Abstract
In flowering plants, double fertilization occurs when the egg cell and the central cell are each fertilized by one sperm cell. In maize, some lines produce pollen capable of inducing in situ gynogenesis thereby leading to maternal haploids that originate exclusively from the female plant. In this paper, we present a genetic analysis of in situ gynogenesis in maize. Using a cross between non-inducing and inducing lines, we identified a major locus on maize chromosome 1 controlling in situ gynogenesis (ggi1, for gynogenesis inducer 1). Fine mapping of this locus was performed, and BAC physical contigs spanning the locus were identified using the rice genome as anchor. Genetic component analysis showed that (a) a segregation distortion against the inducer parent was present at this locus, (b) segregation resulted only from male deficiency and (c) there was a correlation between the rate of segregation distortion and the level of gynogenetic induction. In addition, our results showed that the genotype of the pollen determined its capacity to induce the formation of a haploid female embryo, indicating gametophytic expression of the character with incomplete penetrance. We propose the occurrence of a gametophytic-specific process which leads to segregation distortion at the ggi1 locus associated with gynogenetic induction with incomplete penetrance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In flowering plants (angiosperms), double fertilization occurs when the egg cell and the central cell are each fertilized by one sperm cell. The embryo that results from the fertilization of the egg cell, and the endosperm that originates from the fertilization of the central cell, are the two major components of the resulting mature seed. The development of male (Twell 2002; McCormick 2004) and female gametophytes (Chevalier et al. 2002; Yadegari and Drews 2004) as well as the resulting fertilization mechanisms (Edlund et al. 2004; Weterings and Russell 2004; Boavida et al. 2005) have been well described in plants. Gametophytic mutants were studied in order to elucidate molecular mechanisms involved in double fertilization. Mutants affected in female reproduction like sirene (srn; Rotman et al. 2003) and feronia (Huck et al. 2003) indicated that the megagametophyte is involved in guidance of the pollen tube and in fertilization. A recent work (Escobar-Restrepo et al. 2007) indicated that these two mutants were disrupted on the same FER gene, a receptor-like kinase that mediates male-female interactions during pollen tube reception. The Ig1 (indeterminate gametophyte 1) spontaneous mutant (Kermicle 1969) was affected during mitotic synchronization of the embryo sac (Huang and Sheridan 1996). The gene involved in the ig1 character encodes a LOB (lateral organ boundaries) protein that is probably involved in the transition between cellular proliferation and differentiation (Evans 2007). Gametophytic mutants affected during pollen development (Procissi et al. 2001; Johnson-Brousseau and McCormick 2004) are surprisingly rare (Bonhomme et al. 1998) and in many cases present incomplete penetrance (Chen and McCormick 1996; Bonhomme et al. 1998; Park et al. 1998). An Arabidopsis mutant showing one sperm cell in the pollen grain has been described (Nowack et al. 2006), allowing viable seed with maternal endosperm to be obtained when a female polycomb mutant (Guitton et al. 2004) was pollinated (Nowack et al. 2007). The GCS1 (Generative Cell Specific 1) gene was identified in Lilium as being essential for angiosperm fertilization (Mori et al. 2006) and encodes a protein directly associated with the generative cell membrane structure. In mammals, gene expression is restricted to diploid cells, but evidence that the sperm cells cannot be considered just as a DNA vector has been reported recently (for reviews see Ainsworth 2005; Krawetz 2005). In maize, sperm cells have a diverse complement of mRNA (Engel et al. 2003). Despite all these recent advances, little is known about the possible role(s) of the plant male gametophyte in the double fertilization process.
Gynogenesis in plants is defined as the ability to produce an embryo originating exclusively from the female parent. This feature was described in situ for Hordeum vulgare (hap mutant, Hagberg and Hagberg 1980), and in vitro for Allium cepa (Martinez et al. 2000) and for Cucumis sativus (Gémes-Juhasz et al. 2002). Gynogenesis was also obtained following interspecific crosses between Hordeum vulgare and Hordeum bulbosum (Subrahmanyam and Kasha 1973), Triticum aestivum and Zea mays (Matzk and Mahn 1994) or Pennisetum glaucum (Gemand et al. 2005). The mechanism proposed for wheat × pearl millet crosses (Gemand et al. 2005) involves a new pathway for paternal chromosomes elimination, including formation of micronuclei, heterochromatinization and final fragmentation of the pearl millet chromatin. In maize, some genotypes produce pollen capable of inducing in situ gynogenesis that results in maternal haploids originating exclusively from the female plant (Chase 1949). Coe (1959) first described the Stock6 line with an induction rate of up to 2.3%. Lashermes and Beckert (1988) obtained the inducer WS14 line (induction rate 3–5%) from a cross between W23ig and Stock6 lines. Recently, Röber et al. (2005) described the RWS line with the highest induction rate (8.1% on average) reported to date. These lines are extensively used to produce doubled haploid (DH) lines (Bordes et al. 1997, 2006; Eder and Chalyk 2002; Röber et al. 2005), but they can also be considered as original mutant lines for the analysis of double fertilization mechanisms. Lashermes and Beckert (1988) suggested a dominant character with nuclear determination, and an oligogenic component for the inducer character of Stock6, with a correlation of 0.59 between F3 lines and F2 plants. In the W23ig × Stock6 cross, Deimling et al. (1997) identified a major QTL (Quantitative Trait Loci) on chromosome 1 (interval 1.03–1.06) and a minor QTL on chromosome 2 (interval 2.04–2.06), which together explained 17.9% of the phenotypic variability. Cytological analyses were performed on the same material by Mahendru and Sarkar (2000) using haploidy inducer lines derived from Stock6. These authors did not identify any nuclear anomaly in the pollen or in the tubes of haploid inducer lines. The hypothesis of double fertilization of the central cell could explain the absence of fertilization of the egg cell, but in fact must be ruled out, since the resulting tetraploid endosperm with two maternal and two paternal chromosome sets was described as being highly defective (Lin 1984). The mechanism of gynogenesis induction in maize Stock6 thus remains unclear.
In this paper, we present the genetic analysis and the identification of a major locus (ggi1) controlling in situ gynogenesis in maize. We showed that a segregation distortion was present at the ggi1 locus, and that this segregation resulted only from pollen deficiency. In addition our results showed that the genotype of the pollen grains determined its capacity to induce the formation of a haploid female embryo, indicating gametophytic expression of the character with incomplete penetrance. Thus, our data suggest a relationship between failure of fertilization and gynogenetic induction.
Materials and methods
Plant materials
PK6 is a non-glossy maize line developed in our laboratory from Stock6, WS14 (Lashermes and Beckert 1988), FIGH1 (Bordes et al. 1997) and MS1334 lines, with high ability to induce in situ gynogenesis (average of 6%). A188 is a well known maize line extensively used for molecular and cellular genetic studies (Brettell et al. 1981). DH99 (non glossy) and DH7 (gl1/gl1; glossy) are doubled haploid lines with high androgenetic response (Barloy et al. 1989). F7 was developed from a Lacaune population (Tarn, France). F564 is a glossy line with agronomic aptitudes originating from the F7 × F64 cross (Argentina PI186223). F564 × DH7 is a hybrid line with a glossy phenotype. Du101 was developed from an Umkirch population. F52 is “Grand Roux Basque”, F10 is “Star of Normandie” and F575 is “Millette Lauragais” (INRA, France). F22 was derived from a “Chavannes” population and F72 from a “Larums” population (INRA, France). F1417 was derived from the F52 × W153R cross. Uh002 is a European flint (French, Spanish and German sources).
Production of highly recombined F2 plants
F1 plants originating from the cross between the DH99 and PK6 lines were self pollinated to produce F2 seeds. Intercrosses were produced between the F2 plants to produce an F2i1 (F2 intercross 1) population. In order to maximize the rate of crossing-over produced in heterozygous regions, the F2i2 population was produced by intercrossing F2i1 plants that were the progeny of four independent F2 plants. Finally, an F2i3 population was produced in the same conditions, allowing us to obtain a population of highly recombined plants.
F2 haploid induction rate determination
For each F2 plant originating from the F1 hybrid DH99 × PK6, two crosses were performed using the glossy F564 × DH7 hybrid as a female. Seeds from each cross were sown and the induction rate was evaluated as a percentage of glossy plants out of the total number of germinated seeds (200 germinated seeds tested).
Detection of haploid plants
The F1 DH99 × PK6 hybrid or the highly recombinant F2i3 population was used as male parents in the cross with the glossy F564 line. In order to avoid fungal attacks that could be deleterious for the quality of the endosperm DNA, the seeds were sown on cotton. For each glossy plant identified, the haploid status was checked using flow cytometer analysis and PCR amplification of the STS CG678277a (forward primer: GCAAATGAATACTCCAGCAA; reverse primer: GGACCATAATCATCATCATC; PCR conditions described below) defined on a BAC end sequence that, on agarose gel, showed a specific allele for F564. The plants validated for the three check points (glossy, haploid peak on flow cytometer and absence of DH99 or PK6 allele) were consider as true in situ gynogenetic induced plants.
Flow cytometer analysis
Flow cytometer analysis was performed as described in Antoine-Michard and Beckert (1997).
Extraction of DNA in leaves, endosperm and BACs
Leaves of each F2 plant were extracted according to the method described by Dellaporta et al. (1983). Endosperm DNA was recovered according to the following protocol. Endosperm from post-germinated seeds were recovered and ground in liquid nitrogen with a pestle and mortar. The powder was transferred on 700 μl of extraction buffer containing Tris base (0.1 M), EDTA (0.05 M), NaCl (0.5 M) and sodium metabisulfite (0.02 M) and incubated for 1 h at 94°C. After 10 min of centrifugation at maximum speed, 600 μl of the supernatant were recovered in 1 vol (600 μl) of isopropanol and 0.125 vol (75 μl) of 7.5 M ammonium acetate. After 5 min on ice, DNA was pelleted by centrifugation at maximum speed, washed with 70% ethanol and resuspended in 200 μl of low TE (10 mM Tris, 0.1 mM EDTA) for PCR analysis. Mini-preparation of DNA from BAC clones was performed as described in Sambrook et al. (1989).
Genetic mapping with SSR and STS markers
Amplification of PCR microsatellites was performed as described in Barret et al. (2004) and resolved on standard 5% agarose gel. Oligonucleotide sequences are provided as supplementary data 1 (Electronic Supplementary Material, ESM).
STS primers were designed using VNTI software on EST, BAC, BAC ends or GSS maize sequences (Oligonucleotides sequences are provided as supplementary data 1, in ESM). The PCR reaction mixture (25 μL) contained 1× Taq polymerase buffer (Quiagen) containing 1.5 mM MgCl2, 150 μM of each dNTP, 10 pmole of each primer, 0.5 unit of Taq polymerase (Quiagen) and 50 ng of plant genomic DNA. PCR was performed in a MJ research PCR apparatus with 1 cycle at 94°C for 3 min, 35 cycles at 94°C for 45 s, at 55°C for 45 s, at 72°C for 2 min and a final extension step at 72°C for 5 min. PCR products were resolved on standard 3% agarose gel.
Non-denaturing acrylamide gel electrophoresis
For SSCP (Single Strand Conformation Polymorphism) analysis, PCR amplifications were denaturated for 5 min at 95°C in 50% formamide and immediately placed on ice before loading. For heteroduplex analysis, PCR amplification was denaturated for 5 min at 95°C and allowed to renature at ambient temperature before loading. DNA fragments were separated by electrophoresis (20 h at 350 V) on standard 30 cm non-denaturing 5% polyacrylamide gels and stained with silver nitrate as described in Bassam et al. (1991).
Sequence analysis (last release June 2007)
In silico walking was performed using nr (non-redundant), EST and GSS maize banks. From an initial sequence (starting point), homologous sequences were identified using BLAST software (Altschul et al. 1997) on NCBI web site. Sequences showing at least 98% identity on 100 bp were assembled using ContigExpress software (Vector NTI package) under standard conditions. Successive rounds of analysis were performed for each position. All the accession numbers of the sequences used in our study for in silico walking are available upon request. Maize SSR markers were recovered on maize Genome Database (maize GDB; Lawrence et al. 2005) web site and physical map/overgo information was recovered on the maize map FTP site. The rice pseudomolecule accessions used were DP000009-2 for chromosome 3 (The rice Chromosome 3 sequencing consortium 2005) and AP008213 for chromosome 7.
Results
Identification of a major locus controlling in situ gynogenesis induction
A total of 471 F2 plants resulting from a cross between DH99, the female parent showing no induction capacities, and PK6, the male parent with a mean induction rate of 6%, were evaluated for in situ gynogenesis induction (Fig. 1). The induction rate ranged from 0 to 13.9%, and the induction of the F1 hybrid was 2.1%. Only 19 F2 plants had an induction rate higher than PK6 (>6%), and 209 F2 showed no induction in our conditions. The hypothesis of a Mendelian segregation 3:1 of the character for a single gene thus cannot be accepted (Chi square = 154; error rate 5%). We tested the hypothesis of monogenetic control with segregation distortion due to partial failure of fertilization of the male gamete triggered by the presence of the PK6 allele. Based on the number of homozygous plants for the female line (=209), we estimated the penetrance (k) of the PK6 allele [with k = H6 (homozygous plants for PK6)/H f (homozygous plants for the female line)]. The theoretical number x of heterozygous plants under the hypothesis of a k distortion for PK6 male gametophytes was estimated using the formula [x = H f (1 + k)]. The χ 2 test performed for residual segregation distortion (with H f/x/H 6 as theoretical data and 1 ddl) indicated that the hypothesis of a k distortion for PK6 male gametes cannot be excluded (k = 0.09; Chi square = 1.0; error rate 5%), therefore suggesting a monogenic trait with distortion.
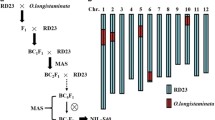
To confirm the monogenic trait controlling the in situ gynogenesis, a set of more than 300 microsatellites was tested for polymorphism between the two parental lines DH99 and PK6. One hundred and one polymorphic microsatellites covering the entire maize genome were chosen for the analysis (supplementary data 1, in ESM). Our hypothesis was that all the 19 high inducer plants are homozygous for the PK6 allele for the loci involved in the character. In that case, segregation distortion should be observed for microsatellites mapping near a locus involved in the control of in situ gynogenesis. Genotyping of the 19 highest F2 inducer plants with the 101 microsatellites identified one locus showing high segregation distortion around bin 1.04, the closest microsatellite being umc1169 (two heterozygous plants among the 19 highest inducers; Chi square = 42.26; error rate 5%; Fig. 2a). As no other microsatellite tested showed significant segregation distortion among the 19 high inducer plants, we conclude that we detected a major locus on chromosome 1 showing a significant transmission bias, thus likely correlated to in situ gynogenesis. This major locus was named ggi1 for “gynogenesis inducer 1”.
Genetic mapping and analysis of the frequency of the gynogenetic induction character using microsatellite markers. a The umc1169 marker, localised on maize chromosome 1, is closely linked to the gynogenesis induction character on the cross [DH99 (female parent showing no induction capacities) × PK6 (male parent, with a mean induction of 6%)]. b Saturation of the genetic map around the 1.04 bin using all the polymorphic microsatellite markers available. The % of linkage was those obtained on the 44 F2 plants analysed. Position of the three more closely linked microsatellites on the IBM neighbour 1 map was given in centimorgans
To increase the resolution of the genetic analysis, 25 F2 plants homozygous for the absence of induction capacities were selected [the absence of induction capacities was checked in the F3 generation (data not shown)]. The previously detected microsatellites linked to the ggi1 locus were mapped on the resulting 44 (25 + 19) F2 plants. Five other microsatellites of bin 1.04 were mapped, allowing us to define an interval of 11.6 cM (from umc1917 (374.8) to bnlg1811 (386.4) on the IBM LHR map) containing the ggi1 locus (Fig. 2b).
Segregation distortion at the ggi1 locus is due to a lower rate of transmission of the male gamete and depends on the female genotype
To check that the parental origin of the PK6 allele does not influence the induction abilities of the resulting hybrid, the two reciprocal crosses DH99 × PK6 and PK6 × DH99 were performed. No differences were observed between the two resulting F1 for the in situ gynogenesis induction capabilities on the glossy F564 × DH7 hybrid (data not shown).
Two reciprocal crosses between the hybrid [A188 × PK6] and the UH002 line were performed to test whether the lack of transmission of the PK6 allele at the ggi1 locus is due to fertilization deficiency of the male gamete. DNA of 93 plants resulting from each cross was extracted and the genotype of each plant was determined for the two microsatellites umc1917 and bnlg1811. When the PK6 gamete was of paternal origin, the two umc1917 and bnlg1811 markers were present in 32 and 34% of the progeny, respectively (χ 2 = 14.1 and 8.7, respectively, indicating distorded segregation with a confidence interval of 0.05). In addition, the markers segregated in a Mendelian way (56 and 57%, respectively) when the PK6 gamete was of maternal origin (χ 2 of 1.3 and 1.6, respectively, indicating Mendelian segregation with a confidence interval of 0.05). These results indicate that the segregation distortion observed on PK6 progenies is the consequence of a transmission failure of the PK6 allele via the male gametophyte. These experiments also demonstrated that the lack of transmission of the PK6 allele at the ggi1 locus is not specific to the cross DH99 × PK6, but also occurs in the A188 × PK6 cross.
To test whether the segregation distortion observed at the ggi1 locus was independent in the female line used, ten different crosses using A188, DH99, F7, F10, F52, DU101, F22, F72, F1417 and UH002 lines as females were evaluated (Table 1). Each line was crossed by PK6 and the resulting hybrid was selfed to produce an F2 population. Each population was genotyped with a polymorphic microsatellite linked to the ggi1 locus. We found that in seven cases (A188, DH99, F7, F52, F10, F72 and DU101 lines), the hypothesis of a Mendelian monogenic segregation was rejected by the Chi square test (Table 1). As three lines did not lead to significant segregation distortion, we concluded that the distortion observed depended on the female genotype used for hybrid production.
The in situ gynogenesis induction rate of each hybrid was assessed (Table 1). The six hybrids showing in situ gynogenetic induction (crosses between F7, F52, F10, DU101, DH99, A188 and PK6) also showed segregation distortion against the PK6 allele at the ggi1 locus (Table 1). Likewise, all hybrids that did not show induction (F22, F1417 and UH002 as female lines) presented no significant distortion, with the exception of the F72 × PK6 hybrid (see “Discussion”). These data indicate a correlation between the induction rate and the segregation distortion at the ggi1 locus.
As described above for the DH99 × PK6 cross, we estimated for each cross the penetrance (k) of the PK6 allele to quantify the distortion (Table 1). The five hybrids with the highest induction rates (F7, F52, F10, DU101 and DH99) displayed the lowest penetrance of PK6 allele (k ≤ 0.41). The five other hybrids showing the lowest rates of induction also showed the highest penetrance (k ≥ 0.42). These data showed a significant correlation (r² = 0.42) between penetrance of the PK6 allele at the ggi1 locus and penetrance of “in situ gynogenesis” induction of the initial F1 hybrid plant. The Uh002 × PK6 cross showed a distortion against Uh002 gametes, indicating a possible failure of fertilization of the Uh002 accession and, as a consequence, false data concerning the estimation of PK6 fertilization capacities in this cross. A new analysis conducted without the Uh002 × PK6 cross data gave a highly significant correlation (r² = 0.61).
PK6 allele at the ggi1 locus is responsible for in situ gynogenesis
To determine whether part of the under-represented male gametes showing the PK6 allele at the ggi1 locus were involved in the production of female haploid plants, the genotypes of the male gamete inducers at the ggi1 locus were checked. If double fertilization takes place, the egg cell of the female gametophyte is induced for the development of the gynogenetic embryo, and the central cell is fertilized by the second nucleus. Thus, it is possible to identify the genotype of the male gametophyte by analysing endosperm DNA. The pollen of the hybrid [DH99 × PK6] was used to pollinate the glossy F564 line. The resulting seeds were sown and putative haploid plantlets (i.e. glossy) were obtained. Haploidy status was checked by flow cytometer and by genotyping with the umc1829 microsatellite. DNA was extracted from the endosperm of 33 post-germinated seeds (Table 2). We first checked that Mendelian segregation was observed for the umc1829 microsatellite that was not linked to the ggi1 locus (data not shown). Four microsatellites (umc1144, umc1917, bnlg1811 and umc1590) spanning the region of the ggi1 locus were then tested (see Fig. 3a for analysis of umc1144). Out of the 33 endosperm analysed, all but one (GA33) showed only the PK6 allele for the four microsatellites tested. The gynogenetic-related endosperm 33 (GA33) showed the PK6 allele for bnlg1811 and umc1590, and the DH99 allele for umc1144 and umc1917 (Table 2). As the interval containing the ggi1 locus was located between umc1917 and bnlg1811, we cannot exclude that GA33 presents the PK6 allele at this locus (see “Discussion”). These results indicate that there is a significant correlation between the presence of the PK6 allele at the ggi1 locus and gynogenesis induction.
F564 female plants were induced by the pollen of the hybrid DH99 × PK6. Endosperm DNA recovered from haploid plants originating from kernels were subjected to genetic analysis. a Analysis was performed using the umc1144 microsatellite, closely linked to the ggi1 locus. All the endosperm genotyped showed the maternal F564 allele, and only the paternal PK6 allele. b Genetic mapping of the STS marker derived from the AY110477 cDNA sequence. All the electrophoreses were performed on 5% standard agarose gels
The ggi1 locus shows colinearity with rice chromosomes 3 and 7
To increase the density of markers, we investigated the colinearity between the maize ggi1 locus region and orthologous regions of the rice genome. To this end, in silico mapping of microsatellites umc1917, umc2390 and bnlg1811 was performed on rice pseudomolecules. Umc1917 and umc2390 were directly mapped in silico on rice pseudomolecule 3 in a proximal region (Fig. 4). As bnlg1811 did not directly match any rice sequence using blastN or TblastX, chromosome walking was conducted using maize GSS (genome survey sequences). Starting from the two bnlg1811 primers, the maize GSS sequence was identified (CG301067). After two rounds of blastN analysis, three other maize GSS sequences were identified, allowing us to construct in silico a sequence of 2,409 bp (see supplementary data 2 for sequence accessions of minimal tailing path contigs, in ESM). TblastX analysis of this contig allowed us to anchor the bnlg1811 locus on rice chromosome 3 (Supplementary data 2, in ESM and Fig. 4). The three microsatellites mapped on the same orthologous region of rice pseudomolecule 3, and in the same order. Moreover, a second orthologous region was identified for umc2390 and bnlg1811 on pseudomolecule 7 (Supplementary data 2, in ESM and Fig. 4). The interval spanning the ggi1 locus between umc1917 and bnlg1811 markers was partially anchored on rice chromosome 7 and completely anchored on rice chromosome 3. The size of the rice chromosome 3 region corresponding to this locus was 2,574 kb (rice pseudomolecule DP000009-2).
In silico alignment of the [umc1917-bnlg1811] interval on rice chromosomes 3 and 7 physical maps. Bold type: Microsatellites shown in Fig. 3. All the maize accessions presented were putatively positioned at the ggi1 locus according to their position on rice, but only underlined accessions were localized on the maize region by genetic mapping
To confirm colinearity between maize and rice at the ggi1 locus, the predicted rice protein sequences on the region were used in a blastX analysis against nr (non-redundant), EST (expressed sequence tagged), GSS and htgs (unfinished high throughput genomic sequences) maize sequences. Using this strategy, 34 non-repeated maize sequences putatively localised at the ggi1 locus were identified (Fig. 4) and used to design PCR primers (see supplementary data 1 for primers sequences, in ESM). Polymorphism was evaluated between the DH99 and PK6 parental lines using electrophoresis on high-percentage agarose gels, or heteroduplex analysis on non-denaturing acrylamide gels. Thirty-two couples of primers resulted in amplification, and nine were polymorphic. The 44 F2 plants from the DH99 × PK6 cross were genotyped using these new markers. All these markers mapped at the ggi1 locus (Fig. 4). These new data confirmed that we had identified a major region that was orthologous to the ggi1 locus on rice chromosome 3. Moreover, the order of markers mapped in maize was conserved in rice, indicating a good colinearity between the two regions. Genetic mapping of the CD972903 marker in maize reduced the size of the genetic interval containing the ggi1 locus (Fig. 4). The size of the orthologous region on rice was reduced from 2,574 to 1,609 kb.
Establishment of a physical contig spanning the ggi1 region in maize
The bnlg1811 and umc1917 microsatellites identified the Cg29 and Cg28 maize contigs, respectively, on the maize FPC map web site. To validate the orthologous position of the contigs, sequence data available for these two contigs (hybridization probes, cDNA associated with overgos) were used for BlastN and TblastX analysis against the rice chromosome 3 pseudomolecule. Eight new in silico linkage points were obtained for Cg28 and four for Cg29 (Fig. 5). Moreover, the markers CD438541 and CF630266 were found on Cg28 and Cg29, respectively, confirming the position of the contigs on the genetic map.
To establish a physical contig spanning the ggi1 locus, we needed to identify a third contig between CD972903 and AY104356 (Fig. 5). Overgo (overlapping oligonucleotides) sequences were recovered from six maize sequences that were previously identified as putatively localised on this region (CF631359, BU037361, AW424491, AA054821, CF636018 and CD972903; Fig. 4). Sequence CF631359 was associated with cDNA AY107563 that contained the overgo PCO093543_ov. This overgo enabled us to identify contig number 1095 (maize FPC map web site). cDNA sequences associated with other overgos present on the Cg1095 and BAC end sequences were also checked for a possible orthologous position on rice chromosome 3 or 7. Three sequences associated with overgos (AY112393/ CL14390-1-ov; AY106910/ PCO124560_ov; AY105275/ PCO143153_ov) and one BAC end (CD440214) were localised on the expected interval on the rice chromosome 3 pseudomolecule (Fig. 5). Moreover, the order of the overgos on the maize contig was the same as those on the rice chromosome, indicating perfect colinearity in this region. This result was confirmed by genetic mapping at the ggi1 locus of the CG22-1 marker (Fig. 4) identified after in silico walking from AY105275 (data not shown) and physically localised by PCR on contig 1095. The anchorage was confirmed by PCR amplification of the CD972903 marker at the end of the Cg1095 on BAC clone c0487O08 (Fig. 5). In conclusion, three maize BAC contigs were anchored on the maize region spanning the ggi1 locus.
Fine mapping of the ggi1 locus
Genetic mapping data obtained herein were based on a F2 population of 44 individual plants. This small population did not allow fine mapping of the ggi1 locus. The 471 F2 plant population originating from the DH99 × PK6 cross could have been used, but the phenotypic data obtained in our conditions were not sufficiently precise to differentiate plants that were potentially homozygous versus heterozygous for gynogenetic induction. Phenotyping of at least 20 F3 plants for each F2 plant would have been necessary to improve the quality of the F2 data, requiring the phenotyping of about 8,500 plants, which was not possible in our conditions.
To overcome this problem, we created a new mapping population based on the data obtained on endosperm recovered from haploid kernels (Table 2). We hypothesize that these types of endosperm were generated by a pollen grain presenting the PK6 allele at the ggi1 locus. Under this hypothesis, the data presented in Table 2 indicate that endosperm GA33 is recombinant and that the ggi1 gene cannot be present on the interval [umc1144-umc1917].
We anticipated encountering two problems in using this particular population to perform fine mapping of the ggi1 gene. First, the wild type standard induction (estimated at up to 0.1% by Chase 1949) can lead to haploid plantlets not related to induction mechanisms of the ggi1 gene. As the induction rate of the DH99 × PK6 hybrid was estimated to be 2.1% (Table 1), the error rate due to the standard induction would be (0.1/2.1)/2 = 2.4%. Second, heterofertilization (which occurs when the egg cell and the central cell are fertilized by sperm cells originating from two different pollen grains) can lead to decorrelation of the genotype of the endosperm of the haploid plantlet and the genotype of the pollen nucleus that carries the ggi1 allele. We estimated the heterofertilization rate to be 3.8% by comparing the genotypes of 124 plantlets and endosperms from the F564 × F1[DH99 × PK6] cross (data not shown). Thus, the error rate due to heterofertilization was 3.8/2 = 1.9%. The sum of error rates was 2.4 + 1.9 = 4.3%. We assumed that we would not find markers more closely linked to the ggi1 gene than ~4.3 cM (Fig. 5). Two mapping populations were constituted: a population of 306 recombinant endosperms originating from the F564 (gl/gl) × F1[DH99(Gl/Gl) × PK6(Gl/Gl)] cross and a population of 259 highly recombinant endosperms originating from the F564 (gl/gl) × F2i3 [DH99(Gl/Gl) × PK6(Gl/Gl)] cross. Each plantlet showing the glossy phenotype was checked for female haploidy (see “Materials and methods” for details). More than 80,000 seeds were sown to obtain these two populations.
Markers previously mapped around the ggi1 locus were mapped on the two populations described (Figs. 3b, 5). One new marker, CL424968-1, derived from a BAC end, was mapped near the bnlg1811 locus. Two other markers, AY111534 and AY110477, derived from overgos positioned on BAC contigs 28 and 1095, respectively, were found closely linked to the ggi1 locus (4.5 and 4.9 cM, respectively). According to the previously calculated confidence interval (4.3 cM), the identification of molecular markers more closely linked to the ggi1 locus in our particular population would be impossible. For this reason, the four non-polymorphic markers identified from the [AY111534-AY110477] interval (Fig. 5) were not subjected to further analysis. The orthologous interval in the rice genome was 135 kbp and did not contain any predicted gene previously described as being directly involved in plant reproduction (data not shown).
We identified the major ggi1 locus for in situ gynogenesis induction in maize. This locus was physically localised on maize chromosome 1. We showed that in situ gynogenetic induction had a gametophytic expression with incomplete penetrance and that this expression was correlated to partial loss of fertilization of the male gamete.
Discussion
Genetic mapping of ggi1, the major locus involved in in-situ gynogenesis in maize
In-situ gynogenesis was assessed in maize. In F2 plants derived from the DH99 × PK6 cross, a high percentage (38%) of non-inducer plants was observed. Deimling et al. (1997) observed also a high percentage (30%) of non-inducer plants on the W23ig × Stock6 cross. Segregation distortion of genes that are important for gametophytic growth and development is a common feature (Howden et al. 1998; Bonhomme et al. 1998). We proposed the hypothesis of a monogenetic control with distorded segregation due to partial fertilization failure of the male gamete carrying the PK6 allele.
Genetic mapping of microsatellite markers enabled us to identify a major locus involved in gynogenesis induction on bin 1.04. This locus is at the same location as the major QTL identified by Deimling et al. (1997) from the W23ig × Stock6 cross. This result completed previous data suggesting an oligogenic component for the inducer character (Lashermes and Beckert 1988; Deimling et al. 1997). A second minor QTL, with an inducing allele coming from the non-inducer parent, was detected by Deimling et al. (1997) but was not identified in our experimental conditions. This second QTL could be specific to the W23ig × Stock6 cross, or due to the above authors’ experimental conditions.
Microcolinearity between rice and maize enabled identification of new markers and physical mapping of previously unassigned BAC contigs
Microsatellite markers enabled us to identify an interval of 11.6 cM containing the ggi1 locus. To obtain a more precise location for the locus, new markers were identified using the rice genome as an anchor. Extensive studies were performed to describe the synthenic relationship between maize and rice (Ahn and Tanksley 1993; Gale and Devos 1998; Wilson et al. 1999; Salse et al. 2004). Orthology between maize chromosome 1 and rice chromosome 3 in our region of interest has been well established (The rice chromosome 3 sequencing consortium 2005) and microcolinearity between grass genomes has been proposed as a helpful tool for gene cloning in species like wheat and maize, even if detailed analysis revealed a number of exceptions depending on the region analysed (Feuillet and Keller 1999, 2002; Keller and Feuillet 2000; Brunner et al. 2003). Our results showed that the level of colinearity between the region containing the ggi1 gene and rice chromosome 3 was sufficient to perform efficient chromosome walking on maize using rice genome sequences. We demonstrated that, in our conditions, the recovery of overgo sequences showing in silico homologies with rice sequences orthologous to the ggi1 region was an original and efficient method to identify BAC contigs spanning the locus.
Fine mapping of the ggi1 locus and partial identification of a physical contig containing the gene
Genetic mapping of the character was performed on endosperm populations corresponding to F2 and highly recombinant lines. In the same way, two genetic maps were constructed on Pinus pinaster (maritime pine) using haploid megagametophyte tissues (Plomion et al. 1995; Costa et al. 2000). The first limitation of our system was the occurrence of heterofertilization events that disconnected the endosperm and embryo genotype. Robertson (1984) estimated a mean of 2% for the standard heterofertilization rate while reporting that both male and female genotypes can influence this percentage, and suggested that some aspects of this process might be under genic control. Choosing male-female couples showing low rates of heterofertilization could minimize the standard error (the rate could be around 1% instead of 3.8% in our conditions). The second limitation was the occurrence of a standard induction rate of 0.1% (Chase 1949). As the error rate depends on the ratio between the standard induction rate and the induction of the cross, choosing a high inducing cross would lead to a lower error rate. According to the lowest heterofertilization rates observed (about 1%) and the highest induction cross (in our case F7 × PK6; 3.4%), we estimated that the error rate can be limited to 2%, but only if heterofertilization and gynogenesis induction are independent events (Kraptchev et al. 2003).
The ratio between genetic distances and physical distances (measured as the approximate number of minimal tailing path BACs on the contig sections) was estimated to be from 0.5 to 1 cM per BAC clone using a F2 and a highly recombinant population, which are similar to the results obtained by Brunner et al. (2003) on barley (0.3 cM for one BAC on an F2 population), except that the ratio is very high due to the relatively small size of the population (565 endosperms). Moreover, a very high ratio was probably present (depending on the size of the gap between contigs 28 and 1095) around the ggi1 gene (up to 5 cM/BAC). The presence of numerous hot spots of recombination in the same region is highly improbable. Another explanation could be that the selection of the genetic material (i.e. the recovery of the endosperm of the haploid plantlets) could have introduced a distortion under the hypothesis that a cluster of genes involved in reproduction with partial deleterious effects on PK6 spans the region around the ggi1 locus. Lee and Sonnhammer (2006) described gene clustering in eukaryotes, with a species-specific pathway of clustering. According to recent advances in maize genomics (Bortiri et al. 2006), map-based cloning of the ggi1 gene will be possible in the near future.
Segregation distortion at the ggi1 locus and possible correlation with the induction level
The PK6 line was crossed with diverse maize lines to obtain F2 populations. Even if more crosses need to be evaluated to confirm the results, our data suggest for the first time that a possible quantitative correlation exists between the rate of gynogenetic induction of a hybrid and the level of segregation distortion observed on the corresponding F2 population. Lu et al. (2002) detected chromosomal regions associated with segregation distortion in maize. They detected a segregation distortion region (SDR 1.1) covering a part of bin 1.04 in one cross out of the four they examined. These results suggest that one distorted marker was observed in one cross near the ggi1 locus, but that segregation distortion is not a general feature in bin 1.04. We can thus conclude that this is the first time a relationship has been established between distorded segregation of the PK6 allele at the ggi1 locus and gynogenetic induction.
The pollen grain genotype at the ggi1 locus is highly correlated with its capacity to induce a haploid female embryo
The results presented here suggest a relationship between the failure of the gametophytes containing the PK6 allele at the ggi1 locus to perform complete fertilization and gynogenetic induction on one hand, and demonstrate a high correlation between the presence of the PK6 allele at the ggi1 locus and the capacity of the resulting pollen grain to induce gynogenesis, on the other hand. Taken together, these results indicate that gynogenetic induction is an phenomenon related to dysfunction(s) of the fertilization mechanism (Fig. 6). The phenotype associated with the ggi1-PK6 allele showed incomplete penetrance, as the resulting gametes could lead to either normal fertilization (plants homozygous for the ggi1-PK6 allele were recovered in the progeny) or absence of fertilization (segregation distortion occurred in the progeny if the male donor was a hybrid). Incomplete penetrance has previously been described for affected mutants in pollen development (Chen and McCormick 1996; Bonhomme et al. 1998; Park et al. 1998) and for the in vivo androgenesis inducer ig1 (Evans 2007). One way to approach the concept of incomplete penetrance would be to consider that pollen is not made up of a single population of cells, but by multiple sub-populations that could be developmentally defined. A character with complete penetrance in one sub-population would be expected to be determined in fine as incompletely penetrant. During androgenesis, another form of gametophytic embryogenesis, only a small proportion of microspores develop in vitro. If microspores were provided by a completely fixed double haploid line, the character could appear as incompletely penetrant. Sub-populations of yeast on the same clone that showed differential responses to stress were described by Madeo et al. (2002). Maraschin et al. (2003a, b) demonstrated that during androgenesis in Hordeum vulgare, one sub-population of microspores underwent embryogenesis while one other sub-population followed a pathway related to PCD (programmed cell death).
Proposed model for induction of gynogenesis. Male gametes with PK6 allele at the ggi1 locus (G6) fertilized (white circles) or failed to fertilize (grey circles) the F564 line with incomplete penetrance. Female haploid embryos could result from non fertilization events. a In the PK6 × F7 hybrid, male sporophytes failed to complement the ggi1-PK6 mutation. Segregation distortion and haploid embryos were recovered in the progeny. b In the PK6 × F22 cross, male sporophytes complement the ggi1-PK6 allele for fertilization. No segregation distortion and no haploids were observed. c PK6 showed reduced fertility due to the PK6 allele at the ggi1 locus. As all the gametes were G6, the gynogenesis induction rate was higher than for hybrids
Our data showed that the presence of the PK6 allele at the ggi1 locus is necessary, but not sufficient, to induce gynogenesis. As the rate of penetrance of the character is related to the genotype of the male sporophyte, a sporophytic pathway capable of complementing the ggi1-PK6 mutation could be involved. Under this hypothesis, F7 and, a fortiori, PK6 genetic backgrounds would not be capable of complementing ggi1-PK6 mutation, although the genetic background of the sporophytic F22 rendered ggi1-PK6 pollen capable of fertilization (Fig. 6). We hypothesize also that failure of fertilization, undetectable in our conditions, occurred in some of the male gametes in the PK6 line. The same kind of feature has been described for the Arabidopsis male gametophytic mutant gem1 (Gemini pollen 1) that showed altered division symmetry (Park et al. 1998). Heterozygous gem1 mutants showed only ~20% aberrant pollen, and reduced penetrance was confirmed in gem1 homozygotes which produced ~40% phenotypically aberrant pollen. Interestingly, like in ggi1, a large proportion of pollen that carries the gem1 mutation, but appears to be of the wild type, fails to produce viable seeds. In the other Arabidopsis male gametophytic mutant scp (sidecar pollen) affected for microspore division during pollen development, the mutant character showed, like in ggi1, differential gametophytic penetrance and variable expressivity in the genetic backgrounds of different Arabidopsis ecotypes (Chen and McCormick 1996).
Complete analysis of the gynogenesis induction phenomena will provide new data for further investigation of double fertilization (Friedman 1998). Future work will need to determine if the development of female haploid embryos is induced by fertilization failure of the egg cell, as observed in some interspecific crosses (Zhang et al. 1996) or is related to the fertilization and/or the development of the endosperm, as complex signals appear to occur between the two tissues (Nowack et al. 2006). This work also opens new prospects for single fertilization of the central cell, one component of apomixis.
References
Ahn S, Tanksley SD (1993) Comparative linkage maps of the rice and maize genomes. Proc Natl Acad Sci USA 90:7980–7984
Ainsworth C (2005) The secret life of sperm. Nature 436:770–771
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Antoine-Michard S, Beckert M (1997) Spontaneous versus colchicines induced chromosome doubling in maize anther culture. Plant Cell Tissue Organ Cult 48:203–207
Barloy D, Denis L, Beckert M (1989) Comparison of the aptitude for anther culture in some androgenetic doubled haploid maize lines. Maydica 34:303–308
Barret P, Brinkman M, Dufour P, Murigneux A, Beckert M (2004) Identification of candidate genes for in vitro androgenesis induction in maize. Theor Appl Genet 109:1660–1668
Bassam BJ, Caetano-Anollés G, Gresshof PM (1991) Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal Biochem 196:80–83
Boavida LC, Vieira AM, Becker JD, Feijo JA (2005) Gametophyte interaction and sexual reproduction: how plants make a zygote. Int J Dev Biol 49:615–632
Bonhomme S, Horlow C, Vezon D, de Laissardière S, Guyon A, Férault M, Marchand M, Berchtold N, Pelletier G (1998) T-DNA mediated disruption of essential gametophytic genes in Arabidopsis is unexpectedly rare and cannot be inferred from segregation distorsion alone. Mol Gen Genet 260:444–452
Bordes J, Dumas de Vaulx R, Lapierre A, Pollacsek M (1997) Haplodiploidization of maize (Zea mays L.) through induced gynogenesis assisted by glossy markers and its use in breeding. Agronomie 17:291–297
Bordes J, Charmet G, Dumas de Vaulx R, Pollacsek M, Beckert M, Gallais A (2006) Doubled haploid versus S1 family recurrent selection for testcross performance in a maize population. Theor Appl Genet 112:1063–1072
Brettell RIS, Thomas E, Wernicke W (1981) Production of haploid maize plant by anther culture. Maydica 26:101–111
Bortiri E, Jackson D, Hake S (2006) Advances in maize genomics: the emergence of positional cloning. Curr Opin Plant Biol 9:1–8
Brunner S, Keller B, Feuillet C (2003) A large rearrangement involving genes and low-copy DNA interrupts the microcollinearity between rice and barley at the Rph7 locus. Genetics 164:673–683
Chase SS (1949) Monoploid frequencies in a commercial double cross hybrid maize, and in its component single cross hybrids and inbred lines. Genetics 34:328–332
Chen Y-CS, McCormick S (1996) sidecar pollen, an Arabidopsis thaliana male gametophytic mutant with aberrant cell divisions during pollen development. Development 122:3243–3253
Chevalier D, Sieber P, Schneitz K (2002) The genetic and molecular control of ovule development. In: O’Neill SD, Roberts JA (eds) Plant Reproduction. Sheffield Academic Press, Sheffield, pp 61–81
Coe EH (1959) A line of maize with high haploid frequency. Am Nat 93: 381–382
Costa P, Pot D, Dubos C, Frigerio JM, Pionneau C, Bodenes C, Bertocchi E, Cervera MT, Remington DL, Plomion C (2000) A genetic map of Maritime pine based on AFLP, RAPD and protein markers. Theor Appl Genet 100:39–48
Deimling S, Röber F, Geiger HH (1997) Methodik und genetic der in-vivo-haploideninduktion bei mais. Vortr Pflanzenzüchtg 38:203–224
Dellaporta SL, Wood J, Hicks JB (1983) A plant DNA minipreparation: version 2. Plant Mol Biol Rep 1:19–22
Eder J, Chalyk S (2002) In vivo haploid induction in maize. Theor Appl Genet 104:703–708
Edlund AF, Swanson R, Preuss D (2004) Pollen and stigma structure and function: the role of diversity in pollination. Plant Cell 16:S84–S97
Engel ML, Chaboud A, Dumas C, McCormick S (2003) Sperm cells of Zea mays have a complex complement of mRNAs. Plant J 34:697–707
Escobar-Restrepo J-M, Huck N, Kessler S, Gagliardini V, Gheyselinck J, Yang W-C, Grossniklaus U (2007) The FERONIA receptor-like kinase mediates male-female interactions during pollen tube reception. Science 317:656–660
Evans MMS (2007) The indeterminate gametophyte 1 gene of maize encodes a LOB domain protein required for embryo sac and leaf development. Plant Cell 19:46–62
Feuillet C, Keller B (1999) High gene density is conserved at syntenic loci of small and large grass genome. Proc Natl Acad Sci USA 96:8265–8270
Feuillet C, Keller B (2002) Comparative genomics in the grass family: molecular characterization of grass genome structure and evolution. Ann Bot 89:3–10
Friedman WE (1998) The evolution of double fertilization and endosperm: an “historical” perspective. Sex Plant Reprod 11:6–16
Gale MD, Devos KM (1998) Plant comparative genetics after 10 years. Science 282:656–659
Gemand D, Rutten T, Varshney A, Rubtsova M, Prodanovic S, Brüb C, Kumiehn J, Matzk F, Houben A (2005) Uniparental chromosome elimination at mitosis and interphase in wheat and pearl millet crosses involves micronucleus formation, progressive heterochromatinization, and DNA fragmentation. Plant Cell 17:2431–2438
Gémes-Juhasz A, Balogh P, Ferenczy A, Kristof Z (2002) Effect of optimal stage of female gametophyte and heat treatment on in vitro gynogenesis induction in cucumber (Cucumis sativus L.). Plant Cell Rep 21:105–111
Guitton AE, Page DR, Chambrier P, Lionnet C, Faure JE, Grossniklaus U, Berger F (2004) Identification of new members of Fertilization Independant Seed Polycomb group pathway involved in the control of seed development in Arabidopsis thaliana. Development 131:2971–2981
Hagberg A, Hagberg G (1980) High frequency of spontaneous haploids in the progeny of an induced mutation in barley. Hereditas 93:341–343
Howden RS, Park K, Moore JM, Orme J, Grossniklaus U, Twell D (1998) Selection of T-DNA-tagged male and female gametophytic mutants by segregation distortion in Arabidopsis. Genetics 149:621–631
Huang B-Q, Sheridan WF (1996) Embryo sac development in the maize indeterminate gametophyte 1 mutant: abnormal nuclear behaviour and defective microtubule organization. Plant Cell 8:1391–1407
Huck N, Moore JM, Federer M, Grossniklaus U (2003) The Arabidopsis mutant feronia disrupts the female gametophytic control of pollen tube reception. Development 130:2149–2159
Johnson-Brousseau SA, McCormick S (2004) A compendium of methods useful for characterizing Arabidopsis pollen mutants and gametophytically-expressed genes. Plant J 39:761–775
Keller B, Feuillet C (2000) Colinearity and gene density in grass genomes. Trends Plant Sci 5:246–251
Kermicle JL (1969) Androgenesis conditioned by a mutation in maize. Science 166:1422–1424
Kraptchev B, Kruleva M, Dankov T (2003) Induced heterofertilization in maize (Zea mays L.). Maydica 48:271–273
Krawetz SA (2005) Paternal contribution: new insights and future challenges. Nat Rev Genet 6:633–642
Lashermes P, Beckert M (1988) Genetic control of maternal haploidy in maize (Zea mays L.) and selection of haploid inducing lines. Theor Appl Genet 76:405–410
Lawrence CJ, Seigfried TE, Brendel V (2005) The Maize Genetics and Genomics Database. The community resource for access to diverse maize data. Plant Physiol 138:55–58
Lee JM, Sonnhammer ELL (2006) Genomic gene clustering analysis of pathways in eukaryotes. Genome Res 13:875–882
Lin B-Y (1984) Ploidy barrier to endosperm development in maize. Genetics 107:103–115
Lu H, Romero-Severson J, Bernardo R (2002) Chromosomal regions associated with segregation distorsion in maize. Theor Appl Genet 105:622–628
Madeo F, Engelhardt S, Herker E, Lehmann N, Maldener C, Proksch A, Wissing S, Fröhlich K-U (2002) Apoptosis in yeast: a new model system with applications in cell biology and medicine. Curr Genet 41:208–216
Mahendru A, Sarkar KR (2000) Cytological analysis of the pollen of haploidy inducer lines in maize (Zea mays L.). Indian J Genet 60:37–43
Maraschin SF, Lamers GEM, de Pater BS, Spaink HP, Wang M (2003a) 14-3-3 isoforms and pattern formation during barley microspore embryogenesis. J Exp Bot 51:1033–1043
Maraschin SF, Lamers GEM, Wang M (2003b) Cell death and 14–3-3 proteins during the induction of barley microspore androgenesis. Biologia 58:59–68
Martinez LE, Agüero CB, Lopez ME, Galmarini CR (2000) Improvement of in vitro gynogenesis induction in onion (Allium cepa L.) using polyamines. Plant Sci 156:221–226
Matzk F, Mahn A (1994) Improved techniques for haploid production in wheat using chromosome elimination. Plant Breed 113:125–129
McCormick S (2004) Control of male gametophyte development. Plant Cell 16:S142–S153
Mori T, Kuroiwa H, Higashiyama T, Kuroiwa T (2006) Generative Cell Specific 1 is essential for angiosperm fertilization. Nat Cell Biol 8:64–71
Nowack MK, Grini PE, Jakoby MJ, Lafos M, Koncz C, Schnittger A (2006) A positive signal from the fertilization of the egg cell sets off endosperm proliferation in angiosperm embryogenesis. Nat Genet 38:63–67
Nowack MK, Shirzadi R, Dissmeyer N, Dolf A, Endl E, Grini PE, Schnittger A (2007) Bypassing genomic imprinting allows seed development. Nature 447:312–315
Park SK, Howden R, Twell D (1998) The Arabidopsis thaliana gametophytic mutation gemini pollen 1 disrupts microspore polarity, division asymmetry and pollen cell fate. Development 125:3789–3799
Plomion C, Bahrman N, Durel C-E, O’Malley DM (1995) Genomic mapping in Pinus pinaster (maritime pine) using RAPD and protein markers. Heredity 74:661–668
Procissi A, de Laissardière S, Férault M, Vezon D, Pelletier G, Bonhomme S (2001) Five gametophytic mutations affecting pollen development and pollen tube growth in Arabidopsis thaliana. Genetics 158:1773–1783
Röber FK, Gordillo GA, Geiger HH (2005) In vivo haploid induction in maize- Performance of new inducers and significance of doubled haploid lines in hybrid breeding. Maydica 50:275–283
Robertson DS (1984) A study of heterofertilization in diverse lines of maize. J Hered 75:457–462
Rotman N, Rozier F, Boavida L, Dumas C, Berger F, Faure J-E (2003) Female control of male gamete delivery during fertilization in Arabidopsis thaliana. Curr Biol 13:432–436
Salse J, Piégu B, Cooke R, Delseny M (2004) New in silico insight into the syntheny between rice (Oryza sativa L.) and maize (Zea mays L.) highlights reshuffling and identifies new duplications in the rice genome. Plant J 39:960–968
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn Cold Spring Harbor Laboratory. Cold Spring Harbor, New York
Subrahmanyam NC, Kasha KJ (1973) Selective chromosomal elimination during haploid formation in barley following interspecific hybridization. Chromosoma 42:111–125
The rice Chromosome 3 sequencing consortium (2005) Sequence, annotation, and analysis of syntheny between rice chromosome 3 and diverged grass species. Genome Res 15:1284–1291
Twell D (2002) The developmental biology of pollen. In: O’Neill SD, Roberts JA (eds) Plant Reproduction. Sheffield Academic Press, Sheffield, pp 86–153
Weterings K, Russell SD (2004) Experimental analysis of the fertilization process. Plant Cell 16:S107–S118
Wilson WA, Harrington SE, Woodman WL, Lee M, Sorrells M, McCouch S (1999) Inferences on the genome structure of progenitor maize through comparative analysis of rice, maize and the domesticated panicoids. Genetics 153:453–473
Yadegari R, Drews GN (2004) Female gametophyte development. Plant Cell 16:S133–S141
Zhang J, Friebe B, Raupp WJ, Harrison SA, Gill BS (1996) Wheat embryogenesis and haploid production in wheat × maize hybrids. Euphytica 90:315–324
Acknowledgments
We would like to thank C. Feuillet (INRA, Clermont-Ferrand) and S. Bonhomme (INRA, Versailles) for their comments on the manuscript, and P. Rogowsky (ENS; Lyon) for helpful discussion. Our thanks also go to the greenhouse and culture chamber team (Richard Blanc, Raphael Giroud, Jean Claude Girard, Christophe Serre and Christophe Troquier) for ensuring excellent conditions for the controlled, reproducible growth of plants. This work was supported by the Institut National de la Recherche Agronomique (INRA) and PROMAIS.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by H. H. Geiger.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Barret, P., Brinkmann, M. & Beckert, M. A major locus expressed in the male gametophyte with incomplete penetrance is responsible for in situ gynogenesis in maize. Theor Appl Genet 117, 581–594 (2008). https://doi.org/10.1007/s00122-008-0803-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-008-0803-6