Abstract
Haploid technology can significantly shorten the time required for inbred line improvement, accelerate the breeding process, and reduce breeding costs. The production of haploids is not only dependent on the genetics of the paternal haploid inducer, but it is also affected by the genetic background of the maternal donor during the process of haploid production. To address the maternal genetic contribution to haploid production, we pollinated a mapping population consisting of 186 F2:3 family lines derived from a cross between Zheng58 and Chang7-2 with the inducer line CAU5 and selected haploid kernels using R1-nj kernel markers. Two quantitative trait loci (QTLs), qmhir1 and qmhir2, which contribute to the maternal genetics of haploid induction, were detected on chromosomes 1 and 3, respectively. The qmhir1 locus is located between the flanking marker loci umc1292 and bnlg1014, and it explained 14.70 % of the phenotypic variation. The qmhir2 locus is located between marker loci umc1844 and umc2277 and it explained 8.42 % of the phenotypic variation. The genetic effect of both QTLs is partial dominance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The use of haploid plants is a rapidly developing technology in maize breeding. The production of doubled haploid lines in maize can accelerate breeding cycles and allow breeders to rapidly generate new inbred lines for evaluation and testing (Hallauer et al. 2010). Haploid breeding strategies have been widely adopted by many maize breeders during the past decade (Schmidt 2003; Seitz 2005). Along with transgenic technology and molecular marker-assisted breeding techniques, maize haploid breeding has gradually become one of the three core technologies of modern maize breeding programs (Chen et al. 2012). The production of maize doubled haploid lines is most commonly performed in vivo using a paternal haploid inducer to pollinate a maternal donor population. One of the challenges associated with producing doubled haploid lines is the limited number of kernels that are successfully induced; this usually depends on the inducer, the genetic background of the donor, and the environment in which the inductions are made.
Currently, researchers have mainly focused on the paternal effects of haploid induction rate (HIR), including selection, QTL mapping, and the mechanisms of induction. On one hand, inducer traits are expressed in pollen (the male gametophyte), and the HIR can gradually be improved through breeding (Sarkar and Coe 1966; Geiger and Gordillo 2009; Prigge et al. 2012). The HIR of the first haploid inducer, Stock6, was reported to be only ~1 % (Coe 1959), but the HIR of currently used inducer lines now exceeds 5 %, and some are >10 % as a result of continuous inducer selection; examples of this include WS14 (Lashermes and Beckert 1988), ZMS (Chalyk 1994), MHI (Eder and Chalyk 2002), RWS (Röber et al. 2005), and UH400 (Kebede et al. 2011). On the other hand, genetic studies have identified QTLs controlling paternal induction ability. Evans (2007) located the in vivo androgenesis inducer gene ig1 (indeterminate gametophyte1) on chromosome 3 between SSR markers umc1311 and umc1973; the work was performed in a BC1F1 segregating population constructed from W23ig and Mo17, and the ig1 gene was cloned. Barret et al. (2008) detected a major QTL related to paternal induction (qhir1) at bin 1.04, and Dong et al. (2013) fine mapped qhir1 within a 243-kb region between flanking markers X291 and X263. Prigge et al. (2012) detected seven additional HIR-related QTLs on five maize chromosomes.
HIR is not only affected by the paternal inducer, but it is also related to the maternal genetic background (Randolph 1940; Chase 1952; Coe 1959). Chase (1969) found that after many generations, the HIR of agronomically improved maternal lines were higher than those of unimproved lines. Liu and Song (2000) used CAUHOI as an inducer to pollinate 16 different inbred and hybrid lines and found that the highest HIR in this material was 9.20 %, and the lowest HIR was only 1.64 %. Cai et al. (2012) also crossed 12 maternal lines from different genetic backgrounds with the inducer line JAAS3, and the results showed that there was a significant difference in HIR among the maternal materials with different genetic backgrounds. In the current study, based on the ability of the paternal inducer line, we introduce the term “inducibility” into maize haploid breeding. This term describes the ability of the maternal parent to produce haploids. In addition, we measured the HIR of maternal lines. Currently, few studies have addressed this area, and inducibility-related QTL mapping has not previously been reported. We therefore investigated the genetic mechanisms of maternal inducibility and QTL mapping in an F2:3 population. The results of this study should enable breeders to focus on the development of doubled haploids (DH) within genetic backgrounds that maximize the number of haploid kernels produced.
Materials and methods
Plant material and field experiments
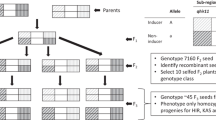
The maize line CAU5 (Xu et al. 2013) was used as the haploid inducer line. The HIR of CAU5 is ~10 %. The maternal donors were 186 F2:3 family lines of the hybrid Zhengdan958, which from the Henan Academy of Agriculture Science, China; the parental lines were Zheng58 and Chang7-2. The cross was performed manually at the Shangzhuang Experimental Station in Beijing, China in 2010. A randomized complete block design was utilized with two replications per genotype. In each block, plants were sown in single-rows, 3 m long, with a density of 60,000 plants/ha. Standard agronomic practices such as irrigation, fertilization, and weeding were followed during the entire growth period to ensure a uniform stand.
Haploid identification and phenotypic value statistics
Line CAU5 is homozygous for the dominant marker gene R1-nj (Nanda and Chase 1966), leading to a purple scutellum and a “purple crown” aleurone in the F1 kernels when crossed with unpigmented donors. These two characteristics were used as embryo and endosperm markers, respectively. Normally, kernels with a haploid embryo and a regular triploid endosperm have colorless embryos and purple endosperm, whereas hybrid kernels with a diploid embryo display purple pigmentation in both the embryo and endosperm. The harvested kernels in our study were classified into two groups: (1) putative haploids, i.e., kernels with purple endosperm and no purple on the embryo (Fig. 1a) and (2) hybrid kernels, i.e., kernels with purple endosperm and a strongly pigmented purple embryo (Fig. 1b). Putative haploids were planted in the field the following year to verify the identification results based on morphological characters; the haploid plants were short with upright leaves, while the hybrid plants had drooping leaves and were of normal stature (Fig. 1c). HIR was then calculated by dividing the number of haploid plants verified in the field generated per plot in 2011 by the total number of kernels harvested in 2010.
Different types of kernels produced from in vivo haploid induction by inducer CAU5. a Hybrid kernels with purple on the endosperm and strongly pigmented purple on the embryo. b Haploid kernels with purple on the endosperm and no purple on the embryo. c Haploid plants in the field. The red triangle denotes haploid plants with upright leaves and short stature, and the yellow triangle denotes hybrid plants of normal size with drooping leaves. (Color figure online)
Molecular data collection and linkage map construction
Young leaves from F2 plants were harvested, flash-frozen in liquid N2, ground to a powder, and stored at −20 °C in individually labeled vials. Genomic DNA was extracted using a CTAB-based method described by Hoisington et al. (1994). SSR analysis was conducted as reported by Senior and Heun (1993) using publicly available primers from the MaizeGDB (http://www.maizegdb.org). In our study, 120 polymorphic SSR markers were chosen for uniform coverage of the maize genome and used in the F2 population to develop a genetic linkage map, which was constructed with MAPMAKER3.0 (Lincoln et al. 1992). Linkage groups were identified using the “Group” command with a logarithm of odds difference (LOD) score of 3.0. Recombination frequency was converted into centiMorgans using the Kosambi mapping function (Kosambi 1944).
QTL analysis
WinQTL Cartographer V2.5 (Basten et al. 1997) was used to detect QTLs. The software was developed based on the composite interval mapping (CIM) method, which is performed to identify QTLs and to estimate their effects (Zeng 1994). A significance threshold for declaring a putative QTL was obtained from 1,000 permutations at p < 0.05 for each data set. A LOD score of 2.5 was used in the model to facilitate identification of putative QTLs that contribute to the maternal genetics of HIR.
Results
Phenotypic data analysis
The HIR of the two parents induced by CAU5 were quite different (Table 1). Zheng58 was the high HIR parent, with an HIR of 11.55 %, and Chang7-2 was the low HIR parent, with an HIR of 5.60 %. The average HIR of the 186 Zhengdan958 F2:3 families was 7.10 %, which is skewed toward the HIR of the low HIR parent. Frequency distribution analysis showed that the trait presents a continuous distribution in this population (Fig. 2). Normal results for the trait obtained with the Kolmogorov–Smirnov test showed that the HIR of the maternal materials in this population presented a normal distribution; the skewness, kurtosis coefficient, and Z value were 0.123, 0.575, and 0.673, respectively, and the significance coefficient was 0.755.
Variance analysis for the trait in the Zhengdan958 F2:3 population showed that for the maternal materials, there was a highly significant difference in HIR among the different families, and there was a significant difference between the two replications at the 5 % level (Table 2). Statistical analysis of the two replications for the trait showed that the overall variability between replications for the maternal genetic contribution to HIR was not large, and the variation coefficient was approximately 30 % (Table 3).
QTL analysis using SSR markers
To construct a genetic linkage map, 528 simple sequence repeat (SSR) markers from across the maize genome were screened against the two parental lines, Zheng58 and Chang7-2; 120 of these SSR markers were polymorphic, and 119 of the polymorphic markers were used to develop the genetic map. The map covered a total length of 1,279.8 centimorgans (cM), with an average distance of 10.75 cM between markers (Fig. 3), and was therefore suitable for QTL detection.
Identification of maternal QTLs for HIR. Red triangles denote the two QTLs, qmhir1 and qmhir2, located at bins 1.01 and 3.08/09, respectively. The position of qmhir1 is between marker loci umc1292 and bnlg1014, and the position of qmhir2 is between marker loci umc1844 and umc2277. Blue triangles denote the inducer genes qhir1 and seven QTLs were identified on six chromosomes (Prigge and Xu 2012). (Color figure online)
Two QTLs for inducibility with partially dominant effects were detected on chromosomes 1 and 3 (Table 4). In general, for each of the inducibility QTLs, the percentage of contribution to the phenotypic variation was <15 %. For the partially dominant effect QTLs, the QTL at bin 1.01 (named qmhir1), flanked by SSR marker loci umc1292 and bnlg1014, had the largest contribution to the phenotypic variance (14.70 %), and the QTL at bin 3.08/09 (named qmhir2), located between SSR marker loci umc1844 and umc2277, contributed 8.42 % to the phenotypic variance. These four SSR markers showed no distortion from the expected segregation ratio of 1:2:1 in this population. For each trait QTL, the parental line Zheng58 donated beneficial alleles for increasing inducibility, which is consistent with the phenotype of Zheng58 having a higher HIR than Chang7-2.
Discussion
Maternal haploid inducibility is genetically controlled
The development of homozygous inbred lines is an important aspect of the breeding of maize and many other crops. Breeders and researchers have traditionally produced inbred lines by selfing heterozygous materials for five to six generations, but this approach is expensive and time consuming (Eder and Chalyk 2002). Breeding with haploids can result in the creation of pure breeding lines (Szarejko and Forster 2007; Chang and Coe 2009; Geiger and Gordillo 2009) in fewer seasons. The production of DH lines makes it easy to carry out genetic studies and significantly shortens the breeding time (Seitz 2005). DH technology has become the routine breeding strategy employed in many commercial maize breeding programs (Geiger and Gordillo 2009; Prigge and Melchinger 2011). The first step in DH technology is the production of a sufficient number of haploids. Since the first inducer, Stock6 (Coe 1959), was reported, increasing the HIR has solely depended on improving the inducer (Lashermes and Beckert 1988; Chalyk 1994; Eder and Chalyk 2002; Röber et al. 2005). However, our results suggest that inducibility represents a maternal influence on HIR, and they reveal two maternal QTLs for in vivo HIR.
The mean and range of the HIR obtained in the present study approached or were slightly higher than those reported in maize by Kebede et al. (2011). This may be due to the fact that we used a haploid inducer with an equally high HIR in our study (Xu et al. 2013), which was selected from CAUHOI × UH400. Furthermore, to exclude false positive haploid kernels from all of the putative haploid kernels, we planted a sufficient number of haploid seeds to verify the haploid number in a field in Hainan, China, which has favorable conditions for haploid germination. Moreover, the haploid verification rate (calculated by dividing the haploid plant number by the putative haploid kernel number) was greater than 99 %. Haploids were previously verified through detection by flow cytometry (Häntzschel and Weber 2010) or through observation by counting chromosome number (Li et al. 2009). Here, we were able to calculate the HIR for a large number of different haploid plants.
In the present study, the mean HIR of Zheng58 was found to be higher than that of Chang7-2, and the mean HIR of the Zhengdan958 F2:3 population was slightly lower than that of the mid-parent (8.58 %). In this population, the highest HIR of a family line was 12.59 %, which is higher than that of either parent, and the lowest HIR of a family line was 1.09 %, which is lower than that of either parent. The CV was 26.63. Our results confirm that the maternal genotype had a significant influence on the frequency of haploids obtained. Lashermes and Beckert (1988) first put forward the notion that the haploid-producing frequency is the result of a genetic interaction between the male and female parents, and they suggested that the inducing ability of the female plant genotype should be considered when choosing a female parent for haploid breeding programs. Eder and Chalyk (2002) found that the haploid frequency ranged from 2.7 to 8.0 % among different maternal genotypes, including dent, flint, and flint × dent maize. Moreover, by comparing the HIR of ten different tropical germplasms, Kebede et al. (2011) also revealed that source germplasm plays an important role in the expression of the HIR. Many researchers believe that HIR is not only affected by the paternal inducer, but it is also influenced by the genotype of the maternal donor (Röber et al. 2005; Hu 2008; Li et al. 2008; Cai et al. 2012). In the current study, we first used a population containing a combination of different genotypes to detect the HIR variance in different family lines, and we also found that the HIR is influenced by genetic factors. The presence of a family line with higher HIR than both parents indicates that there is potential for further improvement of this trait through the use of materials with different HIR.
We performed our evaluation in only one environment, as the HIR is not highly dependent on environmental effects, as evidenced by the following: First, the heritability of this trait under our conditions was 0.58, indicating that the HIR does not highly depend on environmental effects. Second, Aman and Sakar (1978) and Sarkar et al. (1994) found that the location and the year did not have significant effects on this trait, but Röber et al. (2005) found that the environmental means for inducer RWS evaluated on the genetically uniform tester donors lglg varied from 2.43 % in Gondelsheim to 22.32 % in Porvenir. However, Röber et al. pointed out that optimizing donor and inducer plant cultivation techniques and choosing a nursery with favorable climatic conditions are likely to reduce environment-caused variations in the induction rate. We also found that the HIR of Zheng58, Chang7-2, and F1 hybrid Zhengdan958 grown in Beijing, China were slightly higher than the HIR of these materials grown in Gansu or Hainan. Therefore, we concluded that Beijing had the most favorable climatic conditions in which to perform the cross; in the year in which we performed the cross, the favorable climatic conditions in Beijing enabled the inducer of pollination and the maternal ears to have high seed setting rates. Moreover, in our experiment, we used appropriate cultivation techniques that led to excellent plant development at all stages, including the use of optimal tillage, timely watering and weeding, and so on. The optimization of these methods helped reduce environment-caused variations in the induction rate.
QTLs for maternal HIR and possible mechanisms in haploid induction
The majority of studies have focused on the paternal genetics of in vivo haploid induction for both inducer selection and inducer gene searching. To date, there are no reports of the identification of maternally-inherited genes that control haploid induction in maize. We constructed an F2:3 population to locate QTLs related to haploid inducibility. Based on kernel phenotype and subsequent field verification, the two QTLs on chromosomes 1 and 3 could explain >20 % of the phenotypic variation for this trait. The QTL for inducibility on chromosome 1 at bin 1.01, which had the largest contribution to phenotypic variation, is on the same chromosome as the qhir1 locus, which is the major paternal inducer QTL (Barret et al. 2008; Prigge et al. 2012; Dong et al. 2013). Other paternal inducer QTLs were also detected in different populations. In the 1,680 × UH400 population, in addition to the major QTL qhir1, qhir2 was also detected on chromosome 3, and in the CAUHOI × UH400 population, seven QTLs were identified on five chromosomes (Prigge et al. 2012). In the current study, we only detected two maternal QTLs for HIR. However, we did not screen a lot of materials to select parents with higher or lower HIR than the parents used in this study. Therefore, there may be other QTLs in other populations.
Previous studies have indicated that for in vivo haploid induction, the production of haploids is mainly affected by male gametes from the inducer line. Possible explanations for HIR include single fertilization (Bylich and Chalyk 1996; Chalyk et al. 2003; Rotarenco and Eder 2003) and chromosome exclusion (Subrahmanyam and Kasha 1973; Laurie and Bennett 1989; Kim et al. 2002; Mochida et al. 2004; Gernand et al. 2005; Li et al. 2009). The haploid inducers in maize can be considered to be original mutant lines when used for the analysis of double-fertilization mechanisms (Li et al. 2009). In this process, the inducer disturbs the original maternal chromosome balance, which leads to kernel abortion; thus, kernel abortion in the maternal material is a phenomenon used to deal with this unbalance (Xu et al. 2013). When the balance of the whole chromosomal complement departs from the standard ratio of two sets of chromosomes of maternal origin to one of paternal origin, development is impaired, often leading to seed failure and the production of sterile grains (Kermicle and Alleman 1990). However, if the maternal chromosomes were suited to this imbalance, haploids would be produced. Therefore, based on the results of our study, we propose that inducibility represents the ability of maternal materials to adapt to the unbalanced, induced genomic environment. This genetic phenomenon is specific to the process of double fertilization.
Therefore, we focused on bin 1.01, bin 3.09, and nearby regions to find potential candidate genes for maternal QTLs of HIR. Our search of the region of bin 1.01 and nearby regions in MaizeGDB for potential candidate genes revealed hydroxylase6 (hyd6), which encodes a beta-carotene hydroxylase. The hyd6 locus was estimated to be located between positions 5,347,015 and 5,383,540 (36,525 base pairs in length) on chromosome 1 based on the map IMBM2 2008 Neighbors 1 (http://www.maizegdb.org/cgi-bin/displaylocusrecord.cgi?id=9021810#a5). The gene hyd6, along with hyd3 and hyd4, play important roles in the carotenoid biosynthetic pathway (Shumskaya and Wurtzel 2013). Many studies (Vallabhaneni et al.2009; Alexandrov et al. 2009; Li et al. 2010) have indicated that the carotenoid biosynthetic pathway serves manifold roles in plants related to development and stress hormones. In another important candidate region, the centromere protein C1 (cenpc1) gene was found at bin 3.09 in MaizeGDB. The locus cenpc1 is estimated to be located between positions 217,200,846 and 217,549,349 (348,503 base pairs in length) on chromosome 3 based on the map IBM2 2008 Neighbors 1 (http://www.maizegdb.org/cgi-bin/displaylocusrecord.cgi?id=223037#a5). Du et al. (2010) found that maize centromere protein (CENPC) plays an important role in DNA binding reactions, and we believe that it may be related to haploid kernel formation. Based on the results of our analyses, hyd6, located at bin 1.01, and cenpc1, located at bin 3.09, are potential candidate genes for maternal QTLs controlling the HIR.
Abbreviations
- DH:
-
Doubled haploids
- HIR:
-
Haploid induction rate
- SSR:
-
Simple sequence repeat
References
Alexandrov NN, Brover W, Freidin S, Troukhan ME, Tatarinova TV, Zhang H, Swaller TJ, Lu YP, Bouck J, Flavell RB, Feldmann KA (2009) Insights into corn genes derived from large scale cDNA sequencing. Plant Mol Biol 69:179–194
Aman MA, Sakar KR (1978) Selection for haploidy inducing potential in maize. Indian J Genet 38:452–457
Barret P, Brinkmann M, Beckert M (2008) A major locus expressed in the male gametophyte with incomplete penetrance is responsible for in situ gynogenesis in maize. Theor Appl Genet 117:581–594
Basten CJ, Weir BS, Zeng ZB (1997) QTL cartographer: a reference manual and tutorial for QTL mapping. Department of Statics, North Carolina State University, Raleigh
Bylich VG, Chalyk ST (1996) Existence of pollen grains with a pair of morphologically different sperm nuclei as a possible cause of the haploid-inducing capacity in ZMS line. Maize Genet Coop Newsl 70:33–37
Cai Z, Guo-liang X, Xianghui L, Ya-lin D (2012) The breeding of JAAS3-haploid inducer with high frequency parthenogenesis in maize. J Maize Sci 15:1–4 (in Chinese)
Chalyk ST (1994) Properties of maternal haloid maize plants and potential application to maize breeding. Euphytica 79:13–18
Chalyk ST, Baumann A, Daniel G, Eder J (2003) Aneuploidy as a possible cause of haploid-induction in maize. Maize Genet Coop Newsl 77:29–30
Chang MT, Coe EH (2009) Doubled haploids. In: Kriz AL, Larkins BA (eds) Molecular genetic approaches to maize improvement. Springer, Berlin
Chase SS (1952) Production of homozygous diploids of maize from monoploids. Agron J 44:263–267
Chase SS (1969) Monoploids and monoploid derivatives of maize (Zea mays L.). Bot Rev 35:117–167
Chen SJ, Li L, Li HC, Xu XX (2012) Maize haploid breeding. China Agricultural University Press, Beijing
Coe EH (1959) A line of maize with high haploid frequency. Am Nat 93:381–382
Dong X, Xu XX, Chen SJ, Melchinger AE (2013) Fine mapping of qhir1 influencing in vivo haploid induction in maize. Theor Appl Genet 126:1713–1720
Du Y, Topp CN, Dawe RK (2010) DNA binding of centromere protein C (CENPC) is stabilized by single-stranded RNA. PLoS Genet 6:e1000835
Eder J, Chalyk S (2002) In vivo haploid induction in maize. Theor Appl Genet 104:703–708
Evans MMS (2007) The indeterminate gametophyte1 gene of maize encodes a LOB domain protein required for embryo sac and leaf development. Plant Cell 19:46–62
Geiger HH, Gordillo GA (2009) Doubled haploids in hybrid maize breeding. Maydica 54:485–499
Gernand D, Rutten T, Varshney A, Rubtsova M, Prodanovic S, Brüß C, Kumiehn J, Matzk F, Houben A (2005) Uniparental chromosome elimination at mitosis and interphase in wheat and pearl millet crosses involves micronucleus formation, progressive heterochromatinization, and DNA fragmentation. Plant Cell 17:2431–2438
Hallauer AR, Carena MJ, Miranda FJB (2010) Quantitative genetics in maize breeding. Springer, New York
Häntzschel KR, Weber G (2010) Blockage of mitosis in maize root tips using colchicine-alternatives. Protoplasma 241:99–104
Hoisington D, Khairallah M, Gonzalez de Leon D (1994) Laboratory protocols, 2nd edn. CIMMYT Applied Molecular Genetics Laboratory, Mexico City
Hu EL (2008) The research of using stock6 induce corn parthenogenesis haploid and doubling. Department of Agricultural, Sichuan Agricultural University, Chengdu
Kebede AZ, Dhillon BS, Schipprack W, Araus JL, Bänziger M, Semagn K, Alvarado G, Melchinger AE (2011) Effect of source germplasm and season on the in vivo haploid induction rate in tropical maize. Euphytica 180:219–226
Kermicle JL, Alleman M (1990) Gametic imprinting in maize in relation to the angiosperm life cycle. Dev Suppl 108:9–14
Kim NS, Armstrong KC, Fedak G, Ho K, Park NI (2002) A microsatellite sequence from the rice blast fungus (Magnaporthegrisea) distinguishes between the centromeres of Hordeum vulgare and H. bulbosum in hybrid plants. Genome 45:165–174
Kosambi DD (1944) The estimation of map distances from recombination values. Ann Eugen 12:172–175
Lashermes P, Beckert M (1988) Genetic control of maternal haploidy in maize (Zea mays L.) and selection of haploid inducing lines. Theor Appl Genet 76:405–410
Laurie DA, Bennett MD (1989) The timing of chromosome elimination in hexaploid wheat × maize crosses. Genome 32:953–961
Li GL, Jun SU, Li CX (2008) Research on haploids frequency of different germplasm and generations in maize by CAUHO inducer 1. J Maize Sci 16:3–6 (in Chinese)
Li L, Xu X, Jin W, Chen SJ (2009) Morphological and molecular evidences for DNA introgression in haploid induction via a high oil inducer CAUHOI in maize. Planta 230:367–376
Li Q, Farre G, Nagvi S, Breitenbach J, Sanahuja G, Bai C, Sandmann G, Capell T, Christou P, Zhu C (2010) Cloning and functional characterization of the maize carotenoid isomerase and Bata-carotene hydroxylase genes and their regulation during endosperm maturation. Transgenic Res 19:1053–1068
Lincoln S, Daly M, Lander E (1992) Mapping genetic mapping with MAPMAKER/EXP3.0. Whitehead Institute Technical Report, Cambridge
Liu ZZ, Song TM (2000) Correlation analysis between induction of maize parthenogenesis haploid in vivo and intersperm nucleus distances in pollen tubes germinating in vitro of male parent. Acta Bot Boreal 20:495–502 (in Chinese)
Mochida K, Tsujimoto H, Sasakuma T (2004) Confocal analysis of chromosome behavior in wheat and maize zygotes. Genome 16:199–205
Nanda DK, Chase SS (1966) An embryo marker for detecting monoploids of maize (Zea mays L.). Crop Sci 6:213–215
Prigge V, Melchinger AE (2011) Production of haploids and doubled haploids in maize. In: Loyola-Vargas VM, Ochoa-Alejo N (eds) Plant cell culture protocols, 3rd edn. Humana Press/Springer, Totowa
Prigge V, Xu XX, Li L, Babu R, Chen SJ, Atlin GN, Melchinger AE (2012) New insights into the genetics of in vivo induction of maternal haploids, the backbone of doubled haploid technology in maize. Genetics 190:781–793
Randolph LF (1940) Note on haploid frequencies. Maize Genet Coop Newsl 14:23–24
Röber FK, Gordillo GA, Geiger HH (2005) In vivo haploid induction in mazie—performance of new inducers and significance of doubled haploid lines in hybrid breeding. Maydica 50:275–283
Rotarenco VA, Eder J (2003) Possible effect of heterofertilization on the induction of maternal haploids in maize. Maize Genet Coop Newsl 77:30–36
Sarkar K, Coe E (1966) A genetic analysis of the origin of maternal haploid in maize. Genetics 54:453–464
Schmidt W (2003) Hybridmaiszü chtung bei der KWS SAAT AG In: Berichtü ber die 54. Tagung der Vereinigung der Pflanzenzu¨ chter und Saatgutkaufleute Ö sterreichs, Gumpenstein, Austria 54 pp 1–6 (in German)
Seitz G (2005) The use of doubled haploids in corn breeding. In: Proc Forty-first Annual Illinois Corn Breeder’s School 2005 Urbana-Champaign, Illinois, pp 1–8
Senior ML, Heun M (1993) Mapping maize microsatellites and polymerase chain reaction confirmation of the targeted repeats using a CT primer. Genome 36:884–889
Shumskaya M, Wurtzel ET (2013) The carotenoid biosynthetic pathway: thinking in all dimensions. Plant Sci 208:58–63
Subrahmanyam NC, Kasha KJ (1973) Selective chromosomal elimination during haploid formation in barley following interspecific hybridization. Chromosoma 42:111–125
Szarejko L, Forster BP (2007) Doubled haploidy and induced mutation. Euphytica 158:359–370
Vallabhaneni R, Gallagher CE, Licciardello N, Cuttriss AJ, Quinlan RF, Wurtzel ET (2009) Metabolite sorting of a germplasm collection reveals the hydroxylass3 locus as a new target for maize provitamin A biofortification. Plant Physiol 151(3):1635–1645
Xu XX, Li L, Dong X, Jin WW, Melchinger AE, Chen SJ (2013) Gametophytic and zygotic selection leads to segregation distortion through in vivo induction of a maternal haploid in maize. J Exp Bot 64:1083–1096
Zeng ZB (1994) Precision mapping of quantitative trait loci. Genetics 136:1457–1468
Acknowledgments
This research was supported by grants from the National 973 project (2009CB118400), National Maize Industrial Technology System (CARS-02-09) and the National 863 Project (2011AA10A103). The authors greatly appreciate the helpful comments and suggestions from Dr. Xu Xiaowei and another anonymous reviewer.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wu, P., Li, H., Ren, J. et al. Mapping of maternal QTLs for in vivo haploid induction rate in maize (Zea mays L.). Euphytica 196, 413–421 (2014). https://doi.org/10.1007/s10681-013-1043-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-013-1043-7