Abstract
Verticillium wilt, caused by Verticillium dahliae, is a soil-borne disease of the cultivated eggplant (Solanum melongena). The accession PI388846 from a wild species S. linnaeanum, shows resistance to Verticillium wilt. The introgression of its disease resistance gene into cultivated eggplants would allow for breeding disease resistant eggplants. In this study, interspecific hybridization and subsequent backcrossing between PI388846 and cultivated eggplants were performed. The results showed that Verticillium wilt resistance was successfully introduced into the cultivated eggplants, and the agronomic traits of the interspecific hybrid progeny were improved by continuous backcrossing with the cultivated eggplants. In addition, a gene specific marker for the Ve homolog in PI388846 was developed to detect Verticillium wilt resistance in the backcross population. The results represent a positive beginning for the genetic enhancement of cultivated eggplants for Verticillium wilt resistance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The cultivated eggplant (Solanum melongena) is a popular vegetable crop worldwide, especially in Asia and Africa (Collonnier et al. 2001), with China being the largest producer and consumer of eggplants. In China, eggplants are planted both in open fields and in greenhouses. In fact, it is one of China’s most widely grown greenhouse vegetables. Verticillium wilt is a soil-borne disease caused by Verticillium dahlia (V. dahlia) that greatly reduces the yield and quality of eggplants, especially in greenhouse production. Verticillium wilt is a vascular wilt disease that cannot be sufficiently controlled by many fungicides, and there are few fungicides that can rid infected eggplants of the disease (Fradin et al. 2009). Other approaches, such as grafting and soil fumigation, are effective. However, they are either expensive or harmful to the environment (Lee 1994; Fradin and Thomma 2006; King et al. 2010). Therefore, the best way to control Verticillium wilt is to develop disease resistant varieties.
Eggplants have diversified genetic resources, including accessions of S. melongena and other closely related Solanum species. S. melongena accessions display varying levels of susceptibility to Verticillium wilt. However, sources of resistance to V. dahliae were found in some wild Solanum species related to eggplants, such as S. linnaeanum, S. aculeatissimum, S. sisymbrifolium and S. torvum (Collonnier et al. 2001). Genetic relationship analysis based on amplified fragment length polymorphisms indicated that S. linnaeanum is closely related to S. melongena (Mace et al. 1999). Due to the cross-incompatibility problem between the above wild Solanum species and S. melongena, fertile F1 hybrids were only obtained from a cross between S. linnaeanum and S. melongena (Collonnier et al. 2001). S. linnaeanum has been used to construct a comparative genetic linkage map and analyze the quantitative trait loci of morphological traits (Doganlar et al. 2002a, b; Frary et al. 2003). By screening a naturally infected field between 1998 and 2001, it was observed that the resistance to V. dahliae of backcrossed progeny of S. linnaeanum and a cultivated eggplant increased by about 60 % (Acciarri et al. 2001; Sunseri et al. 2003). Therefore, it is possible to use S. linnaeanum to improve the resistance of eggplants to Verticillium wilt.
Verticillium wilt also affects other economically important crops such as tomato (S. lycopersicum), potato (S. tuberosum), cotton (Gossypium spp.), alfalfa (Medicago sativa) and mint (Mentha piperita) (Tjamos and Beckman 1989). Two genes, Ve1 and Ve2, were cloned in tomatoes, and further research showed that Ve1, but not Ve2, provides resistance in tomatoes against race 1 strains of V. dahliae and V. albo-atrum (Diwan et al. 1999; Kawchuk et al. 2001; Fradin et al. 2009). In potatoes, cleaved amplified polymorphic sequence markers were generated according to the homologs of tomato Ve1 and Ve2, and were successfully used to test Verticillium wilt resistance of potato germplasm (Bae et al. 2008; Uribe et al. 2014). In addition, three Ve homologs, mVe1, Vr1 and Gbve1, were responsible for Verticillium wilt resistance in mint, lettuce and cotton, respectively (Vining and Davis 2009; Hayes et al. 2011; Zhang et al. 2012). These results indicated that Ve gene-mediated resistance might play an important role for plant resistance to Verticillium wilt.
Eggplant belongs to the Solanaceae family, which includes some other economically important crops such as potato (S. tuberosum), tomato (S. lycopersicum), and pepper (Capsicum annuum). Comparative genomic studies showed that the genomes of Solanaceae species are relatively conserved, and few genome rearrangements and duplications have occurred during their evolution (Doganlar et al. 2002a, b; Mueller et al. 2005; Daniell et al. 2006; Wu et al. 2009a, b). Genome information about eggplants and their related species is limited, but the gene information of other Solanaceae species can provide important references. The Ve genes are responsible for Verticillium wilt resistance in tomato and other species. Therefore, the Ve homolog in PI388846 may have a similar function.
Here, we reported an interspecific hybridization followed by backcrossing between S. linnaeanum and S. melongena. By screening the disease resistance and agronomic traits, the Verticillium wilt resistance was successfully transferred to cultivated eggplants. Additionally, a gene specific marker for the Ve homolog in PI388846 was developed to test Verticillium wilt resistance in the backcross population. The result will be useful for the genetic enhancement of cultivated eggplants for Verticillium wilt resistance.
Materials and methods
Plant materials
S. linnaeanum accession PI388846 introduced from the United States Department of Agriculture (USDA) and four V. dahliae susceptible inbred lines of S. melongena (EP12, EP15, EP21 and D13) were used as the hybridization parents. Twenty-one Chinese landraces including ‘Suzhouniujiao’, ‘Xuzhouchangqie’, ‘Qiyeqie’, ‘Yangzhoutangqie’, ‘Chengdumoqie’, ‘Duanbahei’, ‘Sanyueqie’, ‘Xianlvqie’, ‘Bengbuqingchangqie’, ‘Wuhubaiqie’, ‘Hangzhouhongqie’, ‘Pingfuxiaoqie’, ‘Ningbotengqie’, ‘Erminqie’, ‘Jinanliuyeqie’, ‘Juxianhongqie’, ‘Panqie’, ‘Zimiantiao’, ‘Anyangdahong’, ‘Wangbuzichangqie’ and ‘Qixianheiyouguan’ were used to verify the gene specific marker for the Ve homolog in S. linnaeanum. All these accessions of S. melongena are susceptible to V. dahliae. A modified cetyltrimethylammonium bromide method (Murray and Thompson 1980) was used to isolate DNA from fresh leaf tissues.
Interspecific hybridization and backcrossing
PI388846 was used as the maternal parent, and four inbred lines of S. melongena were used as paternal parents. Hybridization was then performed. Briefly, PI388846 flowers were manually emasculated and bagged 2 days before anthesis. Pollination was repeated every day from 2 days before anthesis to the day of anthesis. For backcrossing, EP15 was used as the recurrent paternal parent because it was the only cross with success. From the BC1 to BC4 generations, the plants were inoculated with V. dahliae, only resistant plants with comparatively better agronomic traits were selected as maternal parents for further backcrossing.
Verticillium wilt resistance screening
Cultivated eggplant lines and the backcross populations were infected at the stage of four true leaves using the root-dip method with an isolate of V. dahliae from Nanjing, China. The trimmed roots were dipped into the spore suspension inoculums (107CFU/ml) for 15 min, and the inoculated plants were kept in a greenhouse (25 ± 2 °C). The disease symptoms of leaf yellowing and wilting were scored after 2–3 weeks on a scale from 0 to 3: 0 = no symptoms; 1 = one leaf shows symptoms; 2 = more than two leaves show symptoms; 3 = all leaves fall or the plant dies. Plants that scored 0 or 1 were considered to be resistant, and the others were considered to be susceptible.
Molecular marker analysis
Primer Premier 5.0 software was used to design a pair of primers, SL01 and SL02, based on the tomato Ve gene, which could amplify expected 1077 bp product. Homologous sequences in PI388846 and three susceptible inbred lines, EP12, EP15 and EP21, were first amplified using primers SL01 and SL02, and then sequenced. DNAMAN software (version 6.0) was used to align the sequences for detecting nucleotide polymorphisms between PI388846 and the three susceptible inbred lines. Three reverse primers, SL181, SL182, and SL183 were designed according to nucleotide polymorphisms with additional different mismatched bases (Table 1).
The PCR was performed in a volume of 20 µl, containing 50 ng template DNA, 2 × Taq Mix and 0.5 µM of each primer. The PCR cycle program was as follow: initial denaturation at 94 °C for 5 min, 35 cycles of 94 °C for 30 s, 60 °C for 1 min, 72 °C for 1 min, and a final extension of 72 °C for 7 min. The PCR was carried out in a veritiTM 96-well thermal cycler. PCR products were visualized on a 1.2 % agarose gel stained with ethidium bromide.
Results
Interspecific hybridization
To overcome potential hybridization barriers, we selected four inbred lines of the cultivated eggplant as paternal parents, and the repeated pollination was done at the immature and mature pistil stages. For each parent combination, more than 50 flowers of the maternal parent were used. Only two hybrid fruits were harvested from the combination ‘PI388846 × EP15’ while the other hybridization combinations failed. In the two hybrid fruits, there were only seven mature seeds in total. Thus, although PI388846 may be crossed with cultivated eggplants, a hybridization barrier still remains.
Improvement of resistance and agronomic traits by backcrossing
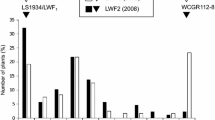
The interspecific hybrid showed strong heterosis and good fertility. Figure 1 showed certain traits of the two parents, the hybrid and the backcross progeny. The hybrid set 2–3 fruits in each inflorescence, and more than 100 seeds could be harvested in each fruit. PI388846 was spiny and bore round, green, striped and small fruits, while EP15 was a non-spiny commercial type inbred line which bears long, dark purple, un-striped and large fruits. The interspecific hybrid expressed the intermediate traits of two parents: spiny with oval, light purple, striped and small fruits. Three interspecific hybrid seedlings and each 10 PI388846 and EP15 seedlings were inoculated with V. dahliae (Fig. 2a). The hybrid seedlings showed disease resistance. Thus, the Verticillium wilt resistance in PI388846 was a dominant trait. Certain agronomic traits, such as spiny, small and striped fruits are not suitable for commercial varieties. Therefore, they must be removed for commercial breeding.
Symptoms on plants of PI388846 (Sl), EP15, and their progenies inoculated with Verticillium dahliae. a Responses of Sl, EP15 and F1 after 2 weeks of inoculation. EP15 showed yellow and wilt leaves, while PI388846 and F1 showed disease resistance. b Response of BC4 population after 3 weeks of inoculation. (Color figure online)
To transfer disease resistance into the cultivated eggplant and improve the agronomic traits, the interspecific hybrid was backcrossed with EP15 for four generations. Backcross progeny were artificially inoculated with V. dahliae, and the resistant seedlings scored as 0 or 1 were picked and planted in a disease nursery for further resistance and agronomic traits screening (Fig. 2b). In the BC1 population, the segregation ratio was 32 resistant: 65 susceptible, which was not in a normal Mendelian fashion. We observed that the first two generations showed that the fruits of many resistant plants were small and striped. However, after four generations of backcrossing, the agronomic traits were improved significantly, with some being similar to the cultivated eggplants (Fig. 1). Therefore, to pyramid disease resistance together with desirable traits, it is necessary to backcross the resistant plants with more eggplant elite inbred lines.
Development of a gene specific marker and analysis in the backcross population
The allele-specific PCR technique is an effective method to develop markers to distinguish different alleles, and the bases at the 3′ end are sensitive for PCR (Zhang et al. 2003; Bundock et al. 2006; Liu et al. 2012). Therefore, the development of an allele-specific PCR marker for the Ve homolog in PI388846 would be useful for marker-assisted selection. Based on the sequence of tomato Ve, a pair of primers, SL01 and SL02 was designed to amplify the homologs from the genome of PI388846 and three cultivated eggplant inbred lines. The sequence alignment showed 9 single nucleotide polymorphisms (SNPs) between PI388846 and cultivated eggplants. Further analysis by Primer Premier 5.0 software indicated that only the sequence near the SNP pos. 829 was suitable for primer design, and three allele-specific PCR primers, SL181, SL182, and SL183 were designed (Fig. 3a; Table 1).
Allele-specific PCR marker analysis. a The SNP pos. 829 between PI388846 and three cultivated eggplant inbred lines, and the underlined sequence was used to design allele-specific PCR primers. b PCR results of primer SL01 combined with three allele-specific PCR primers SL181, SL182, and SL183. 1–4 represent PI388846, EP12, EP15, and EP21, respectively. c PCR detection of marker SlVR844 in BC4 population. 1–10 are resistant plants and 11–20 are susceptible plants
Primer SL01 combined with different allele-specific PCR primers was used to amplify the genomic DNA of PI388846 and three cultivated eggplant inbred lines. The result showed that only PI388846 generated PCR products of expected size using primer combination ‘SL01 × SL181’, while no PCR products were amplified in the cultivated eggplants (Fig. 3b). The above results indicated that primer SL181 can detect the Ve allele in PI388846, and adding another base mismatch at the 3′ end led to failed PCR under the same PCR conditions. Therefore, the primer combination ‘SL01 × SL181’ can be used to detect the Ve homolog in PI388846. The marker was named SlVR844 according the product size.
Marker SlVR844 was tested using the BC4 population. Based on the results of disease evaluation, 10 resistant progenies scored as 0 and 10 susceptible progenies scored as three were selected. The PCR detection results indicated that the marker could correctly distinguish the resistant/susceptible genotype dominated by the Ve gene (Fig. 3c). The marker was tested for another 140 BC4 plants. The result showed that all 67 plants with the marker were resistant. However, the marker could not be detected in seven resistant plants scored as 1 (failed PCR was excluded). In addition, SlVR844 was not detected in any of the 21 susceptible Chinese landraces of S. melongena, (data not shown). The above results demonstrated that SlVR844 is a gene specific marker for the Ve homolog in PI388846, and could be used for marker-assisted selection for Ve gene-mediated Verticillium wilt resistance originating from PI388846.
Discussion
A difficulty of eggplant breeding is the absence of resistance gene(s) towards Verticillium wilt in S. melongena germplasm. Some wild Solanum species related to eggplants, such as S. linnaeanum, S. aculeatissimum, S. sisymbriifolium and S. torvum, exhibit resistance to Verticillium wilt (Daunay 2008). Therefore, the introgression of resistance genes from those wild eggplants into cultivated eggplants is a powerful tool for improving the resistance of eggplants to Verticillium wilt. S. linnaeanum is considered resistant to Verticillium wilt when only leaf wilting is used as the resistance assessment tool, but it does show mild symptoms of Verticillium wilt, including browning of vessels and plant growth reduction. However, studies have shown that the resistance of cultivated eggplants to V. dahliae was increased significantly by interspecific hybridization with S. linnaeanum (Acciarri et al. 2001, 2007; Sunseri et al. 2003). In this study, Verticillium wilt resistance in S. linnaeanum accession PI388846 was transferred into cultivated eggplants by interspecific hybridization, and the agronomic traits of interspecific hybrid progeny were improved by subsequent backcrossing. To facilitate screening for resistant plants, a gene specific marker for the Ve homolog in PI388846 was successfully developed. The result will aid Verticillium wilt resistant eggplant breeding.
In cultivated tetraploid potato, the inheritance of resistance was considered to be polygenic and complex (Bae et al. 2008). However, in tomato, it was reported the resistance is controlled by Ve1 gene (Diwan et al. 1999; Kawchuk et al. 2001; Fradin et al. 2009). Recent studies indicated that a homolog of the DAHP (3-deoxy-D-arabinoheptulosonate-7-phosphate) gene in S. torvum was related to V. dahliae resistance. Also, the expression of S. torvum ribosomal protein L13a in potatoes enhanced resistance to V. dahliae (Wang et al. 2010; Yang et al. 2013). Our data showed an unexpected ratio of disease resistance in BC1 generation. The distorted segregation could be one of possible reasons, as the initial cross (F1) was made between wild species and a cultivated line. Also, genetic background in which the resistance gene was present may also play an important role and influence the expression of the resistance. In addition, the resistance in PI388846 may be explained by the quantitative nature considered by Acciarri et al. (2007), but more data are still needed for this possibility.
Artificial inoculation is not a high throughput method for disease resistance screening, so the development of a marker-assisted selection is necessary. Based on the nucleotide polymorphisms of the Ve homolog in PI388846 and susceptible eggplants, we developed a gene specific marker SlVR844 and tested it in the BC4 backcross population. The data showed that all plants with this marker were disease resistant. However, the marker could not be detected in a few resistant plants showing mild disease symptoms, which may be resulted from some escapes in artificial inoculation. In this study, 9 SNPs between PI388846 and cultivated eggplants changed the amino acid composition of predicted protein, but further work is needed to explore whether these SNPs can cause loss of function of the gene in susceptible lines.
References
Acciarri N, Rotino GL, Valentino D, Vitelli G, Sunseri F, Martelli G, Tamietti G (2001) Improvement of eggplant for resistance to Verticillium through interspecific hybridization with Solanum sodomeum L. In: Proceedings of the 11th Eucarpia meeting, pp 327–338
Acciarri N; Sabatini E; Voltattorni S, Ciriaci T, Tamietti G, Valentino D, Mennella G, Cavallanti F, Tacconi MG, Toppino L, Grazioli G, Pedretti R, Alberti P, Rotino GL (2007) First eggplant pure lines of different typologies derived from sexual and somatic hybridization with S. sodomaeum and S. aethiopicum gr. aculeatum and gr. gilo. In: Proceedings of the 13th Eucarpia meeting, pp 327–338
Bae J, Halterman D, Jansk S (2008) Development of a molecular marker associated with Verticillium wilt resistance in diploid interspecific potato hybrids. Mol Breed 22:61–69
Bundock PC, Cross MJ, Shapter FM, Henry RJ (2006) Robust allele-specific polymerase chain reaction markers developed for single nucleotide polymorphisms in expressed barley sequences. Theor Appl Genet 112:358–365
Collonnier C, Fock I, Kashyap V, Rotino GL, Daunay MC, Lian Y, Mariska IK, Rajam MV, Servaes A, Ducreux G, Sihachakr D (2001) Applications of biotechnology in eggplant. Plant Cell Tissue Org 65:91–107
Daniell H, Lee S, Grevich J, Saski C, Quesada-Vargas T, Guda C, Tomkins J, Jansen RK (2006) Complete chloroplast genome sequences of Solanum bulbocastanum, Solanum lycopersicum and comparative analyses with other Solanaceae genomes. Theor Appl Genet 112:1503–1518
Daunay MC (2008) Eggplant. In: Prohens J, Nuez F (eds) Handbook of crop breeding., Vegetables: Fabaceae, Liliaceae, Umbelliferae, and SolanaceaeSpringer, New York, pp 163–220
Diwan N, Fluhr R, Eshed Y, Zamir D, Tanksley SD (1999) Mapping of Ve in tomato: a gene conferring resistance to the broad-spectrum pathogen Verticillium dahliae race 1. Theor Appl Genet 98:315–319
Doganlar S, Frary A, Daunay MC, Lester RN, Tanksley SD (2002a) A comparative genetic linkage map of eggplant (Solanum melongena) and its implications for genome evolution in the Solanaceae. Genetics 161:1697–1711
Doganlar S, Frary A, Daunay MC, Lester RN, Tanksley SD (2002b) Conservation of gene function in the solanaceae as revealed by comparative mapping of domestication traits in eggplant. Genetics 161:1713–1726
Fradin EF, Thomma BPHJ (2006) Physiology and molecular aspects of Verticillium wilt diseases caused by V. dahliae and V. albo-atrum. Mol Plant Pathol 7:71–86
Fradin EF, Zhang Z, Juarez Ayala JC, Castroverde CDM, Nazar RN, Robb J, Liu CM, Thomma BPHJ (2009) Genetic dissection of verticillium wilt resistance mediated by tomato Ve1. Plant Physiol 150:320–332
Frary A, Doganlar S, Daunay MC, Tanksley SD (2003) QTL analysis of morphological traits in eggplant and implications for conservation of gene function during evolution of solanaceous species. Theor Appl Genet 107:359–370
Hayes RJ, McHale LK, Vallad GE, Truco MJ, Michelmore RW, Klosterman SJ, Maruthachalam K, Subbarao KV (2011) The inheritance of resistance to Verticillium wilt caused by race 1 isolates of Verticillium dahliae in the lettuce cultivar La Brillante. Theor Appl Genet 123:509–517
Kawchuk LM, Hachey J, Lynch DR, Kulcsar F, van Rooijen G, Waterer DR, Robertson A, Kokko E, Byers R, Howard RJ, Fischer R, Prüfer D (2001) Tomato Ve disease resistance genes encode cell surface-like receptors. Proc Natl Acad Sci USA 98:6511–6515
King SR, Davis AR, Zhang XP, Crosby K (2010) Genetics, breeding and selection of rootstocks for Solanaceae and Cucurbitaceae. Sci Hortic-Amsterdam 127:106–111
Lee JM (1994) Cultivation of grafted vegetables I. Current status, grafting methods, and benefits. Hortscience 29:235–239
Liu J, Huang S, Sun M, Liu S, Liu Y, Wang W, Zhang X, Wang H, Hua W (2012) An improved allele-specific PCR primer design method for SNP marker analysis and its application. Plant Methods 8:34
Mace ES, Lester RN, Gebhardt CG (1999) AFLP analysis of genetic relationships among the cultivated eggplant, Solanum melongena L., and wild relatives (Solanaceae). Theor Appl Genet 99:626–633
Mueller LA, Solow TH, Taylor N, Skwarecki B, Buels R, Binns J, Lin C, Wright MH, Ahrens R, Wang Y, Herbst EV, Keyder ER, Menda N, Zamir D, Tanksley SD (2005) The SOL genomics network: a comparative resource for solanaceae biology and beyond. Plant Physiol 138:1310–1317
Murray HG, Thompson WF (1980) Rapid isolation of higher weight DNA. Nucleic Acids Res 8:4321
Sunseri F, Sciancalepore A, Martelli G, Acciarri N, Rotino GL, Valentino D, Tamietti G (2003) Development of RAPD-AFLP map of eggplant and improvement of tolerance to verticillium wilt. Acta Hort (ISHS) 625:107–115
Tjamos EC, Beckman CH (1989) Vascular wilt diseases of plants: Basic studies and control., NATO ASI Series H: Cell BiologySpringer-Verlag, Berlin
Uribe P, Jansky S, Halterman D (2014) Two CAPS markers predict Verticillium wilt resistance in wild Solanum species. Mol Breed 33:465–476
Vining K, Davis T (2009) Isolation of a Ve homolog, mVe1, and its relationship to verticillium wilt resistance in Mentha longifolia (L.) Huds. Mol Genet Genomics 282:173–184
Wang Z, Xie C, Jue DW, Huang LP, Huang QS, Yang Q (2010) Cloning and expression analysis of Verticillium wilt Pathogenesis related gene StDAHP in Solanum torvum. China Biotechnol 30(6):48–53
Wu F, Eannetta NT, Xu Y, Tanksley SD (2009a) A detailed synteny map of the eggplant genome based on conserved ortholog set II (COSII) markers. Theor Appl Genet 118:927–935
Wu F, Eannetta NT, Xu Y, Durrett R, Mazourek M, Jahn MM, Tanksley SD (2009b) A COSII genetic map of the pepper genome provides a detailed picture of synteny with tomato and new insights into recent chromosome evolution in the genus Capsicum. Theor Appl Genet 118:1279–1293
Yang L, Xie C, Li W, Zhang RJ, Jue DW, Yang Q (2013) Expression of a wild eggplant ribosomal protein L13a in potato enhances resistance to Verticillium dahliae. Plant Cell Tissue Organ Cult 115:329–340
Zhang W, Gianibelli MC, Ma W, Rampling L, Gale KR (2003) Identification of SNPs and development of allele-specific PCR markers for gamma-gliadin alleles in Triticum aestivum. Theor Appl Genet 107:130–138
Zhang B, Yang Y, Chen T, Yu W, Liu T, Li H, Fan X, Ren Y, Shen D, Liu L, Dou D, Chang Y (2012) Island cotton Gbve1 gene encoding a receptor-like protein confers resistance to both defoliating and non-defoliating isolates of Verticillium dahliae. PLoS ONE 7(12):e51091
Acknowledgments
This research is partially supported by the National High Technology Research and Development Program of China (2012AA100103), Natural Science Foundation (BK20131335) and Independent Innovation Foundation of Agricultural Sciences [CX (12)5046] of Jiangsu Province, China.
Author information
Authors and Affiliations
Corresponding author
Additional information
Jun Liu and Zisong Zheng contributed equally to this study.
Rights and permissions
About this article
Cite this article
Liu, J., Zheng, Z., Zhou, X. et al. Improving the resistance of eggplant (Solanum melongena) to Verticillium wilt using wild species Solanum linnaeanum . Euphytica 201, 463–469 (2015). https://doi.org/10.1007/s10681-014-1234-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-014-1234-x