observed as CMA+ bands in d, g, j, DAPI− bands in h, k, and deeply stained regions after FISH denaturing in e, i, l (arrows). The bar in l represents 10 μm
After CMA/DAPI double-staining, some chromosome regions were CMA+/DAPI− (i.e., brighter with CMA and duller with DAPI), whereas others were CMA−/DAPI+, and sometimes neutral for one of the fluorochromes (CMA0 or DAPI0). Figure 2a shows the secondary constriction of A. serpentaria, which is barely visualized with CMA, whereas in A. baetica (Fig. 2c), the large secondary constriction is CMA0. The number and position of bands varied among species, with CMA+ bands predominantly located at the centromeric or pericentromeric region in species of the section Gymnolobus and also possibly in A. clematitis (Figs. 2f, 3c, d, f, g, j).
On the other hand, the number of 5S and 45S rDNA sites was remarkably stable, with only one pair of sites of each rDNA family. The only exception was the polyploid A. paucinervis, which displayed two pairs of 5S rDNA and three pairs of 45S rDNA sites (Fig. 2g, h). The location of the 45S rDNA site always coincided with one of the terminal CMA+/DAPI− bands, except in A. serpentaria, where the 45S rDNA site seemed to be colocalized with a non-terminal CMA+/DAPI− band (Fig. 2a, b). The 45S rDNA site was located on the largest chromosome of all species, except for A. baetica and A. paucinervis. On the contrary, the 5S rDNA sites were always located in euchromatic CMA0/DAPI0 regions, almost always on proximal or interstitial positions.
Figures 1–3 illustrate a few other noteworthy details besides the CMA/DAPI bands and rDNA sites. In several species, the secondary constriction was very distended (Fig. 2a), and in some of them, the labeling with 45S rDNA was typically diffuse and spread over a large area in the center of the nucleus. Figure 2g illustrates the diffuse labeling found in every cell from interphase to prometaphase and most metaphase cells of A. paucinervis. Although all secondary constrictions were CMA+, when distended, they often became CMA0/DAPI− (arrows in Fig. 2c, d).
DAPI+ bands were only observed in A. rotunda and A. clematitis, which also display relatively asymmetric karyotypes. Besides the DAPI+ bands observed with CMA/DAPI staining, two other DAPI-brilliant chromosome regions were observed, which could be confused with DAPI-positive heterochromatin. One such region was observed in the prophase or prometaphase chromosomes of A. baetica and A. paucinervis, which display a sharp differentiation between more condensed and less condensed chromatin (Fig. 2d, g). However, in completely condensed metaphases, such differentiation was never observed, indicating that it is not a kind of heterochromatin but, rather, an early condensed chromatin. Some other DAPI-brilliant regions were observed in several species only after in situ hybridization. In A. brasiliensis, A. loefgrenii, and A. cymbifera, the centromeric and pericentromeric regions, which were DAPI− after CMA/DAPI staining, became more brilliant than the rest of the chromosomes after FISH (Fig. 3d, e, h, i, k, l). In these cases, the brilliant blocks are heterochromatic but not necessarily AT-rich regions. Even some CMA+ bands were observed as DAPI-brilliant blocks after in situ hybridization (Fig. 3d, e, g, i, j, l).
Discussion
The variability of chromosome numbers found in Aristolochia species in the present work is in agreement with that reported by other authors (revised by Sugawara et al. 2001; Ohi-Toma et al. 2006). Chromosome numbers from most species investigated were previously known, except for A. birostris and A. loefgrenii, which were reported here for the first time. Only the diploid numbers found for A. serpentaria (2n = 32) and A. paucinervis (2n = 34) were different from that previously registered (2n = 28 and 2n = 36, respectively) by other authors (Gregory 1956; Nardi 1984).
The chromosome number found here in A. serpentaria is in agreement with recent analyses for other species of the subgenus Siphisia. The species of this subgenus were initially admitted as having a stable chromosome number of 2n = 28 (Gregory 1956; Ma 1989). However, Sugawara and Murata (1992) and Sugawara et al. (2001) observed the number 2n = 32 in 23 species of this subgenus, including six of those previously counted as 2n = 28. The present finding of 2n = 32 in A. serpentaria, which was previously reported by Gregory (1956) as having 2n = 28, further support the conclusion that 2n = 32 is characteristic to this group (Sugawara et al. 2001).
The chromosome number 2n = 32 of Siphisia is at least twice as high as those previously found in the other two Aristolochia subgenera: Pararistolochia, with 2n = 12, and Aristolochia, with 2n = 6, 12, 14, 16 (Ohi-Toma et al. 2006). Therefore, the subgenus is most probably of polyploid origin, although the exact ploidy level is not evident, since the base number of the genus is unknown. The occurrence of two paralogous copies of the nuclear encoded genes phytochrome A, APETALA3, and PISTILLATA in some Siphisia species and a single homolog of each of these genes in diploid species of other subgenera is further evidence of the tetraploid origin of subgenus Siphisia (Stellari et al. 2004; Jaramillo and Kramer 2004; Ohi-Toma et al. 2006).
The diploid number observed here for A. paucinervis, 2n = 34, differed from that reported by Nardi (1984) for the same species (2n = 36). The different results may be due to intraspecific variation, species misidentification, or chromosome number miscount. No other species with 2n = 34 or 2n = 36 is known in the genus (Sugawara et al. 2001; Ohi-Toma et al. 2006), but its phylogenetic relationship with other species of section Diplolobus has been demonstrated (De Groot et al. 2006; Fig. 1). This species is included together with A. baetica and A. clematitis in the section Diplolobus, subsection Aristolochia, a monophyletic clade mainly with 2n = 14 and basic number x = 7 (González 1999; Ohi-Toma et al. 2006). Since 2n = 34 has been observed only in this subsection and most species of the subgenus Aristolochia have 2n = 14, the polyploidy in this case should be a derived condition of recent origin. Analysis of the ideogram of A. paucinervis depicted in Fig. 1 reveals that the chromosomes can be organized by size in groups of three (except for the two smallest ones), as would be expected for a hexaploid. The presence of three pairs of 45S rDNA and two pairs of 5S rDNA sites suggests that they have been conserved since the polyploidization event, because all diploid species exhibited only one site of each rDNA family. In general, old and well established polyploid lineages display strong reduction in the number of 5S and 45S rDNA sites as part of the diploidization process, as seems to be the case of A. serpentaria, whereas young polyploids, such as A. paucinervis, tend to conserve the additive number of rDNA sites (de Melo and Guerra 2003; Clarkson et al. 2005). These data suggest that A. paucinervis is most probably a hexaploid based on x = 6 which suffered a dysploid reduction from 2n = 36 to 2n = 34 or, alternatively, it could be a hexaploid based on x = 7, displaying a strong dysploid reduction.
Variation in chromosome size among Aristolochia species has been previously demonstrated by Ohi-Toma et al. (2006), who observed that Pararistolochia species have larger chromosomes than those of the other two subgenera. In general, the chromosomes of Aristolochia species are much smaller than those of other Aristolochiaceae genera, such as Asarum, Hexastylis, and Thottea (Soltis 1984; Morawetz 1985). In the present work, we refine the karyotypic analysis and found out that chromosomes of Aristolochia also display a considerable structural variation, both in heterochromatic bands detected by fluorochrome staining and in the position of 5S and 45S rDNA sites.
The five species of section Gymnolobus were relatively stable in relation to chromosome size and morphology, banding pattern, and rDNA sites position. The small amount of CMA-positive heterochromatin detected varied between species of this section, but it is quite possible that most chromosomes have some kind of proximal heterochromatin, as observed with DAPI after FISH. It is known that heterochromatic blocks only visible after FISH with DAPI should correspond to C-bands but are not necessarily AT-rich (Besendorfer et al. 2002). Differently from the karyotype variation found in Aristolochia, specially within section Diplolobus, the other two genera of the family investigated by C-banding, Thottea and Asarum, displayed a fairly stable banding pattern (Morawetz 1985; Na and Kondo 1994).
The 5S rDNA site was localized adjacent to the 45S rDNA site in only three species of section Gymnolobus and in no other species investigated. It seems quite possible that the 5S–45S rDNA linkage observed in A. cymbifera, A. brasiliensis, and A. loefgrenii is a synapomorphy, indicating that these three species are more closely related to each other than to the rest of the section. In agreement with this result, the molecular phylogeny of Aristolochia species based on nuclear and plastidial DNA sequences suggests that at least A. cymbifera and A. brasiliensis are closer to each other than to A. gigantea, which has the 5S and 45S rDNA sites in different chromosome pairs (Fig. 1; Ohi-Toma et al. 2006).
Differently from the 45S rDNA site, which often seems to change the position in the karyotype of closely related species, the 5S rDNA site is most commonly a stable chromosome marker (Fregonezi et al. 2006; Pedrosa-Harand et al. 2006; Weiss-Schneeweiss et al. 2008). In Aristolochia, however, the 5S rDNA site position was only conserved in three species of the Gymnolobus section. Although translocations, inversions, and other chromosomal rearrangements may explain the different positions of these sites, increasing evidence indicates that interactions between transposable elements and rDNA repeats may be involved in the redistribution of ribosomal sites (Raskina et al. 2004). The meaning of such dynamics for species differentiation is not clear, but it has been argued that they may be a significant indicator of genomic changes (Raskina et al. 2008). In the case of subgenus Aristolochia, changes in the chromosome number and morphology, as well as in the position of several chromosome marks, such as rDNA sites and different types of heterochromatic blocks, are evidence of different degrees of genome diversification between species and sections of this group.
References
Almeida CCS, Carvalho PCL, Guerra M (2007) Karyotype differentiation among Spondias species and the putative hybrid Umbu-cajá (Anacardiaceae). Bot J Linn Soc 155:541–547
Besendorfer V, Samardzija M, Zoldos V, Solic ME, Papes D (2002) Chromosomal organization of ribosomal genes and NOR-associated heterochromatin, and NOR activity in some populations of Allium commutatum Guss. (Alliaceae). Bot J Linn Soc 139:99–108
Brasileiro-Vidal AC, dos Santos-Serejo JA, Soares Filho WS, Guerra M (2007) A simple chromosomal marker can reliably distinguishes Poncirus from Citrus species. Genetica 129:273–279
Cabral JS, Felix LP, Guerra M (2006) Heterochromatin diversity and its co-localization with 5S and 45S rDNA sites in chromosomes of four Maxillaria species (Orchidaceae). Genet Mol Biol 29:659–664
Clarkson JJ, Lim KY, Kovarik A, Chase MW, Knapp S, Leitch AR (2005) Long-term genome diploidization in allopolyploid Nicotiana section Repandae (Solanaceae). New Phytol 168:241–252
De Groot H, Wanke S, Neinhuis C (2006) Revision of the genus Aristolochia (Aristolochiaceae) in Africa, Madagascar and adjacent islands. Bot J Linn Soc 151:219–238
de Melo NF, Guerra M (2003) Variability of the 5S and 45S rDNA sites in Passiflora L. species with distinct base chromosome numbers. Ann Bot 92:309–316
de Moraes AP, Soares Filho WS, Guerra M (2007) Karyotype diversity and the origin of grapefruit. Chromosome Res 15:115–121
Fregonezi JN, Fernandes T, Torezan JMD, Vieira AOS, Vanzela ALL (2006) Karyotype differentiation of four Cestrum species (Solanaceae) based on the physical mapping of repetitive DNA. Genet Mol Biol 29:97–104
González F (1999) Inflorescence morphology and the systematics of Aristolochiaceae. Syst Geogr Plant 68:159–172
González F, Stevenson DW (2000) Perianth development and systematics of Aristolochia. Flora 195:370–391
Grant V (1982) Periodicities in the chromosome numbers of the angiosperms. Bot Gaz 143:379–389
Gregory MP (1956) A phyletic rearrangement in the Aristolochiaceae. Am J Bot 43:110–122
Guerra M (2000) Patterns of heterochromatin distribution in plant chromosomes. Genet Mol Biol 23:1029–1041
Guerra M (2008) Chromosome numbers in plant cytotaxonomy: concepts and implications. Cytogenet Genome Res 120:339–350
Jaramillo MA, Kramer EM (2004) APETALA3 and PISTILLATA homologs exhibit novel expression patterns in the unique perianth of Aristolochia (Aristolochiaceae). Evol Dev 6:449–458
Kelly LM, González F (2003) Phylogenetic relationships in Aristolochiaceae. Syst Bot 28:236–249
Ma JS (1989) A revision of Aristolochia Linn. from E. & S. Asia. Acta Phytotaxon Sin 27:321–364
Morawetz W (1985) Beiträge zur Karyologie und Systematik der Gattung Thottea (Aristolochiaceae). Bot Jahrb Syst 107:329–342
Moscone EA, Matzke MA, Matzke AJM (1996) The use of combined FISH/GISH in conjunction with DAPI counterstaining to identify chromosomes containing transgene inserts in amphidiploid tobacco. Chromosoma 105:231–236
Na H, Kondo K (1994) A comparison of chromosome variability in Asarum asperum and A. hexalobum (Aristolochiaceae). Cytol 59:165–173
Nardi E (1984) The genus Aristolochia L. (Aristolochiaceae) in Italy. Webbia 38:221–300
Neinhuis C, Wanke S, Hilu KW, Müller K, Borsch T (2005) Phylogeny of Aristolochiaceae based on parsimony, likelihood, and Bayesian analyses of trnL–trnF sequences. Plant Syst Evol 250:7–26
Ohi-Toma T, Sugawara T, Murata H, Wanke S, Neinhuis C, Murata J (2006) Molecular phylogeny of Aristolochia sensu lato (Aristolochiaceae) based on sequences of rbcL, matK, and phyA genes, with special reference to differentiation of chromosome numbers. Syst Bot 31:481–492
Pedrosa-Harand A, de Almeida CCS, Mosiolek M, Blair MW, Schweizer D, Guerra M (2006) Extensive ribosomal DNA amplification during Andean common bean (Phaseolus vulgaris L.) evolution. Theor Appl Genet 112:924–933
Raskina O, Belyayev A, Nevo E (2004) Quantum speciation in Aegilops: Molecular cytogenetic evidence from rDNA cluster variability in natural populations. Proc Natl Acad Sci 101:14818–14823
Raskina O, Barber JC, Nevo E, Belyayev A (2008) Repetitive DNA and chromosomal rearrangements: speciation-related events in plant genomes. Cytogenet Genome Res 120:351–357
Sakai S (2002) Aristolochia spp. (Aristolochiaceae) pollinated by flies breeding on decomposing flowers in Panama. Am J Bot 89:527–534
Schweizer D (1976) Reverse fluorescent chromosome banding with Chromomycin and DAPI. Chromosoma 58:307–324
Soltis DE (1984) Karyotypes of species of Asarum and Hexastylis (Aristolochiaceae). Syst Bot 9:490–493
Stellari GM, Jaramillo MA, Kramer EM (2004) Evolution of the APETALA3 and PISTILLATA lineages of MADS-box-containing genes in the basal angiosperms. Mol Biol Evol 21:506–519
Sugawara T, Murata H (1992) Chromosome numbers of eight species of Aristolochia (Aristolochiaceae) from east Asia. Acta Phytotaxon Geobot 43:27–30
Sugawara T, Murata J, Wu S, Ohi T, Nakanishi T, Murata H (2001) A cytological analysis of 24 taxa in Aristolochia subgenera Siphisia and Aristolochia (Aristolochiaceae). Acta Phytotaxon Geobot 52:149–158
Vaio M, Speranza P, Valls JF, Guerra M, Mazzella C (2005) Localization of the 5S and 45S rDNA sites and cpDNA sequence analysis in species of the Quadrifaria group of Paspalum (Poaceae, Paniceae). Ann Bot 96:191–200
Wanke S, González F, Neinhuis C (2006) Systematics of pipevines: combining morphological and fast-evolving molecular characters to investigate the relationships within subfamily Aristolochioideae (Aristolochiaceae). Int J Plant Sci 167:1215–1227
Weiss-Schneeweiss H, Tremetsberger K, Schneeweiss GM, Parker JS, Stuessy TF (2008) Karyotype diversification and evolution in diploid and polyploid South American Hypochaeris (Asteraceae) inferred from rDNA localization and genetic fingerprint data. Ann Bot 101:909–918
Acknowledgments
The authors wish to thank Conselho Nacional de Desenvolvimento Científico e Tecnológico (Brazil), Fundação de Amparo à Ciência e Tecnologia de Pernambuco (Brazil), and Ministerio de Ciencia e Innovación (Spain) for their financial support, and to Dr. Leonardo Felix, Dr. Juliano Cabral, and Dr. Santiago Castroviejo for their kind help in collecting the plant material.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Berjano, R., Roa, F., Talavera, S. et al. Cytotaxonomy of diploid and polyploid Aristolochia (Aristolochiaceae) species based on the distribution of CMA/DAPI bands and 5S and 45S rDNA sites. Plant Syst Evol 280, 219–227 (2009). https://doi.org/10.1007/s00606-009-0184-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00606-009-0184-6