Abstract
Stormwater infiltration systems (SIS) have been developed to limit surface runoff and flooding in urban areas. The impacts of such practices on the ecological and biological quality of groundwater ecosystems remain poorly studied due to the lack of efficient methodologies to assess microbiological quality of aquifers. In the present study, a monitoring method based on the incubation of artificial matrices (clay beads) is presented to evaluate microbial biomass, microbial activities, and bacterial community structure. Four microbial variables (biomass, dehydrogenase and hydrolytic activities, bacterial community structures) were measured on clay beads incubated in three urban water types (stormwater surface runoffs, SIS-impacted and non-impacted groundwaters) for six SIS. Analyses based on next-generation sequencing (NGS) of partial rrs (16S rRNA) PCR products (V5-V6) were used to compare bacterial community structures of biofilms on clay beads after 10 days of incubation with those of waters collected from the same sampling points at three occasions. Biofilm biomass and activities on clay beads were indicative of nutrient transfers from surface to SIS-impacted groundwaters. Biofilms allowed impacts of SIS on groundwater bacterial community structures to be determined. Although bacterial communities on clay beads did not perfectly match those of waters, clay beads captured the most abundant bacterial taxa. They also captured bacterial taxa that were not detected in waters collected at three occasions during the incubation, demonstrating the integrative character of this approach. Monitoring biofilms on clay beads also allowed the tracking of bacterial genera containing species representing health concerns.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Groundwater constitutes a crucial resource of drinking water for world populations. For example, about 75% of the inhabitants of the EU depend upon groundwater resources for their water supply (Danielopol et al. 2004). Therefore, groundwater ecosystems are more and more considered for their ecological services, and valued for their capacity to provide goods (e.g., safe drinking water supply) and services (e.g., purification of water through microbial and physicochemical processes) (European Groundwater Directive, EU-GWD 2006). At the same time, urbanization threatens these services and alters groundwater resources through chemical pollution, and by reducing aquifer recharge. Indeed, one of the major landscape modifications associated with urbanization is an increase in the impervious surface coverage (Shuster et al. 2005). The sealing of urban surfaces alters the natural water cycle by increasing stormwater runoff during rainfall events (Niemczynowicz 1999). In this context, stormwater management practices have been developed for 30 years to limit surface runoff and their side effects such as urban flooding, surface water pollution (Marsalek and Chocat 2002; Fletcher et al. 2015), and reduced groundwater recharge (e.g., more than 10% of groundwater volume in the East aquifer of Lyon originates from infiltration practices using stormwater infiltration basins, Foulquier 2009). However, aquifer recharge with runoff stormwater in urban area may lead to groundwater contamination with hydrophilic compounds and microbial agents found in stormwaters (Sébastian et al. 2014; Mermillod-Blondin et al. 2015; Marti et al. 2017).
In front of these threats, indicators must be developed to evaluate the responses of groundwater ecosystems to stormwater infiltration (SI) practices. Indeed, efficient indicators of ecosystem health and functioning are rare for groundwater ecosystems despite their crucial importance for human populations (e.g. Danielopol et al. 2004; Steube et al. 2009; Griebler and Avramov 2015). Historically, the assessment of groundwater status was based exclusively on chemical parameters, hydrogeological data, or basic microbiological tests (e.g., Escherichia coli total counts). Monitoring the quality of groundwater could be considered a relatively simple task, but the wide variety of contaminants (chemicals and their by-products, allochtonous microbes, and predators such amoebae) and high number of point (ditches, infiltration basins, rain gardens) and diffuse (connected gardens and parks) sources make it a difficult, time consuming, and expensive task. Therefore, few authors (Hahn 2006; Steube et al. 2009; Stein et al. 2010; Korbel and Hose 2011, 2017) proposed multimetric approaches to determine the ecological status and the health of groundwater ecosystems using abiotic and biotic (microbial activity and diversity, abundance and richness of invertebrates) variables. Then, biological indicators which could sense or build genetic imprints representative of changing groundwater status over time appear as promising tools for ecological monitoring (e.g., Griebler and Avramov 2015) and for the evaluation of health hazards.
Over the last decades, descriptors of general microbial features such as overall biomass, respiratory activities, and genetic diversity fingerprints (e.g., Goldscheider et al. 2006; Griebler et al. 2010) have been proposed to assess groundwater ecosystem health, but they were applied on water samples that did not consider microorganisms living attached to sediments (through biofilm development). Incorporating groundwater biofilms in ecosystem health assessment is greatly needed because biofilms are recognized as the main reactors of biogeochemical and water purification processes occurring in groundwater (e.g., Alfreider et al. 1997; Goldscheider et al. 2006; Griebler and Avramov 2015). Nevertheless, sampling aquifer sediments and attached biofilms after pumping or coring are technically difficult, expensive, and challenging in deep alluvial aquifers (i.e., when the groundwater table is located at 10 m below the soil surface, Chapelle 2001). To circumvent this problem, we aimed at developing an alternative monitoring approach based on the incubation of artificial solid matrices to assess the growth and diversity of micro-organisms among biofilms developing in groundwater impacted or not by stormwater infiltration (SI) practices. This strategy has been rarely used in underground environments, but previous experiments performed in the hyporheic zone of the Rhône River (Claret 1998), in an alluvial gravel aquifer system in New Zealand (Williamson et al. 2012), and in urban groundwaters (Mermillod-Blondin et al. 2013) gave promising results. For instance, Mermillod-Blondin et al. (2013) showed that a monitoring of bacterial growth on glass beads incubated in piezometers could be used to infer the trophic status of groundwater. To extend the uses of such a trapping system, improvements had to be performed by comparing different types and sizes of solid surfaces (Voisin et al. 2015, 2016). Clay beads with a diameter of 8 mm were found more efficient than glass beads of the same diameter for biofilm development and for capturing the bacterial diversity of three water types (wastewater, stormwater runoff water, and groundwater) (Voisin et al. 2016). Nevertheless, these experiments had been done under laboratory conditions without considering the hydrodynamic conditions encountered in the field.
The present study aimed at testing the efficacy of clay beads at capturing representative bacterial communities developing in the field. Clay beads were incubated during 10 days in three “water types” (stormwater runoff entering infiltration basins, and SIS-impacted and not impacted groundwaters) at six sites. Bacterial community structures were obtained from incubated clay beads by metabarcoding using next-generation sequencing datasets built from rrs gene (16S rRNA) PCR products. In parallel, waters were collected three times from the same sampling points to have an assessment of their bacterial community structures over the incubation period. Comparisons of bacterial communities between those of waters and incubated clay beads were done to evaluate the trapping efficacy of clay beads. Differences between the genetic structures of bacterial communities from the runoff waters, and of the underground waters impacted or not by SIS, and their matching clay beads biofilms profiles, were investigated. Furthermore, the relations between general indicators of biofilm biomass and activities, and the concentrations of nutrients, such as dissolved organic carbon (a key variable according to Mermillod-Blondin et al. (2013)) were evaluated. Biofilm and biomass activities were expected to be positively correlated with the availability of dissolved organic matter in waters (Mermillod-Blondin et al. 2013). According to Voisin et al. (2018), bacterial community structures in waters and attached on clay beads were expected to be distinct among the three water types.
Materials and methods
Experimental sites
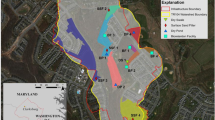
Stormwater infiltration systems (SIS) were located on the eastern main aquifer (catchment area 314 km2) of the Lyon metropolitan area, France (Fig. 1). They have been built in three aquifer corridors of highly permeable glaciofluvial sediments (hydraulic conductivity 10−3–10−2 m/s) (Foulquier et al. 2009; Voisin et al. 2018). These SIS are detention and infiltration basins collecting stormwater runoff from residential, commercial, or industrial areas (Voisin et al. 2018). Foulquier (2009) estimated that the recharge of the aquifer by SIS can represent 10% of the annual flux of groundwater.
The stormwater infiltration systems (SIS) in the Lyon conurbation investigated in this work. (a) Geographical localization of the 6 SIS. (b) Position of sampling points in each SIS. DJR Django-Reinhardt, IUT Campus of the University Lyon 1, RAQ Raquin, MIN Minerve, GB Grange Blanche, FEY Feyzin. Figure modified from Foulquier (2009) and Voisin et al. (2018)
Experimental design
Six SIS (DJR = Django-Reinhardt, IUT = Campus of the University Lyon 1, RAQ = Raquin, MIN = Minerve, GB = Grange Blanche, and FEY = Feyzin) were selected to evaluate the efficacy of clay beads (8 mm in diameter) for monitoring changes in microbial biomass, activity, and genetic diversity that could be observed among the runoff waters reaching the infiltration basins and the connected groundwaters (Fig. 1). Descriptions of these six sites are given in Voisin (2017). Each SIS was equipped with a well located in the aquifer zone impacted by stormwater runoffs (impacted well) and one reference well located in a zone non-impacted by the stormwater runoff plume (non-impacted well). As sampling in wells was previously found to give, in some cases, a biased view of the chemistry and biological life in an aquifer (Claassen 1982; Griebler et al. 2002; Korbel et al. 2017), only sites where water chemistry in the wells (specific conductance, temperature, and dissolved oxygen (DO) concentration) did not differ from that of groundwaters in the aquifer (Foulquier et al. 2010; Voisin 2017) were selected. For each SIS, clay beads were incubated 10 days with stormwater runoffs entering the basin, and in the SIS-impacted and non-impacted wells. The incubation period covered six successive rainfall events (Fig. 2). This kind of rainy period shows clear impacts of infiltrated runoff water on the aquifer. Indeed, a marked decrease in electrical conductivity was monitored in impacted wells due to the transfer of poorly mineralized stormwater from the basin to the aquifer (Fig. 2). In parallel, non-impacted wells were not influenced by stormwater infiltration during the same incubation period. For each SIS, three nylon bags (4 cm × 10 cm) filled with 30 clay beads were incubated in each water type (stormwater runoff, SIS-impacted and non-impacted groundwaters). In parallel with clay bead incubation, water samples were collected three times over the 10-days incubation period (day 0, day 5, and day 10). Grab samples of stormwater runoff were performed at the inlet of the infiltration basins. Groundwaters were sampled using a submerged pump (PP36 inox, SDEC, Reignac-sur-Indre, France) previously cleaned with 70% ethanol. The first 50 pumped liters were used to rinse the sampling equipment and discarded. The following 6 L were used for chemical and microbiological analyses. All samples were collected and stored in pre-combusted glass bottles and brought back to the laboratory in an isotherm box at 4 °C.
Continuous recordings of electrical conductivity in non-impacted (in green) and in SIS-impacted (in red) groundwaters of the DJR site during the incubation period of clay beads. This period was characterized by six rainfall events (in blue). Decrease in electrical conductivity following each rainfall event highlights the presence of low-mineralized stormwater in SIS-impacted groundwater
Water physical and chemical measurements
Electrical conductivity (EC), pH, temperature, and dissolved oxygen (DO) were measured instantly from the stormwater runoff grab samples and groundwater samples collected on day 0, day 5, and day 10 with portable EC (electric conductivity) and DO meter (dissolved oxygen, HQ20, HACH, Dusseldorf, Germany) probes. Inorganic nutrients (NH4+, NO3−, and PO43−) were measured on filtered samples (0.7 µm, Whatman GF/F Glass microfiber filters) by standard colorimetric methods (Grasshoff et al. 1999) using an automatic analyzer (Smartchem200, AMS, Frepillon, France). Dissolved organic carbon (DOC) concentrations were measured on filtered samples with a multi N/C ® 3100 device (Analytik Jena, Jena, Germany) following Foulquier et al. (2010). Biodegradable DOC (BDOC) was determined by the method of Servais et al. (1989) following Mermillod-Blondin et al. (2015). Briefly, DOC concentrations were measured before and after an incubation of the samples with their native bacterial content over 30 days in the dark and at 20 °C. For each water sample, BDOC was then calculated as the difference in DOC concentrations before (initial concentrations of DOC) and after (fraction of refractory DOC, RDOC) the 30 days incubation period.
Microbial analysis
Microbial biomass
Microbial biomass on clay beads was assessed from Lowry’s total protein assays. Total proteins were measured using the Sigma Protein Assay Kit P-5656 (Sigma Diagnostics, St Louis, MO, USA) according to Peterson (1977). From each nylon bag, five clay beads were treated with 4 mL of a 50% solution of Lowry’s reagent for 20 min. Then, 0.5 mL of Folin reagent was added to obtain a color reaction lasting 30 min. Afterward, the absorbance of the solution was measured at 750 nm using an Aquamate spectrophotometer (ThermoSpectronic, Cambridge, UK). Obtained values were compared with a calibration curve performed with bovine serum albumin (quality BioReagent, Sigma-Aldrich, Saint-Quentin-Fallavier, France) to determine protein content. Total proteins were then expressed in micrograms of proteins per square centimeter of clay bead, after subtracting values measured from sterile clay beads.
Microbial activity
Hydrolytic activity of biofilm was estimated using the fluorescein diacetate (FDA) hydrolysis method (Fontvieille et al. 1992). From each nylon bag, five clay beads were incubated with 3 ml of phosphate buffer solution (pH = 7.6) and 0.15 ml of 4.8 mM FDA solution. The incubation was performed at 15 °C for 3 h until a green fluorescein coloration appeared. The reaction was stopped by addition of 3 ml of acetone (Battin 1997). Fluorescein concentration was estimated from the absorbance of the filtered supernatant (0.45 μm, HAWP, Millipore, Billerica, MA, USA) measured at 490 nm, and hydrolytic activity was expressed in micromoles of FDA hydrolyzed per hour and per square centimeter of clay bead, after subtracting values measured from sterile clay beads.
The 2-(p-iodophenyl)-3-(p-nitrophenyl)-5-phenyl tetrazolium chloride (INT) was used to measure dehydrogenase activity (respiratory activity) as modified from Houri-Davignon et al. (1989). From each nylon bag, five clay beads were incubated in 4 mL of a 0.02% INT solution for 2 h at 15 °C. Then, the supernatant was filtered on a nylon membrane (0.22 μm, Millipore, Billerica, MA, USA) to retain free micro-organisms. Extraction of INT-formazan from clay beads and nylon membrane was made in vials containing 5 ml of methanol. The INT-formazan was measured by colorimetry with a spectrophotometer adjusted at 490 nm. The quantity of INT-formazan was expressed in micromoles of INT-formazan produced per hour and per square centimeter of clay bead, after subtracting values measured from sterile clay beads.
16S rRNA gene bacterial community meta-barcoding
For water samples, 5 L of each water sample was filtered with 0.22 μm polycarbonate filters. From each nylon bag , microbial cells were detached from 15 beads by shaking at 2500 rpm for 2 min in 10 mL of 0.8% NaCl. A 0.22-μm filtration was performed to retrieve cells and aggregates. DNA was extracted from filters using the FastDNA spin kit for soil (MP Biomedicals France). For clay beads, the amount of extracted DNA was quantified by spectrophotometry using a ND1000 Nanodrop (Thermo Fisher Scientific, USA).
The sequencing and bioinformatics analyses were performed following the procedure described in Voisin et al. (2018). Sequencing of the V5-V6 16S rRNA gene (rrs) PCR products was performed by MrDNA Company (Shallowater, TX, USA) with Illumina MiSeq technology. Primers 799F (5′-ACCMGGATTAGATACCCKG-3′) and 1193R (CRTCCMCACCTTCCTC) were used, and 20,000 reads (i.e., 2 × 300-bp paired-end reads) were obtained per sample. Data processing was realized with the MOTHUR software version 1.39.5 (Schloss et al. 2009) following the pipeline described in Voisin et al. (2018). Reads were discarded if they were (1) shorter than 200 bp and longer than 400 pb, (2) containing homopolymers longer than 8 bp, (3) having more than one sequence mismatch to barcode DNA tags, (4) having more than two mismatches with the sequencing primer, (5) containing ambiguous bases, and (6) having average sequencing quality score lower than 35 (Schloss et al. 2011). CHIMERA UCHIME (Edgar et al. 2011) was used to detect and remove chimeric sequences. DNA sequences were clustered into operational taxonomic units (OTUs) using a threshold of 97% identity (Schloss and Westcott 2011). Singletons were removed from the dataset. A random resampling was used to make equal the number of sequences per sample. The resulting OTU contingency table reporting OTU abundances (= number of sequences) for each sample was then used to infer bacterial community structures. OTU taxonomic allocations were performed by comparisons with the SILVA v128 database (Quast et al. 2013) and a 80% minimum bootstrap cutoff (Wang 2007). Sequences classified as Archaea, Eukaryota, chloroplasts, mitochondria, or not related to the V5-V6 rrs region were deleted from the dataset.
Statistical analyses
For each SIS, measurements of electrical conductivity, temperature, pH, dissolved oxygen, NH4+, NO3−, PO43−, DOC, and BDOC concentrations performed at the three sampling dates were averaged to obtain one value per water type (stormwater runoff, SIS-impacted, and non-impacted groundwaters). The three replicated analyses of microbial biomass and activities measured on clay beads (corresponding to the three nylon bags incubated per water type in each SIS) were also averaged in each SIS to obtain one value per water type. Physical and chemical variables measured in water were compared among the three water types using a mixed model with SIS as random factor and water types as fixed factor to take into account the statistical dependence of waters collected on the same SIS. Biomass and activities of biofilms developed on clay beads were also compared among water types by the same mixed model. To evaluate the relationship between chemical variables and biofilms, Pearson’s correlations were computed to determine the relation between DOC and BDOC and clay bead biofilm variables (i.e., biomass and activities). When mixed models gave significant results for water type differences, post hoc Tukey’s HSD tests were performed to determine whether significant pairwise differences occurred among water types. For all variables, the normality and the homoscedasticity of the residues were tested using the Shapiro-Wilk’s test and the Bartlett’s test, respectively. When these assumptions were not met, data were Log-transformed before statistical analyses.
The sequence abundances of bacterial OTUs (defined at > 97% identities) collected at the three dates in waters were added for each sampling point to obtain a whole bacterial community per water type and SIS. Similarly, the sequence abundances of bacterial OTUs obtained from the three replicated batches of biofilms developing on clay beads (at day 10 from the three incubated nylon bags) were added for each sampling point to obtain a whole bacterial biofilm community per “water type” and SIS. A non-metric multidimensional scaling (nMDS) analysis was performed based on Bray-Curtis dissimilarity distances. Differences in bacterial community structures obtained from waters and clay beads were compared for the three water types (stormwater runoff, SIS-impacted and non-impacted groundwaters) using permutational multivariate analysis of variance (PERMANOVA; Anderson 2001). Statistical tests were based on 999 permutations of the Bray-Curtis matrix, and significance was accepted at p value < 0.05.
For each sampling point, the percentages of OTUs detected on clay beads from the whole number of OTUs detected (including water and clay beads) and from the number of OTUs detected in waters were computed. The relative proportion in the number of sequences per OTU between water and clay bead biofilms was also computed. The percentage of reads per OTU reported from total reads obtained in waters that were detected or not on clay beads was computed to estimate the relationship between the abundance of an OTU in water and an effective trapping by the clay beads. All these variables used to assess the efficacy of clay beads to capture OTUs being in waters were compared among water types using one-way ANOVAs after data transformation (Logit) to satisfy the normality criterion. Then, the normality and the homoscedasticity of the residues were verified using the Shapiro-Wilk’s test and the Bartlett’s test, respectively.
Statistical tests were performed using the R software (R Development Core Team 2008). The ade4 and Vegan packages were used for nMDS and PERMANOVA analyses (Oksanen et al. 2007).
Data deposition
Sequence data were deposited at the European Nucleotide Archive (https://www.ebi.ac.uk/ena) under the project accession #PRJEB29925. Sample accession #ERS2912776 (SAMEA5128391) = run1. Sample accession #ERS2912777 (SAMEA5128392) = run2. See supplementary material Table S1 for barcode information.
Results
Water chemistry at the sampling sites
Chemical datasets from the six sites were grouped into categories defined according to the sampling point: surface runoff, and SIS-impacted or non-impacted groundwaters. Water types per category showed distinct chemistries (Fig. 3). There was a decreasing gradient in electrical conductivity and NO3− concentrations from the non-impacted groundwater to the low-mineralized stormwater runoff (Fig. 3a, b, mixed models, F2,10 > 20, p < 0.001). Intermediate electrical conductivity and NO3− concentrations were observed for the SIS-impacted groundwaters. The opposite gradient was observed for NH4+, DOC, and BDOC (Fig. 3c–e, mixed models, F2,10 = 8.6 and p < 0.01 for NH4+ concentrations, and F2,10 > 30 and p < 0.0001 for DOC and BDOC concentrations) with highest concentrations measured in stormwater runoff. For example, mean concentrations of DOC varied from 0.9 mg/L in non-impacted groundwater and 1.5 mg/L in SIS-impacted groundwater to 5.1 mg/L in stormwater runoff. The same trend was also detected for PO43− concentrations but was not supported by significant statistical tests (Fig. 3f, mixed model, F2,10 = 3.8, p > 0.05). pH values ranged between 7.1 and 8.2 and were significantly higher in stormwater runoff than in groundwater (Fig. 3g, mixed model, F2,10 = 5.2, p < 0.05). In contrast, temperature and dissolved oxygen concentrations were not significantly different among the three water types (Fig. 3h, i, mixed models, F2,10 < 2.0, p > 0.1). Nevertheless, dissolved oxygen concentrations tended to be lower and were more variable in SIS-impacted groundwater than in other water types (Fig. 3i).
Boxplots illustrating the chemical datasets from the six SIS according to the sampling points: stormwater runoff, SIS-impacted (I) or non-impacted (NI) groundwaters (GW). (a) electrical conductivity. (b) NO3-. (c) NH4+. (d) PO43-. (e) dissolved organic carbon (DOC). (f) biodegradable dissolved organic carbon (BDOC). (g) pH. (h) water temperature. (i) dissolved oxygen. Each boxplot represents the distribution of mean values (calculated from the three sampling dates per SIS) for the six SIS. Upper case letters displayed on graphs indicate significant differences between water types determine by HSD Tukey’s tests (a same letter indicates no significant difference)
Microbial biomass and activities on clay beads
The biomass (protein content, Fig. 4a) and activities (hydrolytic and dehydrogenase activities, Fig. 3b, c) of 10-day-old biofilms developing on clay beads were more than threefold higher after incubation in runoff waters than after incubation in groundwater (mixed models, F2,10 > 25, p < 0.001 for the three microbial variables). Biofilm biomass and activities followed the same trend than the concentrations of dissolved organic matter (DOC and BDOC). Consequently, the most significant and positive correlations between biofilm global characters and chemical variables were measured with the concentration of BDOC (Pearson’s correlations, R2 > 0.6, p < 0.0001 for all correlation tests between BDOC and biofilm variables).
Boxplots illustrating the (a) biomass (total protein content), (b) hydrolytic and (c) dehydrogenase activities of the biofilms recovered on clay beads at the six SIS and exposed to stormwater runoff, and to SIS-impacted (I) or non-impacted (NI) groundwaters (GW) for a 10-day period. Each boxplot represents the distribution of mean values (calculated from the three replicated analyses performed per SIS) for the six SIS. Upper case letters displayed on graphs indicate significant differences between water types determine by HSD Tukey’s tests (a same letter indicates no significant difference)
Bacterial community structures of waters and biofilms on clay beads
The meta-barcoding approach using V5-V6 rrs PCR product sequences (a total of 2,713,392 sequences were generated and analyzed) was affiliated to 29,991 OTUs. The NMDS ordination of OTU numbers showed significant differences in bacterial structures between water type categories (stormwater runoff, SI-impacted (I) or non-impacted (NI) groundwaters (GW)) for the V5-V6 rrs fingerprints of both water and clay bead samples (Fig. 5, PERMANOVA, effect of water type categories, p < 0.001).
NMDS ordination of V5-V6 rrs OTU profiles from waters of the three categories (stormwater runoff—SW (black centroid), SIS-impacted (gray centroid) and non-impacted (white centroid) groundwaters—GW-) investigated in this study over six SIS, and from biofilms developing on clay beads after exposure to these three water types (stormwater runoff (black centroid), SIS-impacted (gray centroid) and non-impacted (white centroid)). Stress test = 0.085
Bacterial V5-V6 rrs OTU patterns from water samples and biofilms were found different. Groundwater OTU profiles were clearly apart from those of stormwater runoff samples (Fig. 5). Mean numbers of OTUs detected in non-impacted (3963 ± 2021, mean ± standard deviation, n = 6) and SIS-impacted (4888 ± 1097) groundwaters were higher than OTU numbers obtained from biofilms developing on clay beads in the same waters (1729 ± 429 OTUs in non-impacted and 2484 ± 876 OTUs in SIS-impacted groundwaters). For stormwater runoff, the mean number of OTUs recovered from clay bead biofilms (3525 ± 367) was comparable to the mean number detected in waters (3277 ± 411). In link with these results, around 25% of OTUs present in groundwaters were detected on the corresponding clay beads, whereas this percentage was between 40 and 50% for stormwater runoff and their matching clay beads (Fig. 6a, one-way ANOVA, F2,15 = 46, p < 0.001). Interestingly, several OTUs recovered on clay beads were not detected in the corresponding water sample: around 20% for groundwater (impacted or not by stormwater infiltration) and around 45% for stormwater runoff (Fig. 6b, one-way ANOVA, F2,15 = 16, p < 0.001). Then, considering all OTUs (except singletons) obtained at each point (addition of OTUs from water and clay bead biofilms), around 35%, 40%, and more than 70%, were detected on clay beads incubated in non-impacted groundwater, SIS-impacted groundwater, and stormwater runoff, respectively (Fig. 6c, one-way ANOVA, F2,15 = 32, p < 0.001).
Boxplots evaluating the efficacy of clay beads at capturing bacteria belonging to a particular OTU. First line of boxplots estimated the ability of clay beads at trapping an OTU without considering their relative number of reads per sample. (a) proportion of OTUs detected in waters that were recovered on clay beads. (b) proportion of OTUs only recovered on clay beads from the total number of OTUs detected (on clay beads and in waters). (c) proportion of OTUs recovered on clay beads from the total number of OTUs detected (on clay beads and in waters). Second line of boxplots estimated the efficacy of clay beads at trapping an OTU according to its relative occurrence (in proportion/percentage of reads) in a water sample. (d) proportion of reads obtained in waters corresponding to OTUs detected on clay beads. (e) mean percentage of reads in waters per OTUs for OTUs detected on clay beads. (f) mean percentage of reads in waters per OTUs for OTUs not detected on clay beads. Boxplots were computed using datasets from the six SIS and were differentiated per water type category (stormwater runoff, SIS-impacted (I) or non-impacted (NI) groundwaters (GW)). Upper case letters displayed on graphs indicate significant differences between water types determine by HSD Tukey’s tests (a same letter indicates no significant difference). Analyses were performed from 25,124 reads per sample
OTUs recovered from clay beads represented a moderate percentage of OTUs found in waters (from 25 to 50%), but these analyses did not consider their representation in number of reads. Considering this point, OTUs that were trapped by clay beads were found to be the most abundant ones per water type. These trapped OTUs represented between 60 and 70% of the rrs reads recovered from groundwaters and around 90% of the reads recovered from stormwater runoff (Fig. 6d, one-way ANOVA, F2,15 = 8.1, p < 0.001). Indeed, OTUs recovered from clay beads corresponded to OTUs having sequence abundances representing more than 0.15% of total sequences obtained from the three water types (Fig. 6e, one-way ANOVA, F2,15 = 3.3, p > 0.05), whereas OTUs not recovered from clay beads had mean percentage of reads lower than 0.04% of sequences in water samples (Fig. 6f, one-way ANOVA, F2,15 = 2.1, p > 0.1).
The rrs OTUs recovered in this investigation were allocated to taxonomic groups by comparison with the SILVA database. A total of 1319 genera (including 210 OTUs not assigned to genera of the SILVA v128 database) could be identified in the dataset, and some of these genera could harbor potential pathogenic lineages such as Bordetella and Legionella. As observed for OTU patterns, distribution analysis of these genera showed that the ones with the highest numbers of rrs reads in the water samples were recovered from clay beads (Fig. 7). Only two genera (Vibrio and Campylobacter) with significant numbers of rrs reads in some water samples were not recovered from clay beads. Interestingly, the clay beads appeared to favor biofilm development of bacteria with variable morphologies going from rods (e.g., Escherichia, Pseudomonas), cocci (e.g., Staphylococcus) to filamentous ones (e.g., Streptomyces, Nocardia). Read numbers allocated to the group of Gram-positive rods like Bacillus and Clostridium were in low number in the water dataset explaining their rare recovery from the biofilm datasets. Some genera (Cupriavidus, Nocardia, and Ralstonia) showed higher relative numbers of rrs reads in biofilms than waters.
Heatmap representing the relative number of V5-V6 rrs sequences (in %) allocated to 26 bacterial genera recovered from the water and clay beads biofilms samples. These genera were selected because they can include pathogenic species. A Log10 scale was used to differentiate the variations in the dataset per compartment (runoffs—SW-, SIS-impacted or non-impacted groundwaters—GW-). A darker color corresponds to a higher number of rrs reads, and thus a higher occurrence of the corresponding genera in the dataset. Analyses were performed from 25,124 reads per sample
Discussion
This study was performed with the aim of testing the ability of clay beads at trapping microorganisms for monitoring the ecological status and microbiological safety of a water system. Clay beads were found, in laboratory conditions, to allow an efficient trapping of bacteria and favor biofilm development after 10 days of incubation (Voisin et al. 2016). Here, these clay beads monitoring systems were tested under field conditions. Their biofilm contents after exposure to different water types (i.e., surface runoffs and SIS-impacted or non-impacted groundwaters) were monitored after 10 days of incubation. It appeared that an incubation of 10 days was long enough to obtain a biofilm development useful for assessing groundwater trophic conditions. Indeed, the total-protein contents and enzymatic activities of biofilms were found correlated with the concentrations of dissolved organic matter (DOC and BDOC) of waters. Thus, these general biofilm descriptors obtained using clay beads could be used to infer transfers of organic matter from the surface to groundwater. The coupled reductions of organic matter concentration and biofilm parameters between the infiltration basin and the SIS-impacted aquifer confirmed previous works highlighting the efficacy of the soil and vadose zone to reduce the amount of organic matter reaching the aquifer (Pabich et al. 2001; Mermillod-Blondin et al. 2015; Voisin et al. 2018).
Biofilms developing on clay beads could also be used to investigate the content of bacterial communities. Although these biofilms were not strictly representative of the bacterial communities present in the tested water grab samples (collected at days 0, 5, and 10), they captured the most abundant taxa (defined as the number of reads per V5-V6 rrs OTU sharing at least 97% identity) recovered from the water samples (each OTU representing around 0.15% of reads obtained from waters). Interestingly, more than 20% of OTUs recovered from clay beads were not detected among the water grab samples. Clay beads thus appeared to have efficiently trapped dominant bacteria over the incubation period (10 days), whereas water grab samples gave a snapshot of the diversity observed which was limited to the sampling time. Artificial matrices such as clay beads are thus likely to be more efficient than water grab samples to monitor microbial communities over infiltration events that can last several days. Indeed, rainfall events produce plumes of stormwater in groundwater ecosystems located below SIS (Fig. 2; Datry et al. 2004; Foulquier et al. 2009) which cannot be easily sampled by short-term approaches. Therefore, the recovery on clay beads of some OTUs that were not detected in the water samples was probably due to the integrative property of the clay bead method. Nevertheless, it could also be due to the selection of bacterial taxa having a greater ability at colonizing solid surfaces under field conditions because of more significant adhesion properties (e.g., Costerton et al. 1995; Crump et al. 1999; Branda et al. 2005; Rösel and Grossart 2012). Bacterial taxa having efficient adhesion properties could be undetectable in water samples but become detectable after colonization and growth on clay beads. The opposite might also have occurred for bacteria having poor adhesion properties. Knowing that the majority (> 90%) of bacteria living in porous aquifers are attached to particles (Alfreider et al. 1997; Griebler et al. 2002; Iribar et al. 2008), assessment of bacterial taxa growing on artificial matrices might be more representative of aquifer bacteria than the assessment of free-living bacteria in groundwater samples.
NMDS analysis of the rrs OTU patterns recovered from the germcatchers and waters was performed. According to our expectations and the results of Voisin et al. (2018) on bacteria from waters, bacterial community structures recovered from water samples and biofilms attached on clay beads were significantly different between water types. The use of clay beads as artificial matrices for trapping bacteria was found an efficient strategy to detect the impact of SIS on groundwater bacterial communities. Voisin et al. (2018) reported on water grab samples that differences in bacterial community structures between non-impacted and SIS-impacted groundwaters were most likely due to (1) modifications of environmental conditions in the aquifer associated with SI practices (e.g., DOC and BDOC concentrations) and/or (2) an increased transport of specific bacteria present in the soil and the vadose zone to the aquifer due to SI practices. By using artificial clay bead matrices for microbial colonization and growth, the present study underlines the fact that differences in bacterial community structures due to SIS practices were not due to free-living bacteria that could be only transitory (i.e., acting as a traveling seed bank; Griebler et al. 2014) in the system (as suggested in Voisin et al. 2018) but were associated with bacterial taxa that can get attached to particles and play key ecosystem functions in porous aquifers (Griebler and Avramov 2015). It is worth noting that the similarity between bacterial community structures of waters and biofilms was higher for stormwater runoffs than for groundwater compartments. These differences could be explained by the higher concentrations of suspended matters (SM) observed in the runoff waters compared with groundwater. These SM have been previously shown to be chemically reactive and to have a tendency at forming aggregates involving micro-organisms and other colloids (Jeng et al. 2005). SM could thus interact more efficiently with the surface of clay beads and increase the trapping efficacy of these matrices. This phenomenon could explain why the highest efficacy of clay beads to capture OTUs was measured in stormwater runoff. In contrast, groundwaters collected in impacted and non-impacted piezometers have been physically and chemically filtered through water transport in the vadose and the saturated zone of the aquifer (Winiarski et al. 2006). Consequently, negligible SM-like elements occurred in groundwater, limiting the number of particles that can favor aggregation and adherence of bacteria on clay beads. It is also possible that bacterial communities from stormwater runoff had many taxa that could grow as biofilm, while a fewer proportion of taxa from groundwater communities was able to do so. Interestingly, the NMDS analysis grouped the rrs OTU patterns for the biofilms coming from groundwaters, supporting the idea of strong and common selective pressures structuring their content.
Although microbes freely suspended in groundwater and those attached to sediments share a core community (e.g., Zhou et al. 2012; Flynn et al. 2013), significant differences between free-living bacteria (collected in waters) and bacteria attached to clay beads were observed. It is important to mention that the majority of microbes living in groundwater ecosystems are associated with particles and form biofilms which play key roles in the aquifer biogeochemical processes (Alfreider et al. 1997; Chapelle 2001; Iribar et al. 2015; Humphreys 2009; Battin et al. 2016). Free-living micro-organisms in groundwaters are considered to be less indicative of their ecological quality (Griebler et al. 2014). Thus, clay beads biofilm monitoring represents a major step forward in the understanding of groundwater microbiological states. For example, quantifying the abundance and expression of genes involved in biogeochemical processes (such as nirK and nirS involved in denitrification, Nogales et al. 2002) on clay bead biofilms would be an interesting perspective to evaluate whether SI practices impact ecosystem services (such as nitrate reduction, Korbel and Hose 2017) played by microbial community in aquifers. Furthermore, these clay beads systems were found to trap genera which can contain species of health concerns, suggesting a potential use in microbiological hazards assessment frames. Nevertheless, the 16S rRNA gene metabarcoding approach developed in the present study was not sensitive enough to allocate reads at the species level. Other approaches will be required to assess the presence of bacterial pathogens in underground waters. This perspective is essential for groundwater quality assessment because previous investigations have shown the presence of significant populations of Aeromonas caviae, Pseudomonas aeruginosa, and of fecal indicator bacteria such as E. coli and intestinal enterococci in a detention basin located upstream of a SIS (Bernardin-Souibgui et al. 2018; Sébastian et al. 2014). In conclusion, our field monitoring approach based on clay beads was found efficient and promising to investigate the incidence of SIS management practices on connected aquifers.
References
Alfreider, A., Krössbacher, M., & Psenner, R. (1997). Groundwater samples do not reflect bacterial densities and activity in subsurface systems. Water Research, 31, 832–840.
Anderson, M. J. (2001). A new method for non-parametric multivariate analysis of variance. Austral Ecology, 26, 32–46.
Battin, T. J. (1997). Assessment of fluorescein diacetate hydrolysis as a measure of total esterase activity in natural stream sediment biofilms. Science of the Total Environment, 198, 51–60.
Battin, T. J., Besemer, K., Bengtsson, M. M., Romani, A. M., & Packmann, A. I. (2016). The ecology and biogeochemistry of stream biofilms. Nature Reviews Microbiology, 14, 251–263.
Bernardin-Souibgui, C., Barraud, S., Bourgeois, E., Aubin, J. B., Bécouze-Lareure, C., Wiest, L., Marjolet, L., Colinon, C., Lipeme-Kouyi, G., Cournoyer, B., & Blaha, D. (2018). Incidence of hydrological, chemical, and physical constraints on bacterial pathogens, Nocardia cells, and fecal indicator bacteria trapped in an urban stormwater detention basin in Chassieu, France. Environmental Science and Pollution Research, 25, 24860–24881.
Branda, S. S., Vik, Å., Friedman, L., & Kolter, R. (2005). Biofilms: The matrix revisited. Trends in Microbiology, 13, 20–26.
Chapelle, F. H. (2001). Ground-water microbiology and geochemistry (2nd ed.). New York: Wiley.
Claassen, H. C. (1982). Guidelines and techniques for obtaining water samples that accurately represent the water chemistry of an aquifer (no. 82–102). US Geological: Survey.
Claret, C. (1998). Hyporheic biofilm development on artificial substrata, as a tool for assessing trophic status of aquatic systems: First results. Annales de Limnologie – International Journal of Limnology, 34, 119–128.
Costerton, J. W., Lewandowski, Z., Caldwell, D. E., Korber, D. R., & Lappin-Scott, H. M. (1995). Microbial biofilms. Annual Reviews in Microbiology, 49, 711–745.
Crump, B. C., Armbrust, E. V., & Baross, J. A. (1999). Phylogenetic analysis of particle-attached and free-living bacterial communities in the Columbia River, its estuary, and the adjacent coastal ocean. Applied and Environmental Microbiology, 65, 3192–3204.
Danielopol, D. L., Gibert, J., Griebler, C., Gunatilaka, A., Hahn, H. J., Messana, G., Notenboom, J., & Sket, B. (2004). Incorporating ecological perspectives in European groundwater management policy. Environmental Conservation, 31, 185–189.
Datry, T., Malard, F., & Gibert, J. (2004). Dynamics of solutes and dissolved oxygen in shallow urban groundwater below a stormwater infiltration basin. Science of the Total Environment, 329, 215–229.
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., & Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics, 27, 2194–2200.
European Groundwater Directive, EU-GWD. (2006). Directive 2006/118 of the European parliament and the council of the 23 October 2006. Official Journal of the European Communities, L372, 371–392.
Fletcher, T. D., Shuster, W., Hunt, W. F., Ashley, R., Butler, D., Arthur, S., Trowsdale, S., Barraud, S., Semadeni-Davies, A., Bertrand-Krajewski, J.-L., Mikkelsen, P. S., Rivard, G., Uhl, M., Dagenais, D., & Viklander, M. (2015). SUDS, LID, BMPs, WSUD and more—The evolution and application of terminology surrounding urban drainage. Urban Water Journal, 12, 525–542.
Flynn, T. M., Sanford, R. A., Ryu, H., Bethke, C. M., Levine, A. D., Ashbolt, N. J., & Santo Domingo, J. W. (2013). Functional microbial diversity explains groundwater chemistry in a pristine aquifer. BMC Microbiology, 13, 146. https://doi.org/10.1186/1471-2180-13-146.
Fontvieille, D. A., Outaguerouine, A., & Thevenot, D. R. (1992). Fluorescein diacetate hydrolysis as a measure of microbial activity in aquatic systems: Application to activated sludges. Environmental Technology, 13, 531–540.
Foulquier, A. (2009). Ecologie fonctionnelle dans les nappes phréatiques: liens entre flux de matière organique, activité et diversité biologiques. Doctoral thesis, University Lyon 1, 259 pp.
Foulquier, A., Malard, F., Barraud, S., & Gibert, J. (2009). Thermal influence of urban groundwater recharge from stormwater infiltration basins. Hydrological Processes, 23, 1701–1713.
Foulquier, A., Malard, F., Mermillod-Blondin, F., Datry, T., Simon, L., Montuelle, B., & Gibert, J. (2010). Change in dissolved organic carbon and oxygen at the water table region of an aquifer recharged with stormwater: Biological uptake or mixing? Biogeochemistry, 99, 31–47.
Goldscheider, N., Hunkeler, D., & Rossi, P. (2006). Review: Microbial biocenoses in pristine aquifers and an assessment of investigative methods. Hydrogeology Journal, 14, 926–941.
Grasshoff, K., Ehrhardt, M., & Kremling, K. (1999). Methods of seawater analysis (3rd ed.). Weinheim; New York; Chiester; Brisbane; Singapore; Toronto: Wiley-VCH.
Griebler, C., & Avramov, M. (2015). Groundwater ecosystem services: A review. Freshwater Science, 34, 355–367.
Griebler, C., Mindl, B., Slezak, D., & Geiger-Kaiser, M. (2002). Distribution patterns of attached and suspended bacteria in pristine and contaminated shallow aquifers studied with an in situ sediment exposure microcosm. Aquatic Microbial Ecology, 28, 117–129.
Griebler, C., Stein, H., Kellermann, C., Berkhoff, S., Brielmann, H., Schmidt, S., Selesi, D., Steube, C., Fuchs, A., & Hahn, H. J. (2010). Ecological assessment of groundwater ecosystems—Vision or illusion? Ecological Engineering, 36, 1174–1190.
Griebler, C., Malard, F., & Lefébure, T. (2014). Current developments in groundwater ecology—From biodiversity to ecosystem function and services. Current Opinion in Biotechnology, 27, 159–167.
Hahn, H. J. (2006). The GW-Fauna-index: A first approach to a quantitative ecological assessment of groundwater habitats. Limnologica-Ecology and Management of Inland Waters, 36, 119–137.
Houri-Davignon, C., Relexans, J.-C., & Etcheber, H. (1989). Measurement of actual electron transport system (ETS) activity in marine sediments by incubation with INT. Environmental Technology Letters, 10, 91–100.
Humphreys, W. F. (2009). Hydrogeology and groundwater ecology: Does each inform the other? Hydrogeology Journal, 17, 5–21.
Iribar, A., Sanchez-Perez, J. M., Lyautey, E., & Garabétian, F. (2008). Differentiated free-living and sediment-attached bacterial community structure inside and outside denitrification hotspots in the river-groundwater interface. Hydrobiologia, 598, 109–121.
Iribar, A., Hallin, S., Sanchez-Pérez, J. M., Enwall, K., Poulet, N., & Garabétian, F. (2015). Potential denitrification rates are spatially linked to colonization patterns of nosZ genotypes in an alluvial wetland. Ecological Engineering, 80, 191–197.
Jeng, H. C., England, A. J., & Bradford, H. B. (2005). Indicator organisms associated with stormwater suspended particles and estuarine sediment. Journal of Environmental Science and Health, 40, 779–791.
Korbel, K. L., & Hose, G. C. (2011). A tiered framework for assessing groundwater ecosystem health. Hydrobiologia, 661, 329–349.
Korbel, K. L., & Hose, G. C. (2017). The weighted groundwater health index: Improving the monitoring and management of groundwater resources. Ecological Indicators, 75, 164–181.
Korbel, K., Chariton, A., Stephenson, S., Greenfield, P., & Hose, G. C. (2017). Wells provide a distorted view of life in the aquifer: Implications for sampling, monitoring and assessment of groundwater ecosystems. Scientific Reports, 7, 40702.
Marsalek, J., & Chocat, B. (2002). International report: Stormwater management. Water Science and Technology, 46, 1–17.
Marti, R., Bécouze-Lareure, C., Ribun, S., Marjolet, L., Souibgui, C. B., Aubin, J. B., Lipeme Kouyi, G., Wiest, L., Blaha, D., & Cournoyer, B. (2017). Bacteriome genetic structures of urban deposits are indicative of their origin and impacted by chemical pollutants. Scientific Reports, 7, 13219.
Mermillod-Blondin, F., Foulquier, A., Maazouzi, C., Navel, S., Negrutiu, Y., Vienney, A., Simon, L., & Marmonier, P. (2013). Ecological assessment of groundwater trophic status by using artificial substrates to monitor biofilm growth and activity. Ecological Indicators, 25, 230–238.
Mermillod-Blondin, F., Simon, L., Maazouzi, C., Foulquier, A., Delolme, C., & Marmonier, P. (2015). Dynamics of dissolved organic carbon (DOC) through stormwater basins designed for groundwater recharge in urban area: Assessment of retention efficiency. Water Research, 81, 27–37.
Niemczynowicz, J. (1999). Urban hydrology and water management—present and future challenges. Urban Water, 1, 1–14.
Nogales, B., Timmis, K. N., Nedwell, D. B., & Osborn, A. M. (2002). Detection and diversity of expressed denitrification genes in estuarine sediments after reverse transcription-PCR amplification from mRNA. Applied and Environmental Microbiology, 68, 5017–5025.
Oksanen, J., Kindt, R., Legendre, P., & O’Hara, B. V. (2007). The vegan package. Community ecology package, R package version 1. pp. 8–5.
Pabich, W. J., Valiela, I., & Hemond, H. F. (2001). Relationship between DOC concentration and vadose zone thickness and depth below the water table in groundwater of Cape Cod, U.S.A. Biogeochemistry, 553, 247–268.
Peterson, G. L. (1977). A simplification of the protein assay method of Lowry et al. which is more generally applicable. Analytical Biochemistry, 83, 346–356.
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., Peplies, J., & Glockner, F. O. (2013). The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Research, 41, D590–D596.
Rösel, S., & Grossart, H.-P. (2012). Contrasting dynamics in activity and community composition of free-living and particle-associated bacteria in spring. Aquatic Microbial Ecology, 66, 169–181.
Schloss, P. D., & Westcott, S. L. (2011). Assessing and improving methods used in operational taxonomic unit-based approaches for 16S rRNA gene sequence analysis. Applied and Environmental Microbiology, 77, 3219–3226.
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., Lesniewski, R. A., Oakley, B. B., Parks, D. H., Robinson, C. J., Sahl, J. W., Stres, B., Thallinger, G. G., Van Horn, D. J., & Weber, C. F. (2009). Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Applied and Environmental Microbiology, 75, 7537–7541.
Schloss, P. D., Gevers, D., & Westcott, S. L. (2011). Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS One, 6, e27310.
Sébastian, C., Barraud, S., Ribun, S., Zoropogui, A., Blaha, D., Becouze-Lareure, C., Lipeme Kouyi, G., & Cournoyer, B. (2014). Accumulated sediments in a detention basin: Chemical and microbial hazard assessment linked to hydrological processes. Environmental Science and Pollution Research, 21, 5367–5378.
Servais, P., Anzil, A., & Ventresque, C. (1989). Simple method for determination of biodegradable dissolved organic carbon in water. Applied and Environmental Microbiology, 55, 2732–2734.
Shuster, W. D., Bonta, J., Thurston, H., Warnemuende, E., & Smith, D. R. (2005). Impacts of impervious surface on watershed hydrology: A review. Urban Water Journal, 2, 263–275.
Stein, H., Kellermann, C., Schmidt, S. I., Brielmann, H., Steube, C., Berkhoff, S. E., Fuchs, A., Hahn, H. J., Thulin, B., & Griebler, C. (2010). The potential use of fauna and bacteria as ecological indicators for the assessment of groundwater quality. Journal of Environmental Monitoring, 12, 242–254.
Steube, C., Richter, S., & Griebler, C. (2009). First attempts towards an integrative concept for the ecological assessment of groundwater ecosystems. Hydrogeology Journal, 17, 23–35.
Voisin, J. 2017. Influence des pratiques de recharge des aquifères par des eaux pluviales sur les communautés microbiennes des nappes phréatiques. Doctoral thesis, University Lyon 1, 196 pp.
Voisin, J., Cournoyer, B., & Mermillod-Blondin, F. (2015). Utilisation de billes de verre comme substrats artificiels pour la caractérisation des communautés microbiennes dans les nappes phréatiques : mise au point méthodologique. La Houille Blanche, 4, 52–57.
Voisin, J., Cournoyer, B., & Mermillod-Blondin, F. (2016). Assessment of artificial substrates for evaluating groundwater microbial quality. Ecological Indicators, 71, 577–586.
Voisin, J., Cournoyer, B., Vienney, A., & Mermillod-Blondin, F. (2018). Aquifer recharge with stormwater runoff in urban areas: Influence of vadose zone thickness on nutrient and bacterial transfers from the surface of infiltration basins to groundwater. Science of the Total Environment, 637-638, 1496–1507.
Wang, Q. (2007). Naïve bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Applied and Environmental Microbiology, 73, 5261–5267.
Williamson, W. M., Close, M. E., Leonard, M. M., Webber, J. B., & Lin, S. (2012). Groundwater biofilm dynamics grown in situ along a nutrient gradient. Groundwater, 50, 690–703.
Winiarski, T., Bedell, J. P., Delolme, C., & Perrodin, Y. (2006). The impact of stormwater on a soil profile in an infiltration basin. Hydrogeology Journal, 14, 1244–1251.
Zhou, Y., Kellermann, C., & Griebler, C. (2012). Spatio-temporal patterns of microbial communities in a hydrologically dynamic pristine aquifer. FEMS Microbiology Ecology, 81, 230–242.
Acknowledgments
This work was supported by l’Agence Nationale de la Recherche [ANR-16-CE32-0006 FROG], Lyon Metropole within the framework of the experimental observatory for urban hydrology (OTHU, http://www.graie.org/othu/), and the French national research program for environmental and occupational health of Anses under the terms of project “Iouqmer” EST 2016/1/120. We thank Félix Vallier, Antonin Vienney, and Laurent Simon for support and advices during field and laboratory work.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary Table 1
(XLSX 8775 kb)
Rights and permissions
About this article
Cite this article
Mermillod-Blondin, F., Voisin, J., Marjolet, L. et al. Clay beads as artificial trapping matrices for monitoring bacterial distribution among urban stormwater infiltration systems and their connected aquifers. Environ Monit Assess 191, 58 (2019). https://doi.org/10.1007/s10661-019-7190-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10661-019-7190-0