Abstract
It is important to detect wheat broad-spectrum powdery mildew resistance (Pm) genes in commercial cultivars with superior agronomic performance. YingBo 700, a Chinese commercial cultivar registered and released in 2012, showed a broad spectrum of resistance to 48 of 49 tested Blumeria graminis f. sp tritici (Bgt) isolates from different regions of China. Genetic analysis of F2 populations from the crosses YingBo 700 × Mingxian 169 and YingBo 700 × Shimai 15 and F2:3 lines from YingBo 700 × Mingxian 169 demonstrated that the resistance was controlled by a single dominant resistance gene, which was temporarily designated PmYB. Using molecular markers and bulked segregant analysis of the F 2:3 lines from YingBo 700 × Mingxian 169, PmYB was mapped to the multi-allelic Pm2 region of chromosome arm 5DS co-segregated with the sequence-characterized amplified region (SCAR) marker SCAR112 and flanked by the simple sequence repeat (SSR) marker Cfd81 and SCAR marker SCAR203 with genetic distance of 0.9 and 1.9 cM, respectively. YingBo 700 showed a unique response pattern to different Bgt isolates compared with the lines with documented Pm2a, Pm2b, Pm48, PmLX66, PmW14, PmZ155 and PmX3986-2 on chromosome arm 5DS. Therefore, PmYB appears to be a novel allele in this multi-allelic region. In order to validate applicability of the closely linked markers for marker-assisted selection, the closely linked markers Cfd81, SCAR112, SCAR203 and Mag6176 were evaluated for applicability in diagnosing the resistance gene in 62 cultivars from different regions of China. The results will contribute to rapidly transfer of PmYB into more cultivars.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Common wheat (Triticum aestivum L.) is one of the most important food crops in the world, but its yield is continuously challenged by various wheat diseases (Costanzo and Bàrberi 2014). Worldwide wheat powdery mildew, caused by Blumeria graminis f. sp. tritici (Bgt), is a devastating and rapidly spreading disease that can cause severe yield losses in a short time (Hao et al. 2014). Of all the defensive measures, host resistance is the most effective, economical and environmentally sound and consistently used method (Petersen et al. 2014; Wang et al. 2015). In the history of breeding for host resistance, some powdery mildew resistance (Pm) genes have played a key role reducing the yield losses of wheat, such as Pm8, one of the most effective and widely popularized Pm gene in the last century that has made great contributions to the control of the wheat powdery mildew for more than two decades (Zeller and Hsam 1996; Huang and Röder 2004; An et al. 2013). Excessive use of this gene led to ‘boom-bust’ cycles of epidemics in the 1990s when new isolates appeared that were capable of overcoming Pm8 (Ren et al. 1997; McIntosh et al. 2011; Ren et al. 2009). Currently, even the most effective and popular resistance gene Pm21 is also suffering severe challenge, since some variant isolates are able to defeat this gene (Shi et al. 2009; Ren et al. 2011). Therefore, gene discovery and allele mining for powdery mildew resistance is a continuous challenge.
Up to now, about 70 formally designated Pm genes/alleles Pm1–Pm54 at 49 loci (Pm8 is allelic to Pm17, Pm18 = Pm1c, Pm22 = Pm1e, Pm23 = Pm4c and Pm31 = Pm21) and an additional about 20 temporarily named Pm genes have been identified on all the chromosomes except 3D in common wheat and its relatives (Hao et al. 2014; McIntosh et al. 2014). Of these genes, some have been not effective to the new virulent Bgt isolates, some are identified in host lines with linkage drags that lead to poor agricultural performance, so are seldom used in resistance breeding, and some need further time before their use (e.g., Zeller and Hsam 1996; Friebe et al. 1994; Ma et al. 2014). However, one class of Pm genes has been identified in commercial cultivars that provide broad-spectrum powdery mildew resistance (e.g., Huang et al. 2012; Zhao et al. 2013). A combination of excellent agronomic performance and broad-spectrum resistance offers an attractive prospect in resistance breeding. In China, some cultivars have been reported to show resistance to powdery mildew by a large number of powdery mildew isolates (Li et al. 2011; Song et al. 2012). On chromosome arm 2AL, two Pm genes PmYm66 linked to Ksum193 and PmHNK54 flanked by Barc5 and Gwm312 were identified in Yumai 66 and Zheng 9754, respectively (Hu et al. 2008a; Xu et al. 2011). In Tangmai 4, the Pm gene PmTm4 was flanked by Gwm611 and EST92 on chromosome arm 7BL (Hu et al. 2008b). Jimai 22 and Liangxing 99 both carry a single Pm gene on chromosome arm 2BL, which are linked to Wmc149 and flanked by BE604758 and Gwm120, respectively (Yin et al. 2009; Zhao et al. 2013). Three Pm genes PmLX66, PmW14 and PmZ155 on chromosome 5DS in Liangxing 66, Wennong 14 and Zhongmai 155, respectively, were responsible for their resistance to powdery mildew (Huang et al. 2012; Song et al. 2014; Sun et al. 2015). PmHNK on chromosome arm 3BL confers resistance to powdery mildew in Zhoumai 22 (Xu et al. 2010).
YingBo700 is a commercial winter wheat cultivar that was registered and released in Hebei Province of China in 2012 that has superior yield performance and excellent powdery mildew resistance. To identify the resistance gene(s) in YingBo 700 and use it in resistance breeding, the following researches were carried out: (1) examine the inheritance of the resistance to powdery mildew; (2) determine the chromosome location of the resistance gene(s) using molecular markers; (3) test the seedling reaction of YingBo 700 to a collection of Bgt isolates from different regions of China and compare its response pattern with the documented Pm genes near it; and (4) validate the applicability of the closely linked markers for use in marker-assisted selection (MAS) in different wheat genetic backgrounds.
Materials and methods
Plant materials
YingBo 700 is a winter wheat cultivar developed from the cross of Taigu genetically male-sterile wheat and the wheat line Ji935031 by the Hebei YingBo Seed Technology Co. Ltd., Xingtai City, Hebei Province, China (Zhang et al. 2012). Two susceptible cultivars Mingxian 169 and Shimai 15 were crossed with YingBo 700 to produce F1 hybrids, F2 populations and F2:3 lines for genetic analysis and molecular mapping of the Pm gene(s) in YingBo 700. Wheat lines or cultivars Ulka/8*Cc (with Pm2a), KM2939 (with Pm2b), Liangxing66 (with PmLX66), Tabasco (with Pm48), Wennong14 (with PmW14), X3986-2 (with PmX3986-2) and Zhongmai155 (with PmZ155) that carry documented Pm genes were used to compare their Pm genes with that of YingBo 700. Wheat cultivars Mingxian 169, Shimai 15 and Huixianhong were used as susceptible controls. A set of 62 wheat commercial cultivars and important breeding lines (Supplementary Table S1) that are widely grown or used in China were selected to survey the applicability of the closely linked markers on chromosome arm 5DS for MAS in different genetic backgrounds.
Disease assessment at the seedling stage
Forty-nine single-spore Bgt isolates with different virulence pattern were collected from different wheat production regions (Supplementary Table S2). They were used to determine the reaction patterns of YingBo 700 and wheat genotypes with documented Pm genes in this study. Bgt isolate B03, which is avirulent on YingBo 700 and virulent on Mingxian 169 and Shimai 15, was selected to inoculate YingBo 700, two susceptible parents Mingxian 169 and Shimai 15 and the derived F1 hybrids, F2 populations from YingBo 700 × Mingxian 169 and YingBo 700 × Shimai 15 and the F2:3 lines of YingBo 700 × Mingxian 169 for genetic analysis and mapping of the gene(s) conferring powdery mildew resistance in YingBo 700.
Evaluation of seedling reactions to different Bgt isolates was carried out in a humidity environment at 18/12 °C (day/night) with a photoperiod of 12–14 h of light per day (Si et al. 1992). Each isolate was placed in an independent and enclosed space to avoid cross-infection between different isolates. When the first leaf was fully unfolded, plants of the susceptible control Mingxian 169 with newly increased conidia were dusted evenly. The infection type (IT) of each plant was scored based on the 0–4 scale when the pustules were fully developed on the first leaf of susceptible controls at about 14–15 days after inoculation. Based on IT, the plants were classified as a resistant (R, ITs 0–2) or a susceptible phenotype (S, ITs 3–4) to powdery mildew (Si et al. 1992).
Marker analysis
The total genomic DNA of YingBo 700, Mingxian 169 and the F2:3 lines from the cross YingBo 700 × Mingxian 169 was extracted after evaluation of their IT scores using phenol/chloroform method (Sharp et al. 1988). Equal amounts of genomic DNA of 10 resistant (with IT of 0) and 10 susceptible (with IT of 4) F2 plants were pooled for bulked segregant analysis (BSA) and compared to the banding patterns of resistant and susceptible parents (Michelmore et al. 1991). Fifty molecular markers linked to the documented Pm genes and about 100 simple sequence repeat (SSR), expressed sequence tag (EST) and sequence-characterized amplified region (SCAR) markers that were distributed on chromosomes 4DL, 5DS and 7BS, were used to test for polymorphisms between the parents and the bulks. PCR performance and resolution and visualization of the PCR products were conducted following the methods of An et al. (2013). Markers showing polymorphisms between the bulks and the parents were tested against the F2:3 lines of YingBo 700 × Mingxian 169 to map the gene conferring powdery mildew resistance in YingBo 700.
Statistical analysis
Deviation of observed phenotypic data from theoretically expected segregation ratios was evaluated by Chi-square (χ 2) test for goodness of fit. Software MAPMAKER 3.0 was used to construct linkage map for molecular markers and the resistance gene locus. A LOD threshold value 3.0 and a maximum map distance 37.5 cM were used to declare a linkage group. (Lincoln et al. 1992). Map distance was calculated from the recombination values using the Kosambi function (Kosambi 1944).
Validation of the applicability of the closely linked markers for marker-assisted selection (MAS)
Sixty-two cultivars or important breeding lines from different wheat production regions of China were used to validate the closely linked markers in different backgrounds for MAS (Supplementary Table S1).
Results
Inheritance of powdery mildew resistance in YingBo 700
YingBo 700 produced an immune reaction with IT 0 when inoculated with Bgt isolate B03, whereas Mingxian 169 and Shimai 15 were both highly susceptible with IT 4. YingBo 700 was crossed with Mingxian 169 and Shimai 15 to produce F1 and F2 populations. The F1 generations of YingBo 700 × Mingxian 169 and YingBo 700 × Shimai 15 were both resistant to Bgt isolate B03 with IT 0, indicating the dominant nature of the powdery mildew resistance in YingBo 700. The F2 populations of YingBo 700 × Mingxian 169 and YingBo 700 × Shimai 15 segregated 112 resistant to 32 susceptible plants (\({\chi^{2}}_{3:1} = 0.45\), df = 1, P = 0.50) and 163 to 63 (\({\chi^{2}}_{3:1} = 1.00\), df = 1, P = 0.32), respectively, both fitting a single dominant gene segregation ratio. In order to confirm these results, F2 population of YingBo 700 and Mingxian 169 were transplanted to the field after evaluating the IT of each F2 plant, and 115 plants survived to produce F3 seeds. Thirty plants of each F2:3 line were tested for their powdery mildew response to the Bgt isolate B03. The F2:3 population segregated as 27 homozygous resistant, 57 segregating and 31 homozygous susceptible, fitting a 1:2:1 segregation ratio (χ 2 = 0.29, df = 2, P = 0.87). Therefore, it was concluded that powdery mildew resistance to Bgt isolate B03 in YingBo700 was controlled by a single dominant gene, which was designated PmYB.
To determine consistency of the resistance of PmYB to other Bgt isolates and the transfer to its progenies, all other ten isolates (B05, B08, B15, B16, B28, B30, B33, B39, B41 and B50) that were avirulent on YingBo 700 and virulent on Mingxian 169 were selected to inoculate five randomly selected F2:3 lines with homozygous resistant (No.21, 28, 32, 69 and 84) and five with segregating phenotype (No.4, 16, 24, 34 and 37) selected by B03, respectively. The results demonstrated that all the five F2:3 lines were homozygously resistant to all the ten Bgt isolates, and the other five F2:3 lines had a segregating phenotype to all of the ten Bgt isolates. Therefore, PmYB confers consistent resistance to different Bgt isolates, and the resistance could be transferred to its progenies.
Molecular mapping of PmYB
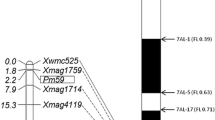
To determine the genetic location of PmYB, the F2 and F2:3 progenies from the cross YingBo 700 to the susceptible cultivar Mingxian 169 were used for BSA. Fifty molecular markers linked to the known Pm genes were first screened for the occurrence of polymorphisms between the parents as well as the resistant and susceptible DNA bulks. Pm2-linked SSR marker Cfd81 showed a polymorphism between the parents and bulks. Cfd81 was further used to genotype the F2:3 lines of YingBo 700 × Mingxian 169 and was mapped 0.9 cM away from PmYB. Since Cfd81 has been mapped to three loci on chromosome arms 4DL, 5DS and 7BS, 13, 32 and 32 SSR markers were subsequently examined for polymorphisms between parents and bulks. Only Cfd40, Cfd78 and Gwm159 which are located on chromosome arm 5DS showed polymorphisms between the parents and bulks, and all the markers on chromosome arms 4DL and 7BS showed no polymorphisms. In order to increase marker density around PmYB locus of the chromosome arm 5DS, a total of 25 EST-derived markers and two SCAR markers SCAR112 and SCAR203 that are located on chromosome arm 5DS were tested for polymorphisms. The EST marker Mag6176 linked to the documented Pm gene PmD57-5D and two SCAR markers SCAR112 and SCAR203 revealed polymorphisms between parents and bulks. Then, Cfd40, Cfd78, Gwm159, SCAR112, SCAR203 and Mag6176 were used to genotype the F2:3 lines. PmYB was linked to these markers at a genetic distance of 13.5, 10.2, 4.8, 0, 1.9 and 2.8 cM, respectively. The loci order of the linked markers was Xcfd78–Xmag6176–Xscar203–PmYB/SCAR112–Xcfd81–Xgwm159–Xcfd40 (Fig. 1). Previous studies showed that Cfd81, SCAR112, SCAR203 and Mag6176 loci on chromosome arm 5DS were all located in the deletion bin 5DS–1–0–0.63 (Ma et al. 1994; Gao et al. 2012; Huang et al. 2012). Therefore, PmYB was delineated in the same physical chromosome bin.
Genetic linkage maps of powdery mildew resistance gene PmYB using the F2:3 lines of YingBo 700 × Mingxian 169 and its locus comparison with the documented Pm genes on chromosome arm 5DS using the marker locus Xcfd81 and the linkage map of linked SSR markers by Somers et al. (2004). Black arrows point to the centromere direction. Genetic distance was shown to the left of the map in cM
Comparison of PmYB and the documented Pm genes on chromosome arm 5DS
Forty-nine Bgt isolates with different virulence were used to test the reaction patterns of YingBo 700 and documented resistant stocks with specific Pm genes on chromosome arm 5DS (Fig. 2; Supplementary Table S2). YingBo 700 showed resistance to 48 of the 49 Bgt isolates, with isolate B56 (with IT 4) being the only exception. Ulka/8*Cc (Pm2a), KM2939 (Pm2b) and Tabasco (Pm48) were all resistant to isolate B56, but susceptible to 13 other isolates (B07, B08, B10, B16, B28, B29, B30, B32, B38, B39, B40, B50 and B51), two isolates (B38 and B40) and two isolates (B29 and B38), respectively. Four other resistant stocks Liangxing 66 (with PmLX66), Wennong 14 (with PmW14), X3986-2 (with PmX3986-2) and Zhongmai 155 (with PmZ155) with tentatively designated Pm genes were also tested for their reaction patterns to these 49 Bgt isolates. They were susceptible to 11 isolates (B07, B28, B29, B30, B32, B38, B40, B50, B51, B69 and B80), three isolates (B28, B29 and B38), 17 isolates (B04, B09, B28, B29, B32, B33, B38, B40, B50, B62, B65, B69, B71, B77, B80, B82 and B83) and two isolates (B38 and B81), respectively. Therefore, YingBo 700 showed a significantly different reaction pattern from those of Ulka/8*Cc, KM2939, Tabasco, Liangxing 66, Wennong 14, X3986-2 and Zhongmai 155 in 14, 3, 3, 12, 4, 18 and 3 of 49 isolates, respectively. Therefore, PmYB is different from the documented Pm2a, Pm2b, Pm48, PmLX66, PmW14, PmX3986-2 and PmZ155 on chromosome arm 5DS.
Reactions of YingBo 700 and the documented wheat lines Ulka/8*Cc (Pm2a), KM2939 (Pm2b), Tabasco (Pm48), Liangxiang 66 (PmLX66), Wennong 14 (PmW14), X3986-2 (PmX3986-2) and Zhongmai 155 (PmZ155) to several selected Blumeria graminis f. sp. tritici (Bgt) isolates selected from 49 Bgt isolates in this study
In order to further compare PmYB-linked markers and their amplification patterns with those of documented Pm genes on chromosome arm 5DS, the closely linked markers Cfd81, SCAR203, SCAR112 and Mag6176 were used to evaluate the documented resistant stocks with Pm2a, Pm2b, Pm48, PmLX66, PmW14, PmX3986-2 and PmZ155. Primer pairs CFD81 amplified a 255 bp polymorphic band which was linked to PmYB in the multi-allelic Pm2 chromosome region in YingBo 700 and other documented resistant stocks carrying Pm2a, Pm2b, Pm48, PmLX66, PmW14, PmX3986-2 and PmZ155, while in the susceptible parent, it produced a 260 bp band (Fig. 3). The three primer pairs SCAR112, SCAR203 and MAG6176 amplified 200, 120 and 550 bp polymorphic bands, while in susceptible parents, there were no band and 202 and 600 bp bands, respectively. Therefore, the amplification patterns of the closely linked markers were all same in YingBo 700 and other documented resistant stocks with Pm2a, Pm2b, Pm48, PmLX66, PmW14, PmX3986-2 and PmZ155.
PCR amplification patterns of YingBo 700, wheat genotypes with documented Pm genes in multi-allelic Pm2 chromosome region and some wheat cultivars or important breeding line using PmYB-linked markers Cfd81. M is DNA marker pUC18 Msp I; Lanes 1–2 is YingBo 700 and Mingxian 169; Lanes 3–11 is documented resistant stocks with sequential order of UlKa/8*Cc (Pm2a), KM2939 (Pm2b), Tabasco (Pm48), Liangxiang 66 (PmLX66), Wennong 14 (PmW14), X3986-2 (PmX3986-2), Zhongmai 155 (PmZ155), Brock (MlBrock) and D57-5D (PmD57-5D); Lanes 12–20 are some wheat cultivars or important breeding line with sequential order of Shixin 828, Kenong 9204, Shimai 15, Lumai 21, Yangmai 158, Zhengmai 366, Zhou 8425B, Liangxing 99 and Jimai 22. The arrows indicate 255 and 260 bp polymorphic bands in YingBo 700 and Mingxian 169, respectively
Validating applicability of the linked markers for marker-assisted selection
From the markers comparison, it was concluded that the closely linked markers could not distinguish PmYB in the wheat phenotypes with Pm genes in the multi-allelic Pm2 chromosome region. In order to survey the applicability of the closely linked markers in other genomic backgrounds, 62 common wheat cultivars or important breeding lines with no documented Pm genes in the multi-allelic Pm2 chromosome region were analyzed using four closely linked markers namely Cfd81, SCAR112, SCAR203 and Mag6176 (Supplementary Table S1; Fig. 3 ). Primer pairs CFD81 amplified PmYB-linked polymorphic band with 255 bp in YingBo 700 and two other wheat cultivars Jimai 22 and Liangxing 99, and in other 60 cultivars it amplified the related bands ranging from 258 to 262 bp. These results implied that Cfd81 could detect PmYB in 60 of 62 cultivar or breeding line backgrounds with no documented Pm genes in the multi-allelic Pm2 chromosome region, except for Liangxing 99 and Jimai 22. The three primer pairs SCAR112, SCAR203 and MAG6176 was also detected PmYB in 60 of 62 cultivar or breeding line backgrounds except for Liangxing 99 and Jimai 22, which was the same as CFD81. These results indicated that these closely linked markers may be used in detecting PmYB in most of the genomic backgrounds with no documented Pm genes in the multi-allelic Pm2 chromosome region except for Liangxing 99 and Jimai 22 when PmYB was transferred into these cultivars through conventional hybridization.
Discussion
YingBo 700 is a commercial wheat cultivar registered and released in Hebei province, China. With its superior agronomic performance, cold hardiness and resistance to several main wheat diseases, such as wheat powdery mildew, stem rust (caused by Puccinia triticina Eriks) and stripe rust (caused by P. striiformis f. sp. tritici Eriks), YingBo 700 is preferable and has attracted the attention of farmers and breeders quickly since it released in 2012. In this study, the powdery mildew resistance in YingBo 700 was shown to be inherited as a monogenic trait. A single dominant gene with resistance to different Bgt isolates, tentatively designated PmYB, was assigned Pm2 locus on chromosome arm 5DS with linked marker loci consistent with the reported linkage map by Somers et al. (2004) (Fig. 1).
In the multi-allelic Pm2 chromosome region, several Pm genes have been reported. Pm2a was the first Pm gene reported on chromosome arm 5DS with an original designation Pm2 and a re-designation Pm2a when the new allele Pm2b in the same locus was discovered (McIntosh and Baker 1970; Ma et al. 2015). Gene Pm2a originated from the common wheat cultivar Ulka and was firstly located on chromosome 5D by monosomic analysis (McIntosh and Baker 1970). Then, Pm2a was mapped to chromosome arm 5DS and linked to the RFLP marker BCD1871 at a genetic distance 3.5 cM by Ma et al. (1994) and SSR marker Cfd81 with genetic distance of 2.0 cM by Qiu et al. (2006) in the near-isogenic line UlKa/8*Cc developed from Ulka and its recurrent parent Chanceller(Cc) (Briggle 1969) (Fig. 1). Two other Pm genes PmD57-5D and MlBrock were also mapped at the same locus as Pm2a in the two wheat lines D57 and Brock. Based on their genetic position and their reaction patterns to different Bgt isolates, PmD57-5D and MlBrock were both considered to be Pm2a (Li et al. 2009; Ma et al. 2011). Over a considerable period of time to the present, Pm2a has been widely used in resistance breeding and remains effective against most of the Bgt isolates in China (Ma et al. 2014). However, more and more Bgt isolates have defeated Pm2a in recent years due to the race-specific nature of resistance and its excessive deployment as a single resistance gene (Ma et al. 2014). In this study, Pm2a was susceptible to 13 of the 49 Bgt isolates tested. PmYB showed significantly broader resistant spectrum with only one virulent isolate (i.e., B56). Therefore, PmYB is significantly different from Pm2a and has much broader resistant spectrum than Pm2a.
Pm2b, the second allele of Pm2, was recently identified in a putatively Agropyron cristatum-derived breeding line KM2939 (Ma et al. 2015). It also showed a broad resistant spectrum with resistance to 47 of 49 Bgt isolates susceptible to B38 and B40. PmYB was resistant to Bgt isolate B38 and B40 but susceptible to B56, showing the reverse response of Pm2b to these three isolates. Also, the IT of PmYB to B28 and B30 were both 0, while Pm2b had a type 2 IT to both isolates. The third difference is the reaction patterns of the resistance phenotype. The most common resistance phenotype of Pm2b to the Bgt isolates (32 of 48) was a hypersensitive response with an IT of 0; while PmYB did not produce a hypersensitive response phenotype to the isolates in total. Therefore, PmYB is also significantly different from Pm2b.
The gene Pm48 was another Pm gene also located in the multi-allelic Pm2 chromosome region in the German cultivar Tabasco (Gao et al. 2012). It was firstly temporarily designated as Pm46 and then formally re-designated Pm48 (McIntosh et al. 2014). Allelism test between Pm48 and Pm2a using a F 2 population of 536 plants showed that Pm48 and Pm2a were closely linked to each other, but were not allelic (Gao et al. 2012). In this study, PmYB was separated by a genetic distance of 2.0 cM from Pm48 and therefore needs to be distinguished from Pm48 (Fig. 1). Disease spectrum analysis indicated that Pm48 was susceptible to B29 and B38 in the 49 Bgt isolates tested, while PmYB was resistant to B29 and B38. Meanwhile, the phenotype of Pm48 to B28 was moderate resistant with an IT of 2, while PmYB was immune to this isolate. Therefore, PmYB is also different from Pm48.
Four temporarily designated Pm genes (i.e., PmLX66, PmW14, PmZ155 and PmX3986-2) were also assigned to chromosome arm 5DS in the three Chinese wheat cultivars Liangxing 66, Wennong 14 and Zhongmai 155 and the Chinese wheat line X3986-2, respectively (Huang et al. 2012; Ma et al. 2014; Song et al. 2014; Sun et al. 2015). Of these four genes, PmLX66 and PmW14 were proven to be alleles of Pm2 (Sun et al. 2015), and PmX3986-2 and PmZ155 were located at a 1 cM from Pm2a, so their allelic relationships need to be further confirmed. Although PmLX66, PmZ155 and PmW14 were also derived from commercial cultivars, they have a narrower resistant spectrum than PmYB, with 11, 3 and 2 virulent Bgt isolates, respectively. PmX3986-2 showed even narrower resistant spectrum with resistance frequency of only 65.3 %. Therefore, PmYB is also different from PmLX66, PmW14, PmZ155 and PmX3986-2 and shows broader resistant spectrum than these four genes.
From the comparison of the resistant spectrum between PmYB and the documented Pm genes on chromosome arm 5DS, PmYB has a broader resistant spectrum, which is different from the documented genes Pm2a, Pm2b, Pm48, PmLX66, PmW14, PmZ155 and PmX3986-2. However, based on the linkage maps, PmYB shares the same linked markers and similar locus with these documented Pm genes (Fig. 1). Amplification patterns of four closely linked markers also were all the same between YingBo 700 and the documented resistant stocks, which further demonstrated that PmYB shared the same locus with the documented Pm genes (Fig. 3). Therefore, PmYB is most likely a novel allele in the multi-allelic Pm2 chromosome region. An allelism test is still required to confirm the allelic relationship between PmYB and the documented genes in the chromosome region.
In this study, PmYB was mapped to a hotspot for resistance alleles, which may be easily contributed to its use for resistance breeding. Other hotspots for resistance to powdery mildew include the Pm1a–1e locus on chromosome 7AL (Hsam et al. 1998; Singrün et al. 2003), Pm3a–3j locus on chromosome 1AS (Zeller and Hsam 1998), Pm4a–4d locus on chromosome 2AL (Schmolke et al. 2012), Pm5a–5e locus on chromosome 7BL (Hsam et al. 2001; Huang et al. 2000a, 2003) and Pm24a–24b locus on chromosome 1DS (Huang et al. 2000b; Xue et al. 2012). More importantly, PmYB originates from a common wheat cultivar associated with superior agronomic performance, which is preferable to breeders unlike genes from wild-related species or lines with poor agronomic performance. The identification of PmYB and its closely linked molecular markers will facilitate the use of PmYB in wheat improvement against powdery mildew. In fact, four closely linked markers of PmYB were validated following their use in MAS of 62 wheat cultivars or important breeding line from different regions of China. If PmYB were transferred to these cultivars through hybridization, PmYB can be detected in most genomic backgrounds tested in this study combined with the four markers, apart from the documented resistant stocks with Pm genes in the multi-allelic Pm2 chromosome region and two cultivars Liangxing 99 and Jimai 22 (Fig. 3; Supplement Table S1). Because of the similar locus, PmYB cannot be detected in these genomic backgrounds and also pyramided with these documented Pm genes. Liangxing 99 and Jimai 22 have single dominant Pm genes Pm52 and PmJM22 both on chromosome arm 2BL (Yin et al. 2009; Zhao et al. 2013). Although PmYB is located on a significantly different genetic locus from those of Pm52 and PmJM22, the amplification pattern by the four closely linked markers of PmYB were same as Liangxing 99 and Jimai 22. This may be related to mutation of this locus that results in the change of base composition and number, but not producing resistance, or other known reasons. Therefore, PmYB cannot be distinguished in Liangxing 99 and Jimai 22 genomic backgrounds by the molecular markers. Apart from closely linked markers, other means were also used to detect PmYB in Liangxing 99 and Jimai 22 backgrounds. Disease spectrum analysis showed that PmYB had a significantly different reaction pattern from those of Pm52 and PmJM22 (Supplement Table S2), which indicated that resistant spectrum may be another mean to distinguish PmYB in Liangxing 99 and Jimai 22 backgrounds.
Abbreviations
- Bgt :
-
Blumeria graminis f. sp. tritici
- BSA:
-
Bulked segregant analysis
- EST:
-
Expressed sequence tag
- IT:
-
Infection type
- MAS:
-
Marker-assisted selection
- Pm :
-
Powdery mildew resistance
- SCAR:
-
Sequence-characterized amplified region
- SSR:
-
Simple sequence repeat
References
An DG, Zheng Q, Zhou YL, Ma PT, Lv ZL, Li LH, Li B, Luo QL, Xu HX, Xu YF (2013) Molecular cytogenetic characterization of a new wheat–rye 4R chromosome translocation line resistant to powdery mildew. Chromosome Res 21:419–432
Briggle LW (1969) Near-isogenic lines of wheat with genes for resistance to Erysiphe graminis f. sp. tritici. Crop Sci 9:70–72
Costanzo A, Bàrberi P (2014) Functional agrobiodiversity and agroecosystem services in sustainable wheat production. A review. Agron Sustain Dev 34:327–348
Friebe B, Heun M, Tuleen N, Zeller FJ, Gill BS (1994) Cytogenetically monitored transfer of powdery mildew resistance from rye into wheat. Crop Sci 34:621–625
Gao HD, Zhu FF, Jiang YJ, Wu JZ, Yan W, Zhang QF, Jacobi A, Cai SB (2012) Genetic analysis and molecular mapping of a new powdery mildew resistant gene Pm46 in common wheat. Theor Appl Genet 125:967–973
Hao YF, Parks R, Cowger C, Chen ZB, Wang YY, Bland D, Murphy JP, Guedira M, Brown-Guedira G, Johnson J (2014) Molecular characterization of a new powdery mildew resistance gene Pm54 in soft red winter wheat. Theor Appl Genet. doi:10.1007/s00122-014-2445-1
Hsam SLK, Huang XQ, Ernst F, Hartl L, Zeller FJ (1998) Chromosomal location of genes for resistance to powdery mildew in common wheat (Triticum aestivum L. em. Thell.). 5. Alleles at the Pm1 locus. Theor Appl Genet 96:1129–1134
Hsam SLK, Huang XQ, Zeller FJ (2001) Chromosomal location of genes for resistance to powdery mildew in common wheat (Triticum aestivum L. em. Thell.). 6. Alleles at the Pm5 locus. Theor Appl Genet 102:127–133
Hu TZ, Li HJ, Liu ZJ, Xie CJ, Zhou YL, Duan XY, Jia X, You MS, Yang ZM, Sun QX, Liu ZY (2008a) Identification and molecular mapping of the powdery mildew resistance gene in wheat cultivar Yumai 66. Acta Agron Sin 34:545–550
Hu TZ, Li HJ, Xie CJ, You MS, Yang ZM, Sun QX, Liu ZY (2008b) Molecular mapping and chromosomal location of the powdery mildew resistance gene in wheat cultivar Tangmai 4. Acta Agron Sin 34:1193–1198
Huang XQ, Röder MS (2004) Molecular mapping of powdery mildew resistance genes in wheat: a review. Euphytica 137:203–223
Huang XQ, Hsam SLK, Zeller FJ (2000a) Chromosomal location of powdery mildew resistance genes in Chinese wheat (Triticum aestivum L. em. Thell.) landraces Xiaobaidong and Fuzhuang 30. J Genet Breed 54:311–317
Huang XQ, Hsam SLK, Zeller FJ, Wenzel G, Mohler V (2000b) Molecular mapping of the wheat powdery mildew resistance gene Pm24 and marker validation for molecular breeding. Theor Appl Genet 101:407–414
Huang XQ, Wang LX, Xu MX, Röder MS (2003) Microsatellite mapping of the powdery mildew resistance gene Pm5e in common wheat (Triticum aestivum L.). Theor Appl Genet 106:858–865
Huang J, Zhao ZH, Song FJ, Wang XM, Xu HX, An DG, Li HJ (2012) Molecular detection of a gene effective against powdery mildew in wheat cultivar Liangxing66. Mol Breed 30:1737–1745
Kosambi DD (1944) The estimation of map distance from recombination values. Ann Eugen 12:172–175
Li GQ, Fang TL, Zhu J, Gao LL, Li S, Xie CJ, Yang ZM, Sun QX, Liu ZY (2009) Molecular identification of a powdery mildew resistance gene from common wheat cultivar Brock (In Chinese). Acta Agron Sin 35:1613–1619
Li HJ, Wang XM, Song FJ, Wu CP, Wu XF, Zhang N, Zhou Y, Zhang XY (2011) Response to powdery mildew and detection of resistance genes in wheat cultivars from China. Acta Agron Sin 37:943–954
Lincoln S, Daly M, Lander E (1992) Constructing genetic maps with Mapmaker/EXP30 Whitehead Institute Technnical Report, 3rd edn. Whitehead Institute, Cambridge
Ma ZQ, Sorrells ME, Tanksley SD (1994) RFLP markers linked to powdery mildew resistance genes Pm1, Pm2, Pm3, and Pm4 in wheat. Genome 37:871–875
Ma HQ, Kong ZX, Fu BS, Li N, Zhang LX, Jia HY, Ma ZQ (2011) Identification and mapping of a new powdery mildew resistance gene on chromosome 6D of common wheat. Theor Appl Genet 123:1099–1106
Ma PT, Xu HX, Luo QL, Qie YM, Zhou YL, Xu YF, Han HM, Li LH, An DG (2014) Inheritance and genetic mapping of a gene for seedling resistance to powdery mildew in wheat line X3986-2. Euphytica 200:149–157
Ma PT, Xu HX, Xu YF, Li LH, Qie YM, Luo QL, Zhang XT, Li XQ, Zhou YL, An DG (2015) Molecular mapping of a new powdery mildew resistance gene Pm2b in Chinese breeding line KM2939. Theor Appl Genet 128:613–622
McIntosh RA, Baker EP (1970) Cytogenetic studies in wheat IV Chromosomal location and linkage studies involving the Pm2 locus for powdery mildew resistance. Euphytica 19:71–77
McIntosh RA, Zhang P, Cowger C, Parks R, Lagudah ES, Hoxha S (2011) Rye-derived powdery mildew resistance gene Pm8 in wheat is suppressed by the Pm3 locus. Theor Appl Genet 123:359–367
McIntosh RA, Dubcovsky J, Rogers WJ, Morris C, Appels R, Xia XC (2014) Catalogue of gene symbols for wheat: 2013–2014 supplement. http://www.shigen.nig.ac.jp/wheat/komugi/genes/symbolClassList.jsp
Michelmore RW, Paran I, Kesseli RV (1991) Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci USA 88:9828–9832
Petersen S, Lyerly JH, Worthington ML, Parks WR, Cowger C, Marshall DS, Brown-Guedira G, Murphy JP (2014) Mapping of powdery mildew resistance gene Pm53 introgressed from Aegilops speltoides into soft red winter wheat. Theor Appl Genet 128:303–312
Qiu YC, Sun XL, Zhou RH, Kong XY, Zhang SS, Jia JZ (2006) Identification of microsatellite markers linked to powdery mildew resistance gene Pm2 in wheat. Cereal Res Commun 34:1267–1273
Ren SX, McIntosh RA, Lu ZJ (1997) Genetic suppression of the cereal rye-derived gene Pm8 in wheat. Euphytica 93:353–360
Ren TH, Yang ZJ, Yan BJ, Zhang HQ, Fu SL, Ren ZL (2009) Development and characterization of a new 1BL.1RS translocation line with resistance to stripe rust and powdery mildew of wheat. Euphytica 169:207–213
Ren TH, Chen F, Zhang HQ, Yan BJ, Ren ZL (2011) Genetic suppression of the powdery mildew resistance gene Pm21 in common wheat. Acta Phytopathol Sin 42:57–64
Schmolke M, Mohler V, Hartl L, Zeller FJ, Hsam SLK (2012) A new powdery mildew resistance allele at the Pm4 wheat locus transferred from einkorn (Triticum monococcum). Mol Breed 29:449–456
Sharp PJ, Kreis M, Shewry PR, Gale MD (1988) Location of bamylase sequences in wheat and its relatives. Theor Appl Genet 75:286–290
Shi YQ, Wang BT, Li Q, Wu XY, Wang F, Liu H, Tian YE, Liu QR (2009) Analysis on the virulent genes of Erysiphe graminis f. sp. tritici and the resistance genes of wheat commercial cultivars in Shaanxi province. J Triticeae Crops 29:706–711
Si QM, Zhang XX, Duan XY, Sheng BQ, Zhou YL (1992) On gene analysis and classification of powdery mildew (Erysiphe graminis f. sp. tritici) resistant wheat varieties. Acta Phytopathol Sin 22:349–355
Singrün CH, Hsam SLK, Hartl L, Zeller FJ, Mohler V (2003) Powdery mildew resistance gene Pm22 in cultivar Virest is a member of the complex Pm1 locus in common wheat (Triticum aestivum L. em Thell.). Theor Appl Genet 106:1420–1424
Somers DJ, Isaac P, Edwards K (2004) A high-density wheat microsatellite consensus map for bread wheat (Triticum aestivum L.). Theor Appl Genet 109:1105–1114
Song FJ, Xiao MG, Huang J, Wang XM, Zhu ZD, Wu XF, Li HJ (2012) Inheritance of resistance to powdery mildew in 12 wheat varieties (lines). Acta Agron Sin 38:1339–1345
Song W, Sun HG, Sun YL, Zhao ZH, Wang XO, Wu XF, Li HJ (2014) Chromosomal localization of the gene for resistance to powdery mildew in the wheat cultivar Wennong14. Acta agron Sin 40:798–804
Sun HG, Song W, Sun YL, Chen XM, Liu JJ, Zou JW, Wang XM, Zhou YF, Lin XH, Li HJ (2015) Resistance to powdery mildew in the wheat cultivar Zhongmai 155: effectiveness and molecular detection of the resistance gene. Crop Sci 55:1017–1025
Wang ZZ, Li HW, Zhang DY, Guo L, Chen JJ, Chen YX, Wu QH, Xie JZ, Zhang Y, Sun QX, Dvorak J, Luo MC, Liu ZY (2015) Genetic and physical mapping of powdery mildew resistance gene MlHLT in Chinese wheat landrace Hulutou. Theor Appl Genet 128:365–373
Xu WG, Li CX, Hu L, Zhang L, Zhang JZ, Dong HB, Wang GS (2010) Molecular mapping of powdery mildew resistance gene PmHNK in winter wheat (Triticum aestivum L.) cultivar Zhoumai 22. Mol Breed 26:31–38
Xu WG, Li CX, Hu L, Wang HW, Dong HB, Zhang JZ, Zan XC (2011) Identification and molecular mapping PmHNK54: a novel powdery mildew resistance gene in common wheat. Plant Breed 130:603–607
Xue F, Wang CY, Li C, Duan XY, Zhou YL, Zhao NJ, Wang YJ, Ji WQ (2012) Molecular mapping of a powdery mildew resistance gene in common wheat landrace Baihulu and its allelism with Pm24. Theor Appl Genet 125:1425–1432
Yin GH, Li GY, He ZH, Liu JJ, Wang H, Xia XC (2009) Molecular mapping of powdery mildew resistance gene in wheat cultivar Jimai 22. Acta Agron Sin 35:1425–1431
Zeller FJ, Hsam SLK (1996) Chromosomal location of a gene suppressing powdery mildew resistance genes Pm8 and Pm17 in common wheat (Triticum aestivum L. em. Thell.). Theor Appl Genet 93:38–40
Zeller FJ, Hsam SLK (1998) Progress in breeding for resistance to powdery mildew in common wheat (Triticum aestivum L.). In: Slinkard AE (ed) Proceedings of the 9th international wheat genet symposium. University Extension Press, Saska-toon, pp 178–180
Zhang WJ, Wu JY, Wang JL, Geng ZS, Dai JJ (2012) Breeding of new wheat cultivar YingBo 700 and its high-yield culture technique. B Agr Sci Technol 10:137–139
Zhao ZH, Sun HG, Song W, Lu M, Huang J, Wu LF, Wang XM, Li HJ (2013) Genetic analysis and detection of the gene MlLX99 on chromosome 2BL conferring resistance to powdery mildew in the wheat cultivar Liangxing 99. Theor Appl Genet 126:3081–3089
Acknowledgments
This research was financially supported by the National High-Tech Research and Development Program of China No. 2011AA1001, the National Natural Science Foundation of China No. 31171550, the National Scientific and Technological Supporting Program of China No. 2013BAD01B02 and the Chinese Academy of Sciences No. XDA08030107.
Author information
Authors and Affiliations
Corresponding author
Additional information
Pengtao Ma and Hongxia Zhang contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ma, P., Zhang, H., Xu, H. et al. The gene PmYB confers broad-spectrum powdery mildew resistance in the multi-allelic Pm2 chromosome region of the Chinese wheat cultivar YingBo 700. Mol Breeding 35, 124 (2015). https://doi.org/10.1007/s11032-015-0320-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-015-0320-7