Abstract
Solanum comprises about 1,400 species of shrubs, trees and vines. This group is cytogenetically interesting because it possesses karyotypes apparently conserved in chromosome number and shape, but with diversity in the repetitive DNA. The objective of this study is to characterize 16 species of Solanum and two species of closely related genera (Lycianthes australe and Vassobia breviflora) using cytogenetic parameters. All the species presented 2n = 24, confirming previous chromosome counting. Additionally, nonreticulated nuclei, proximal condensation in prophase-metaphase and little variation in the karyotype symmetry were observed. Solanum corymbiflorum exhibited chromosomes approximately three times bigger in relation to the other species. GC-rich heterochromatin was preferentially located at terminal regions and AT-rich blocks always appear in the centromeric regions. The 45S rDNA sites were coincident with C/CMA3 + regions (satellites) and found in just one pair, except in S. corymbiflorum which presented two pairs. FISH with 5S rDNA showed signals in the paracentromeric region of one chromosome pair, except in S. trachytrichium and S. gemellum which showed two hybridization signals. The results point out to different ways of karyotype differentiation in Solanum and closely related genera and bring important issues on the value of the cytogenetical information for taxonomic studies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Solanaceae posses about 90 genera and 3,000 species, which have great economical importance, as Solanum tuberosum L. (potato), Solanum melogena L (eggplant) and several peppers (Capsicum L.), used as food in several cultures (D’Arcy 1991). Members of the family Solanaceae show basic chromosome number varying from x = 7 to x = 13 and occurrence of symmetrical karyotypes with predominance of meta- and submetacentric chromosomes. These features are quite evident in Capsicum L. (Moscone 1993; Moscone et al. 1993) and Nicotiana L. (Nakamura et al. 2001; Kitamura et al. 2001). The genus Solanum L., with about 1,400 species of herbs, subtrees and trees (Nee 1999; Child and Lester 2001; Bohs 2005), possess species with 2n = 2x = 24, besides polyploids with 2n = 4x = 48, 2n = 6x = 72 and 2n = 8x = 96 (Hunziker 2001).
Chromosome banding and FISH studies show that the species of Solanaceae have a wide variation of repetitive DNA occurrence and distribution. We can mention the occurrence of AT-rich heterochromatin in the largest chromosomes of Nicotiana kawakamii Y. Ohashi (Nakamura et al. 2001), the predominance of centromeric C-bands in some species of Capsicum in relation to other that are more terminal (Moscone et al. 1993), and different heterochromatin types in Cestrum L. (Fregonezi et al. 2006). The positioning of the rDNA segments has been described in S. bulbocastanum Dunal (Dong et al. 2000) and S. tuberosum L. (Komarova et al. 2004), but scarcely documented in other native species of Solanum.
Molecular phylogeny studies have changed substantially the taxonomic organization in groups close to Solanum, e.g., through the inclusion of Cyphomandra Mart. ex Sendtn. as a section of Solanum L., at the same time that genera which are morphologically closer [such as Lycianthes (Dunal) Hassl.] have been maintained out of Solanum (Olmstead and Palmer 1997; Bohs and Olmstead 1997, 1999; Olmstead et al. 1999; Bohs 2004, 2005). The cytogenetic data do not support the above mentioned taxonomic arrangements, which are mainly based in sequences of few genes. In order to contribute to a better understanding of the taxonomic relationships among Solanum and related genera, this study aimed the karyotype characterization of some species of Solanum, as compared to Lycianthes australe and Vassobia breviflora (Sendtn.) Hunz.
Materials and methods
Plant material
Seeds and samples of Solanum, Lycianthes and Vassobia were collected in different localities of Brazil: Mato Grosso do Sul, Paraná and Rio Grande do Sul states (Table 1), and cultivated in pots at Laboratório de Biodiversidade e Restauração de Ecossistemas––LABRE, CCB, UEL, Londrina, Brazil. At least three individuals from each species were used for the karyotype analyses. Vouchers are kept in the herbarium of the Universidade Estadual de Londrina (FUEL). The species were organized in sections according to Nee (1999).
Conventional staining
The method described by Guerra (1983) with modifications was used for the study of somatic chromosomes. Roots were pretreated with 2 mM 8-hydroxyquinoline for 24 h and fixed in absolute ethanol: glacial acetic acid (3:1, v:v) for 12 h and kept at −20°C until used. Root tips were digested for 1 h in a mixture of 4% (w/v) cellulase and 40% pectinase (v/v), further hydrolyzed in 1 M HCl at 60°C for 11 min, dissected in a drop of 45% acetic acid and squashed. The cover slips were removed after freezing in liquid nitrogen. The material was stained with 2% Giemsa and permanent slides mounted in Entellan. The chromosome measurements performed using the MicroMeasure 3.3 software (http://www.biology.colostate.edu), allowed to calculate the: (1) haploid set length and (2) centromere position and (3) karyotype formulae. Chromosome types were classified according to Guerra (1986).
Chromosome banding
The banding was performed as described by Vanzela et al. (2002), based in the procedure of Schwarzacher et al. (1980). Root tips were digested in an enzyme solution composed of 4% cellulase (w/v) and 40% pectinase (v/v) at 37°C and dissected in a drop of 45% acetic acid. After removal of the cover slips, the slides were aged for 3 days, and then incubated in 45% acetic acid for 10 min at 60°C, 5% barium hydroxide for 10 min at room temperature and 2 × SSC, pH 7.0, for 10 min at 60°C. The samples were washed in distilled water, air dried and stained with the fluorochromes: 0.5 mg/mL CMA3 for 1.5 h and 2 μg/mL DAPI for 30 min. Slides were mounted with a medium composed of glycerol/McIlvaine buffer (pH 7.0) 1:1, plus 2.5 mM MgCl2.
FISH
The in situ hibridization was performed according Heslop-Harrison et al. (1991) and Cuadrado and Jouve (1994), with modifications. Slides were prepared as described for banding and immediately used for FISH. The pTa71 probe containing the 45S rDNA sequence (Gerlach and Bedbrook 1979) was labeled with biotin-14-dATP by nick translation. The pTa794 probe containing the 5S rDNA sequence (Gerlach and Dyer 1980) was labeled with digoxigenin-11-dUTP by nick translation. Each slide was treated with 34 μL of hybridization mixture containing 100 ng of labeled probe (2 μL of each probe), 100% formamide (15 μL), 50% polyethylene glycol (6 μL), 20 × SSC (3 μL), 100 ng of calf thymus DNA (1 μL) and 10% SDS (1 μL). The material was denatured at 90°C for 10 min and hybridization was performed overnight at 37°C in a humidified chamber. Post-hybridization washes were carried out in 2 × SSC, 20% formamide in 0.1 × SSC, 0.1 × SSC and 4 × SSC/0.2% Tween 20, all at 42°C. The probes were simultaneously detected with a solution composed of 5% BSA, avidin-FITC conjugated and anti-dig-rhodamine conjugated (100:1:1, v:v:v), and the post-detection baths were conducted in 4 × SSC/0.2% Tween 20 at room temperature. Slides were mounted with 25 μL of a medium composed of 23 μL of DABCO solution (1,4-diaza- bicyclo (2.2.2)-octane (2,3%), 20 mM Tris HCl, pH 8.0, (2%) and glycerol (90%), in distilled water), 1 μL of 2 μg/mL DAPI and 1 μL of 50 mM MgCl2.
All the images were acquired with a Leica DM 4500 B microscope equipped with a DFC 300FX camera and the Leica IM50 4.0 software. All the images were optimized for best constrast and brightness with iGrafx Image software.
Results
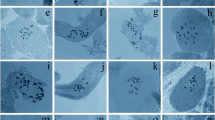
The conventional analysis showed that the species of Solanum, Lycianthes australe and Vassobia breviflora presented nonreticulated interphase nuclei, proximal condensation at prophase (Fig. 1e, q), 2n = 2x = 24, chromosomes measuring around 2 μm and predominance of meta- and submetacentrics (Figs. 1, 4; Table 1), with the exception of Solanum corymbiflorum (Sendtn.) Bohs which exhibited chromosomes with up to 6 μm (Figs. 1r, 4i; Table 1).
Conventional staining in a Solanum americanum, b S. gemellum, c S. atropurpureum, d prometaphase of S. viarum, e S. granuloso-leprosum, f S. palinacanthum, g S. pseudocapsicum, h prometaphase of S. scuticum, i prometaphase of S. variabile, j S. trachytrichium, k S. paniculatum, l S. mauritianum, m S. sisymbriifolium, n prometaphase of Vassobia breviflora, o S. guaraniticum, p S. amygdalifolium, q Lycianthes australe and r S. corymbiflorum. Arrows indicate satellites. Bar = 10 μm
In the subgenera Leptostemonum Dunal (Bitter) and Solanum there was a predominance of metacentrics in relation to submetacentrics (Fig. 4; Table 1). However, the more frequent karyotype formulae was 6 m + 6 sm, observed in S. atropurpureum Schrank, S. sisymbriifolium Lam., S. trachytrichium Bitter, S. granuloso-leprosum Dunal, S. corymbiflorum and Lycianthes australe Barboza et Hunz. (Fig. 4c, d, l, m, i, q; Table 1). In spite of similar chromosome types in these species, the haploid set size was variable, from 13.08 to 27.9 μm, except to S. corymbiflorum that showed 77.88 μm (Table 1).
Karyotypes containing a larger number of metacentrics were observed in S. paniculatum L., S. pseudocapsicum L. and S. mauritianum Scop., with 7 m + 5 sm (Fig. 4e, k, n), S. guaraniticum St.-Hil., S. scuticum M. Nee and S. amygdalifolium Steud., with 8 m + 4 sm (Fig. 4f, g, p) and in Vassobia breviflora (Sendtn.) Hunz., with 9 m + 3 sm (Fig. 4r). In those species, the haploid set size varied from 13.08 to 25.87 μm (Table 1). Only three species were not grouped: Solanum viarum with 11 m + 1 sm and 18.14 μm of haploid set size, S. americanum Mill. with 10 m + 2 sm and 22.39 μm and S. variabile Mart. with only metacentrics and 20.50 μm (Fig. 4a, j, h, respectively; Table 1). Solanum palinacanthum Dunal and S. gemellum Sendtn. also stood out because they exhibited more submetacentrics (3 m + 9 sm, 5 m + 7 sm) and higher haploid set sizes with 27.90 and 25.74 μm (Fig. 4b, o, respectively; Table 1).
The chromosome banding showed four heterochromatin distribution types. In Vassobia breviflora (Figs. 2q, 4r) and in the subgenera Leptostemonum and Solanum (S. pseudocapsicum, S. sisymbriifolium, S. americanum, S. guaraniticum and S. variabile), terminal C-CMA3 + bands in the largest chromosomes and major blocks associated to satellites were found (Figs. 2e, f, i, l, m, 4k, d, j, f, g, h). In other species, as S. atropurpureum (Figs. 2a, 4c), S. viarum Dunal (Figs. 2b, 4a), S. mauritianum (Figs. 2c, 4n), S. trachytrichium (Figs. 2d, 4l), S. granuloso-leprosum (Figs. 2h, 4m), S. amygdalifolium (Figs. 2k, 4p), S. gemellum (Figs. 2p, 4o) and Lycianthes australe (Figs. 2r, 4q), only C-CMA3 + bands associated with satellites were observed. Solanum paniculatum and S. palinacanthum, besides of C-CMA3 + associated with satellites, exhibited proximal, terminal and interstitial dots (Figs. 2n, o, 4b, e). Solanum corymbiflorum, of the section Pachyphylla Dunal (subgenus Bassovia (Aubl.) Bitter), with the largest chromosomes, showed only two pairs with terminal C-CMA3 + bands (Figs. 2t, 4i). Except Lycianthes australe (Figs. 2s, 4q) which showed centromeric C-DAPI+ bands, the remaining species did not present this heterochromatin type.
Chromosome banding in species of Solanum, Vassobia breviflora and Lycianthes australe. C-CMA3 banding in Solanum artropurpureum (a), S. viarum (b), S. mauritianum (c), S. trachytrichium (d), S. pseudocapsicum (e), S. sisymbriifolium (f), S. granuloso-leprosum (h), S. americanum (i), S. amygdalifolium (k), S. guaraniticum (l), S. variabile (m), S. paniculatum (n), S. palinacanthum (o), S. gemellum (p), Vassobia breviflora (q), Lycianthes australe (r), S. corymbiflorum (t). C-DAPI banding in S. sisymbriifolium (g), S. pseudocapsicum (j), S. corymbiflorum (u), see negative bands (arrows), Lycianthes australe (s), with centromeric C-DAPI positive bands. Bar = 10 μm
The double FISH with 45S and 5S rDNA probes always showed two terminal sites of 45S rDNA and two terminal or pericentromeric hybridization sites of 5S rDNA (Fig. 3). The exceptions were Vassobia breviflora (Figs. 3p, 4r) and S. corymbiflorum (Figs. 3r, 4i), which exhibited two pairs with terminal 45S rDNA sites and two signals of 5S rDNA. In all cases, the hybridization sites with 45S rDNA probes were coincident with satellites detected by C-CMA3 + banding (Fig. 4). However, the pair bearer of 45S rDNA varied among species. These sites were located in the pair ten in Solanum sections Acanthophora, Pachyphylla and Brevantherum, in the pair 11 at section Torva and Vassobia breviflora and in different pairs at the remaining groups (Fig. 4). The hybridization signals obtained with 5S rDNA probe were always located in the pair nine in Solanum viarum, S. atropurpureum, S. palinacanthum, S. scuticum, S. guaraniticum, S. americanum and S. pseudocapsicum. However, the pair nine was not constant in shape, being meta or submetacentric, and the 5S rDNA location was terminal or interstitial, independent of the chromosome shape (Figs. 3a–e, g, i, 4a–c, f–g, j–k). The remaining species showed hybridization signals in different chromosome pairs, e.g., Vassobia breviflora in the pair six (Figs. 3p, 4r), Solanum amygdalifolium and Lycianthes australe in the pair seven (Figs. 3j, q, 4p, q), S. paniculatum and S. corymbiflorum in the pair eight (Figs. 3f, r, 4e, i), S. sisymbriifolium and S. variabile in the pair ten (Figs. 3h, k, 4d, h), S. granuloso-leprosum and S. mauritianum in the pair 12 (Figs. 3n, o, 4m, n). On the other hand, Solanum trachytrichium and S. gemellum exhibited four 5S rDNA hybridization signals. In the first one, in the interstitial position of the pair five and in the terminal position of the pair eight (Figs. 3l, 4l). In Solanum gemellum, in the interstitial position of the pair eight and in the terminal position of the pair ten (Figs. 3m, 4o).
Double FISH with rDNA probes in species of Solanum, Vassobia breviflora and Lycianthes australe. Note two and four 45S rDNA sites (the largest signals) always located in terminal regions of the chromosomes. Arrows indicate always two 5S rDNA hybridization signals in almost species, excepted for S. trachytrichium (l) and S. gemellum (m). In S. gemellum only two sites of 5S rDNA are indicate. FISH in Solanum viarum (a), S. artropurpureum (b), S. palinacanthum (c), S. scuticum (d), S. guaraniticum (e), S. paniculatum (f), S. americanum (g), S. sisymbriifolium (h), S. pseudocapsicum (i), S. amygdalifolium (j), S. variabile (k), S. trachytrichium (l), S. gemellum (m), S. granuloso-leprosum (n), S. mauritianum (o), Vassobia breviflora (p), Lycianthes australe (q) and S. corymbiflorum (r). Bar = 10 μm
Idiograms with the physical location of repetitive DNA segments in Solanum species, Lycianthes australe and Vassobia breviflora. The species of Solanum are organized according to Nee (1999). Subgenus Leptostemonum section Acanthophora: Solanum viarum (a), S. palinacanthum (b), S. artropurpureum (c). Section Melongena: S. sisymbriifolium (d). Section Torva: S. paniculatum (e), S. guaraniticum (f), S. scuticum (g), S. variabile (h). Subgenus Bassovia section Pachyphylla: S. corymbiflorum (i). Subgenus Solanum section Solanum: S. americanum (j). Section Geminata: S. pseudocapsicum (k), S. trachytrichium (l): Section Brevantherum: S. granuloso-leprosum (m), S. mauritianum (n). Section Extensum: S. gemellum (o). Section Dulcamara: S. amygdalifolium (p). Lycianthes australe (q) and Vassobia breviflora (r). Bar = 10 μm
Discussion
The chromosome number 2n = 24 was found in all the studied species and is in accordance with those available at Index to Plant Chromosome Numbers (http://mobot.mobot.org/W3T/Search/ipcn.html), except to Solanum scuticum, S. gemellum, S. amygdalifolium and Lycianthes australe, which are new records. About 50% of the species of Solanaceae present chromosome numbers multiple of x = 12, which is considered the basic chromosome number (Hunziker 2001). This number is predominant in the subfamily Solanoideae, in the genera Triguera, Lycopersicon, Lycianthes and Vassobia (Hunziker 2001), as well as in Solanum (Olmstead et al. 1999; Hunziker 2001; Acosta et al. 2005). Only two species of the section Acanthophora showed 2n = 22, Solanum mammosum L. and S. platense Dieckman (Chiarini and Bernardello 2006). Numbers different from x = 12 have been reported in other groups, as in the subfamily Cestroideae (cf. Hunziker 2001).
The conventional analysis also showed invariably nonreticulated nuclei and proximal condensation at prophase. This is in accordance with Guerra (2000) that suggests that small chromosomes (<3 μm) are associated with these features. In relation to chromosome sizes, our results showed that except S. corymbiflorum, whose haploid set size was 77.8 μm, the remaining species presented small chromosomes with haploid set size varying from 13.08 to 27.9 μm, about three times smaller than in S. corymbiflorum. A wide variation in the haploid complement size of Solanum and Lycianthes (from 16.30 to 38.75 μm) has been also reported by Bernardello and Anderson (1990), Bernardello et al. (1994), Acosta et al. (2005) and Chiarini and Bernardello (2006). All the species studied here exhibited predominance of meta and submetacentric chromosomes, with the smallest being always submetacentric, except Solanum variabile and S. americanum that showed the smallest metacentric. The tendency of chromosome size reduction accompanied by a change in the centromere position was also observed in the idiograms reported by Bernardello and Anderson (1990), Pringle and Murray (1991), Bernardello et al. (1994), Acosta et al. (2005) and Chiarini and Bernardello (2006). However, our results showed that there is no correlation between karyotype features and groupings and section of Nee (1999).
Differences on the chromosome size have been useful to delimit some groups in Solanaceae. The section Pachyphylla of Solanum possess larger chromosomes in relation to other groups of the genus. Besides, some morphological features, such as anthers with thickened and glandular connectives, were also considered important to maintain this group as an independent genus, Cyphomandra (Bohs 1989, 1994; Hunziker 2001; Child and Lester 2001). However, phylogenetical analysis based in the sequence of conserved chloroplast genes, ndhF and rbcL, focusing Solanum and closely related genera, indicate that Cyphomandra can be included in Solanum (Bohs 1995; Olmstead and Palmer 1997; Bohs and Olmstead 1997, 1999; Olmstead et al. 1999; Bohs 2004, 2005). Solanum corymbiflorum, previously named Cyphomandra corymbiflora Sendtn. exhibits much bigger chromosomes and the largest haploid set size. The conventional cytogenetic analysis indicate that this species can be maintained in a genus separated of Solanum (Cyphomandra), as proposed by Child and Lester (2001), Moscone (1992) and Hunziker (2001). If Cyphomandra is included in Solanum, as pointed out by the above mentioned phylogenetic studies, therefore we will have to admit that the karyotypes of the section Pachyphylla become differentiated from the other species of Solanum by a substantial DNA accumulation, without important changes in the chromosome number and shape (see Fig. 4i).
The location of 45S ribosomal genes follows the common distribution pattern found in most plant species, always occurring in the terminal chromosome regions. This has been also reported in other genera of Solanaceae as Solanum bulbocastanum (Dong et al. 2000), Capsicum (Moscone et al. 1995), Nicotiana (Lim et al. 2000), and Cestrum (Fregonezi et al. 2006). In spite of the constancy of 45S rDNA location, the number of sites was variable, since in S. corymbiflorum and Vassobia breviflora four sites were observed. However, this is not an innovation in Solanaceae, as long as the number of hybridization sites in Cestrum using of the same probe also varied (Fregonezi et al. 2006). FISH with 5S rDNA probe showed that in almost all cases the signals appear in the short arm of the smallest chromosomes of the complement, except S. trachytrichium and S. scuticum where signals in the large arm were found. Solanum trachytrichium and S. gemellum exhibited four hybridization sites and the remaining species two signals of 5S rDNA. In all cases, the signals appeared in the smaller chromosomes of the set. On the other hand, Dong et al. (2000) showed paracentromeric 5S rDNA sites in the major chromosome of the set in S. bulbocastanum. In other genera, as Nicotiana (Lim et al. 2000; Kitamura et al. 2001), Lycopersicon (Lapitan et al. 1991) and Cestrum (Fregonezi et al. 2006) the 5S rDNA were also located in the paracentromeric region. These data suggest that the 45S and 5S rDNA segments do not present a conserved location and number of sites among species of Solanum and other Solanaceae. This feature seems to be common to other plant groups, e.g., Sapindaceae (Urdampilleta et al. 2006).
The C-banding followed by CMA3/DAPI staining, as described by Vanzela and Guerra (2000), was useful to determinate different heterochromatin type distribution (see also Fregonezi et al. 2006). The different bands can be grouped in: (1) terminal C-CMA3 + bands in all the chromosomes, besides a block associated to satellites, (2) only C-CMA3 + bands associated to satellites, (3) proximal, terminal and interstitial C-CMA3 + dots, besides those C-CMA3 + associated to satellites, and (4) centromeric C-DAPI+ bands. The presence of C-CMA3 + terminal dots can approximate species of distinct groups, as S. sisymbriifolium, S. paniculatum, S. guaraniticum, S. scuticum, S. variabile (sections Melongena and Torva), S. americanum (section Solanum), S. pseudocapsicum (section Geminata) and Vassobia breviflora. The occurrence of interstitial dots in S. palinacanthum indicates that the section Acanthophora is apparently more diverse. Fregonezi et al. (2006) studied species of Cestrum and detected different repetitive DNA families (dots), occupying different equilocal/equidistant regions, as well as observed in Solanum and Vassobia studied here. A similar behavior of bands distribution was reported to Solanum section Petota (Pijnaker and Ferwerda 1984), Capsicum (Moscone et al. 1993) and Lycopersicon (Peterson et al. 1996).
To conclude, the results of chromosome banding and FISH with rDNA probes show that they can be useful to characterize species and possibly populations and that the cytogenetical tools can complement the phylogenetical studies. We considered here the karyotypes of Vassobia breviflora and Lycianthes australe as external members for comparison with Solanum. Vassobia breviflora presents anthers with longitudinal dehiscence and ovary with a nectary, and possess karyotype features similar to species of subgenera Melongena, Torva and Solanum. Lycianthes and Solanum are considered separate genera, in spite of possessing similar morphologic features as poricidal anthers and ovary without a nectary (Hunziker 2001). They also share similar cytogenetic features, as 2n = 24, nonreticulated nuclei, proximal condensation at prophase, small chromosomes, similar karyotype structure and occurrence of two terminal 45S rDNA sites. If the same comparisons are made between Cyphomandra and Solanum, where there are more morphological differences and cytogenetical contrasts that in Solanum and Lycianthes (in spite of the occurrence of proximal C-DAPI bands in Lycianthes), therefore Cyphomandra should be also maintained separated of Solanum, as Lycianthes is. In spite of the great importance and contributions of the molecular phylogeny studies, we have doubts about the sustentation of the recent arrangement in Solanum, which is based on only a few chloroplast genes. We consider that molecular data should be applied along with other tools, including morphological, anatomical, chemical and cytogenetical information.
References
Acosta MC, Bernardello G, Guerra M, Moscone EA (2005) Karyotype analysis in several South American species of Solanum and Lycianthes rantonnei (Solanaceae). Taxon 54:713–723
Bernardello LM, Anderson GJ (1990) Karyotypic studies in Solanum section Basarthrum (Solanaceae). Amer J Bot 77:420–431
Bernardello LM, Heiser CB, Piazzano M (1994) Karyotype studies in Solanum section Lasiocarpa (Solanaceae). Amer J Bot 81:95–103
Bohs L (1989) Solanum allophyllum (Miers) Standl. and the generic delimitation of Cyphomandra and Solanum (Solanaceae). Ann Missour Bot Gard 76:1129–1140
Bohs L (1994) Cyphomandra (Solanaceae). Flora Neotropica, New York, vol 63, Monograph, pp 1–175
Bohs L (1995) Transfer of Cyphomandra (Solanaceae) and its species to Solanum. Taxon 44:583–587
Bohs L (2004) A Chloroplast DNA Phylogeny of Solanum Section Lasiocarpa. Syst Bot 29:177–187
Bohs L (2005) Major clades in Solanum based on ndhF sequence data. In: Keating RC, Hollowell VC, Croat TB (eds) A festschrift for William G. D’Arcy: the legacy of a taxonomist. Monographs in systematic botany from the Missouri Botanical Garden, vol 104. Missouri Botanical Garden Press, St. Louis, pp 27–49
Bohs L, Olmstead R (1997) Phylogenetic relationships in Solanum (Solanaceae) based on ndhF sequences. Syst Bot 22:5–17
Bohs L, Olmstead R (1999) Solanum phylogeny inferred from chloroplast DNA sequence data. In: Nee M, Symon DE, Lester RN, Jessop JP (eds) Solanaceae IV: advances in biology and utilization. The Royal Botanic Gardens, Kew, pp 97–109
Chiarini F, Bernardello G (2006) Karyotype Studies in South American species of Solanum subgen. Leptostemonum (Solanaceae). Plant Biol 8:486–493
Child A, Lester RN (2001) Synopsis of the genus Solanum L. and its infrageneric taxa. In: van der Berg RG, Barendse GWN, van der Weerden GM, Mariani C (eds) Solanaceae V: advances in taxonomy and utilization. Nijmegen University Press, Nijmegen, pp 39–52
Cuadrado A, Jouve N (1994) Highly repetitive sequences in B chromosomes of Secale cereale revealed by fluorescence in situ hybridization. Genome 37:709–712
D’Arcy WG (1991) The Solanaceae since 1976, with a review of its biogeography. In: Hawkes JG, Lester RN, Nee M, Estrada N (eds) Solanaceae III––taxonomy, chemistry, evolution. The Royal Botanic Garden and The Linnean Society of London, Kew, pp 75–137
Dong F, Song J, Naess SK, Helgeson JP, Gebhardt C, Jiang J (2000) Development and applications of a set of chromosome-specific cytogenetic DNA markers in potato. Theor Appl Genet 101:1001–1007
Fregonezi JN, Fernandes T, Torezan JMD, Vieira AO, Vanzela ALL (2006) Karyotype differentiation of four Cestrum species (Solanaceae) based on physical mapping of repetitive DNA. Gen Mol Biol 29:97–104
Gerlach WL, Bedbrook JR (1979) Cloning and characterization of ribosomal RNA genes from wheat and barley. Nucleic Acids Res 7:1869–1885
Gerlach WL, Dyer TA (1980) Sequence organization of the repeated units in the nucleus of wheat which contain 5S-rRNA genes. Nucleic Acids Res 8:4851–4865
Guerra MS (1983) O uso do Giemsa em Citogenética Vegetal: comparação o entre a coloração simples e o bandamento. Ciência e Cultura 35:190–193
Guerra M (1986) Reviewing the chromosome nomenclature of Levan et al. Rev Bras Gen 9:741–743
Guerra M (2000) Patterns of heterochromatin distribution in plant chromosomes. Gen Mol Biol 23:1029–1041
Heslop-Harrison JS, Schwarzacher T, Anamthewat-Jonsson K, Leitch AR, Shi M, Leitch IJ (1991) In situ hybridization with automated chromosome denaturation. Technique 3:109–116
Hunziker AT (2001) Genera Solanacearum. The genera of Solanaceae illustrated, arranged according to a new system. Ruggell ARG. GantnerVerlag, Liechtenstein. 500 p, 136 fig
Kitamura S, Inoue M, Shikazono N, Tanaka A (2001) Relationship among Nicotiana species revealed by the 5S rDNA spacer sequence and fluorescence in situ hybridization. Theor Appl Genet 103:678–686
Komarova NY, Grabe T, Huigen DJ, Hemleben V, Volkov RA (2004) Organization, differential expression and methylation of rDNA in artificial Solanum allopolyploids. Pl Mol Biol 56:439–463
Lapitan NLV, Ganal MW, Tanksley SD (1991) Organization of the 5S ribosomal RNA genes in the genome of tomato. Genome 34:509–514
Lim KY, Matyasek R, Lichtenstein CP, Leitch AR (2000) Molecular cytogenetic analyses and phylogenetic studies in the Nicotiana section Tomentosae. Chromosoma 109:245–258
Moscone EA (1992) Estudios de cromosomas meióticos en Solanaceae de Argentina. Darwiniana 31:261–297
Moscone EA (1993) Estudios cromosómicos en Capsicum (Solanaceae) II. Analisis cariotipico de C. parvifolium y C. annum var. annum. Kurtiziana 22:9–18
Moscone EA, Lambrou M, Hunziker AT, Ehrendorfer F (1993) Giemsa C-banded karyotypes in Capsicum (Solanaceae). Pl Syst Evol 186:213–229
Moscone EA, Loidl J, Ehrendorfer F, Hunziker AT (1995) Analysis of active nucleolus organizing regions in Capsicum (Solanaceae) by silver staining. Amer J Bot 82:276–287
Nakamura R, Kitamura S, Inoue M, Ohmido N, Fukui K (2001) Karyotype analysis of Nicotiana kawakamii Y. Ohashi using DAPI banding and rDNA FISH. Theor Appl Genet 102:810–814
Nee M (1999) Synopsis of Solanum in the New World. In: Nee M, Symon DE, Lester RN, Jessop JP (eds) Solanaceae IV: advances in biology and utilization. The Royal Botanic Gardens, Kew, pp 285–333
Olmstead R, Palmer J (1997) Implications for the phylogeny, classification and biogeography of Solanum from cpDNA restriction-site variation. Syst Bot 22:19–29
Olmstead R, Sweere JA, Spangler RE, Bohs L, Palmer JD (1999) Phylogeny and provisional classification of the Solanaceae based on chloroplast DNA. In: Nee M, Symon DE, Lester RN, Jessop JP (eds) Solanaceae IV: advances in biology and utilization. The Royal Botanic Gardens, Kew, pp 111–137
Peterson DG, Stack SM, Price HJ, Johnston JS (1996) DNA content of heterochromatin and euchromatin in tomato (Lycopersicon esculentum) pachytene chromosomes. Genome 39:77–82
Pijnaker LP, Ferwerda MA (1984) Giemsa C-banding of potato chromosomes. Can J Gen Cyt 26:415–419
Pringle GJ, Murray BG (1991) Karyotype diversity and nuclear DNA variation in Cyphomandra. In: Hawkes JG, Lester RN, Nee M, Estrada N (eds) Solanaceae III––taxonomy, chemistry, evolution. The Royal Botanic Garden and The Linnean Society of London, Kew, pp 247–252
Schwarzacher TP, Ambros P, Schweizer D (1980) Application of Giemsa banding to orchid karyotype analysis. Pl Syst Evol 134:293–297
Stebbins GL (1971) Chromosomal evolution in higher plants. Addison-Wesley, Reading, 216 p
Urdampilleta JD, Ferrucci MS, Torezan JMD, Vanzela ALL (2006) Karyotype relationships among four South American species of Urvillea (Sapindaceae: Paullinieae). Pl Syst Evol 258:85–95
Vanzela ALL, Guerra M (2000) Heterochromatin differentiation in holocentric chromosomes of Rhynchospora (Cyperaceae). Gen Mol Biol 23:453–456
Vanzela ALL, Ruas CF, Oliveira MF, Ruas PM (2002) Characterization of diploid, tetraploid and hexaploid Helianthus species by chromosome banding and FISH with 45S rDNA probe. Genetica 114:105–111
Acknowledgments
The authors thank the Brazilian agencies Fundação Araucária, IAP-SEMA and CNPq for financial support, Prof. Dra. Lilian Auler Mentz for species identification and Dra. Maria Socorro González-Elizondo for comments on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Rego, L.N.A.A., da Silva, C.R.M., Torezan, J.M.D. et al. Cytotaxonomical study in Brazilian species of Solanum, Lycianthes and Vassobia (Solanaceae). Plant Syst Evol 279, 93–102 (2009). https://doi.org/10.1007/s00606-009-0149-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00606-009-0149-9