Abstract
This study assessed the functional significance of attached and free-living bacterial communities involved in the process of denitrification in a shallow aquifer of a riparian zone (Garonne River, SW France). Denitrification enzyme activity (DEA), bacterial density (BD) and bacterial community composition (BCC) were measured in two aquifer compartments: the groundwater and the sandy fraction of the sediment deposit. Samples were collected in wells located inside (IHD) and outside (OHD) identified hotspots of denitrification. Despite high BD values (up to 1.14 × 1012 cells m−3), DEA was not detected in the water compartment (< 0.32 mg N–N2O m−3 d−1). The sandy fraction showed detectable DEA (up to 1,389 mg N–N2O m−3 d−1) and, consistent with BD pattern, higher DEA values were measured in IHD zones than in OHD zones. The BCC assessed by 16S rDNA polymerase chain reaction-denaturing gradient gel electrophoresis (PCR-DGGE) partly supported this result: attached and free-living communities were significantly different (< 30% similarity) but patterns of BCC did not cluster according to IHD and OHD zones. Targeting the denitrifying communities by means of a culture enrichment step prior to 16S rDNA PCR-DGGE showed that the free-living and sediment attached communities differed. Most sequences obtained from DGGE profiles of denitrifying communities were affiliated to Proteobacteria and showed low genetic distance with taxa that have already been detected in aquifers (e.g., Azoarcus sp., Acidovorax sp. and Pseudomonas spp.). This study confirms that in the aquifer the sediment-attached fraction exhibits different functions (DEA) from free-living communities and suggests that this functional difference is related to the communities’ structure.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Nitrogen cycling has received considerable attention in riparian zones (Burt et al., 1999; Hill et al., 2000; Clément et al., 2002; Duff et al., 2007). The river–groundwater interface has been shown to contribute to nitrogen retention and/or transformation in the land-river continuum (Sabater et al., 2003). This interface participates in the purification of water thanks to its ability to eliminate nitrate during their infiltration through the vegetation-soil system to the groundwater, and through diffusion from the groundwater to the river (Pinay et al., 1998; Takatert et al., 1999; Maître et al., 2002). Several studies have focused on the processes influencing nutrient retention or elimination from the alluvial groundwater in riparian systems (Peterjohn & Correll, 1984; Pinay et al., 1998; Hill et al., 2000; Sánchez-Pérez & Tremolieres, 2003) and denitrification appears to be the most important process for nitrate elimination (Haycock & Burt, 1993; McMahon & Böhlke, 1996; Hinkle et al., 2001; Brettar et al., 2002) ranging from 1 to 40 g N m−2 yr−1 (Brüsch & Nilsson, 1993; Van Cleemput et al., 2007) unless other microbial processes such as dissimilatory nitrate reduction to ammonium (DNRA) or anaerobic ammonium oxidation (Anammox) are taken into account (Burgin & Hamilton, 2007).
The process of bacterial denitrification is nearly exclusively a facultative trait in bacteria. By means of denitrification, nitrate (NO3 −) and nitrite (NO2 −), which are highly oxidized forms of nitrogen, are reduced into gaseous forms such as nitric oxide (NO) and nitrous oxide (N2O), which may themselves be further reduced into dinitrogen (N2). This process generally requires an anoxic environment and a source of both organic matter and nitrate (Mosier et al., 2002). In previous work in the Garonne River floodplain, Sánchez-Pérez et al. (2003b) demonstrated that the organic matter fuelling denitrification is provided by the river. Spatial variations in organic carbon and nitrate contents in groundwater at the studied site are correlated with exchanges between the groundwater and the river. These results suggest that nitrate derived from alluvial groundwater influenced by agricultural practices is denitrified by bacteria in the presence of organic carbon derived from river water.
In aquifers, the major proportion of the bacterial biomass in groundwater is attached to solid material and only a small fraction is free-living (Harvey et al., 1984; Bekins et al., 1999; Lehman et al., 2001). As assessed by fingerprinting techniques, the bacterial community composition (BCC) differs between sediment-attached and free-living communities (Besemer et al., 2005) consistent with differences in extracellular enzyme activities (Lehman & O’Connell, 2002). However, no study has clarified whether differences between free living and sediment attached communities applied to denitrification and denitrifying communities.
Our objective was to study the denitrification capability of sediment-attached and free-living bacteria in alluvial groundwater (Garonne River, France). Our hypothesis was that denitrification would differ between sediment and groundwater habitat, and would be explained by differences in the BCC of sediment-attached and free-living assemblages. Five wells were selected, two inside identified denitrification hotspots (IHD) and three outside hotspots (OHD), and in each well, denitrification enzyme activity (DEA), total bacterial density (BD) and BCC of both the total and cultivable denitrifying fraction of sediment attached and free-living assemblages were assessed with the aim to explore their relationships. By combining structural and functional descriptors of contrasted bacterial communities (free-living vs. sediment attached), exhibiting contrasted denitrification activity (IHD vs. OHD), this experiment was designed to approach the question of the link between biodiversity and functioning for the denitrifying assemblages in the river–groundwater interface.
Materials and methods
Study site
The study site is a riparian zone located within a meander of the Garonne River (43°53′ N, 1°43′ E), 50 km northern Toulouse city (France) (Fig. 1). The Garonne River is a 8th order river, 625 km long, with an annual mean discharge of 600 m3 s−1. In the study site, annual mean discharge is 200 m3 s−1; low-water period discharge is 50 m3 s−1, flood discharge up to 4,000 m3 s−1 and bank-full discharge 600 m3 s−1. The average water depths are 1 m during low flow, 3.5 m for the bank-full discharge and up to 8 m during floods. Throughout the studied zone, which represents about 0.49 km2, the groundwater flow is governed by the slope of the valley (1:1,000), the water table levels in floodplain terraces and the water level of the Garonne River. The groundwater flows from south-east to north-west, with an average gradient of 1.5 10−3 m m−1. The water table is at 2–6 m depth in low-water periods, but can rise rapidly to the soil surface during floods. In the floodplain, between 4 and 7 m thick layer of Quaternary sand and gravel overlie impermeable molasses deposits. The riparian zone is covered by a stand of Populus alba and riparian vegetation (Salix alba, Fraxinus excelsior). Beyond the riparian zone lies agricultural land cropped with corn, sunflower and wheat. Nitrogen leaching of the fertilised soil leads to concentrations exceeding 15 mg N–NO3 − l−1 in the groundwater. The floodplain is equipped with wells, constructed of 6.3 cm diameter polyvinyl chloride pipes, which intersect the alluvium down to the molasse (Pinay et al., 1998; Lambs, 2000; Sánchez-Pérez et al., 2003a, b; Baker & Vervier, 2004; Fig. 1).
Sampling and environmental variables
Five wells were selected (Fig. 1): well p17 is drained by freshly infiltrated river waters, wells p9, p10, and p11 are drained by a variable mix of river waters and groundwaters and well p26 is considered to be representative of the groundwaters of the unconfined adjacent aquifer (Weng et al., 2003). The water mixing in this system follows a two end-member mixing model (Lambs, 2000; Sánchez-Pérez & et al., 2003b; Baker & Vervier, 2004). Using this model together with in situ denitrification measurements, previous studies demonstrated that wells p9 and p17 are located inside an hotspot of denitrification (IHD)—60% of nitrate depletion and in situ denitrification rates ranging from 0.1 to 2.7 g N–N2O m3 d−1—while the wells p10, p11 and p26 are located outside hotspot of denitrification (OHD)—no nitrate depletion and in situ denitrification rates ranging from 0.05 to 1.2 g N–N2O m3 d−1 (Sánchez Pérez et al. 2003a, b). In the present study, the assignment of each well to the IHD or OHD group was confirmed by calculating nitrate variation using the two end-member mixing model and by measuring DEA on the sampling dates (21 and 22 October 2003).
Mixing of surface and subsurface water masses was estimated for each well by two-end-member mixing model (EMMA) using chloride (Cl-) concentrations observed in river water and the agricultural well (p26). EMMA solves the equations: [Cl- well] = [Cl- SW]f SW + [Cl- GW]f GW; 1 = f SW + f GW where f SW is the surface water fraction and f GW is the groundwater fraction. This model assumes that there is no precipitation input and that Cl- is chemically and biologically conservative. Measured river chloride concentrations (Cl- SW) were 10.5 and 63.7 mg l −1 in agricultural well (Cl- GW). Predicted N–NO3 − concentrations were calculated based on a similar EMMA, substituting observed surface water and agricultural well N–NO3 − concentrations (1.1 and 9.5 mg l−1 respectively) for Cl-. This allowed the estimation of N–NO3 − depletion (negative values) via uptake or denitrification or input (positive values) compared to dilution by low N–NO3 − content surface water.
Water physical and chemical conditions were characterized every 3 months from 2000 to 2004 at 16 sampling dates representing diversified hydrological conditions (min = 50 m3 s−1 max = 764 m3 s−1 mean = 227 m3 s−1). Groundwater and sediment samples for BD, BCC and DEA analysis were recovered on 21 and 22 October 2003, between 3.5 and 5.0 m depth, during a river low-water period (100 m3 s−1). This hydrological condition occurred with a frequency of 40% in the last 35 years (Direction Régionale de l’Environnement www.hydro.eaufrance.fr/accueil.html) and of 45% among the sampled dates from 2000 to 2004. Water samples were collected from each well by pumping with an electric pump (8 l min−1), while sandy sediment (<2 mm) samples were recovered by pumping with a motor pump designed for sediment (30–120 l min−1). Sediment layer composition is sandy down to 3 m depth (Ruffinoni 1994).
Dissolved oxygen was measured using a WTW Oxical-SL Cell Ox 325, pH and temperature using a WTW 197i and electrical conductivity using a WTW Tetacon 325. Redox potential was measured using platinum electrodes against Ag/AgCl reference electrodes (Schott Gerate PT737 A). Measures were corrected adding 217 mV for the field (for an average temperature of 10°C) as indicated by the manufacturer (Mettler-Toledo GmbH, Steinbach, Germany).
Water and sediment analysis
Water samples for determination of dissolved inorganic nitrogen (nitrate, nitrite, and ammonium) and chloride concentrations were filtered on the field using 0.45 μm-pore-size cellulose acetate membranes. Dissolved inorganic nitrogen was measured in a segmented flow analyzer (ALPKEM model Flow solution IV) and chloride by ionic chromatography (DIONEX model DX500) according to standardized methods for water analysis (APHA, 1992). For dissolved organic carbon (DOC) analysis, water was filtered in the field using pre-combusted (450°C for 4 h) GF/F filters, and analyzed using a platinum catalyzer at 650°C (Shimadzu, Model TOC 5000).
Sediment samples (20 g) recovered the 21 and 22 October 2003 were successively exposed to three treatments with 50% H2O2 and one with 110% H2O2 to remove organic matter (Jackson, 1985). Flocculants were eliminated by adding 40 ml of HCl at 10% and dispersed by adding 40 ml of a 0.55% aqueous solution of sodium hexametaphosphate. Samples were stirred magnetically for 6 h and granulation was determined using a Coulter TM LS230 laser granulometer equipped with a variable-speed fluid module, which measures the size of waterborne particles by diffraction at the laser light at 750 nm wavelength into 126 photodiode detectors. The instrument measures particles in the size range 0.375–2,000 μm.
Organic matter was determined as loss on ignition by combustion of dried samples (pre-dried at 105°C to constant weight) at 450°C for 8 h. The loss on ignition organic matter values were then converted to organic carbon using a conversion factor of 0.5 as proposed by Brinson et al. (1984).
Denitrification enzyme activity
DEA in water and sediment collected on the 21 and 22 October 2003 was measured by the acetylene block technique (Yoshinari & Knowles, 1976). Triplicate of groundwater (400 ml) and/or sediment (mix of 300 g sediment plus 100 ml groundwater) samples were incubated in 500 ml glass bottles adding Na-acetate (50 mg C l−1) and NaNO3 (100 mg N l−1). Oxygen in the liquid and gaseous phase was removed by sparging with helium (Helium U 99.99% purity, containing 20 ppmv of N2, Air Liquide, France). Acetone-free acetylene was added (AAS27 quality, Air Liquide, France) at 10% v/v. Incubations were performed in the dark at 14°C for 20 h. The gaseous phase was sampled (3 ml) at initial and final incubation for N2O analysis using a GIRDEL gas chromatograph equipped with an electron capture detector (63Ni), Porapak Q columns (2 m long packed columns) using argon-methane (90–10%, Air Liquide, France) as carrier gas.
Bacterial density and community composition analyses
For total bacteria counts, 10 ml water and 1.5 g sediment subsamples (wet weight) from larger collected samples were used. An ultrasonic cleaner (ELMA model Transonic T460-35 kHz, Germany) was used to recover bacteria from the sediment without lysing them. Cell staining using DAPI (4′, 6-diamino-2-phenylindole-dihydrochloride) was performed as described previously (Garabétian et al., 1999). Counting was made with a Leitz Dialux 22 microscope (×1250 magnification) fitted for epifluorescence: HBO 100 W mercury light source, with an excitation filter for 270–380 nm and barrier filter of 410–580 nm.
Comparable units were obtained for BD and DEA by transforming cell g−1 sediment and cell ml−1 water into cell m−3 using porosity (12%) and bulk sediment density (1.4 g cm−3).
For total BCC analysis, DNA was extracted from a filter (3 l groundwater; 0.22 μm polycarbonate membranes, Millipore, Ireland) and/or 10 g sediment samples. DNA extraction was performed using the phenol–chloroform method described previously (Mouné et al. 2003). Amounts of extracted DNA were quantified by fluorimetry (Fluroskan Ascent II; Labsystems, Helsinki, Finland) using SYBR Green (Sigma). Extracted DNA was amplified by PCR using the universal Bacteria primers 341F-GC and 907R (Muyzer et al., 1997) and 20 ng (water and sediment) or 50 ng (enrichment samples from water and sediment) of template DNA using a protocol described in Lyautey et al. (2005). Denaturing gradient gel electrophoresis (DGGE) was carried out using D-Code Universal Mutation Detection System (BioRad). About 500 ng (natural samples from water and sediment) and 700 ng (enrichment samples from water and sediment) of amplified DNA were set down in an acrilamide gel containing a 35–70% urea and formamide gradient (100% corresponding to 7 M of urea and 40% of deionized formamide). The electrophoresis was run for 18 h at 100 V at 60°C. The gel was stained for 30 min with SYBR Green (Sigma) and gel image was captured using a CCD camera and Biocapt software (Vilber Lourmat) and analyzed using Bio-1D++ software (Vilber Lourmat).
For approaching the BCC of the denitrifying bacteria, an enrichment culture step was used to target this group by increasing their relative abundance in the groundwater and sediment samples using a cascade of dilution and culture (Soriano & Walker, 1968). As proposed by Tiedje (1994), a 40 ml enrichment solution (0.22 μm filtered well water containing 50 mg C l−1 Na-acetate, and 100 mg N l−1 NaNO3) was inoculated with bacteria from a 20 ml portion of the supernatant of the DEA assay extended for another 7 days. The culture was deoxygenated by sparging with helium and lasted 7 days at 14°C in the dark. This last step was repeated once. Then 20 ml of culture were harvested by centrifugation (14,000×g, 20 min, 4°C). The BCC of these enrichment cultures representing the cultivable fraction of the denitrifying bacteria from the groundwater or sediment samples was then assessed by DGGE as described above, except for DNA extraction, which was performed using UltraClean Soil DNA kit (Bio101, Vista CA, USA) as described by manufacturer. Predominant DGGE bands were sequenced as described elsewhere (Lyautey et al., 2005). Sequence analysis and phylogenetic tree was constructed with ARB software (www.arb-home.de) (Ludwig et al., 2004). A total of 14 partial 16S rRNA gene sequences were deposited in the GenBank sequence database under accession numbers AY816197 to AY816210.
Data analysis
DGGE profiles were compared using presence and absence of different bands using Jaccard coefficients. A matrix from the Jaccard similarity index J = [c/(a+b-c)] was calculated, where a is the number of bands in sample A, b in sample B and C the number of bands shared by sample A and B, and dendrogram was constructed using UPGMA (Unweighted Pair Group Method with Average means).
DEA and bacterial richness were compared by Mann-Whitney tests using the SPSS software 11.0, statistical significance level at P < 0.05. Statistical tests based on cluster were realized using Monte-Carlo test (Kropf et al., 2004), statistical significance level at P < 0.001.
Results
Environmental conditions
Physical and chemical characteristics of groundwater for the wells selected in the present study are summarized in Table 1. The different wells from IHD and OHD zones showed no marked differences considering physical and chemical characteristics except the p17 which presented low dissolved oxygen (1.2 mg l−1) and nitrate (0.9 mg l−1) concentrations. Lower conductivity and nitrate concentration and higher dissolved oxygen and DOC concentrations were recorded in river compared to groundwater. Sediment collected in these wells had a sandy texture (98–99% sand and 1–2% clay) and exhibited comparable organic carbon content between 1.5 and 2 g C kg−1 of sediment.
In the present experiment, we ensured that hotspots of denitrification activity were sampled by calculating the percentage of the nitrate variation rates based on water mixing on the 21 and 22 October 2003 (Table 2). The results confirmed that in two wells (p9 and p17) out of five, marked nitrate depletion indicate intense denitrification activity.
Bacterial density and denitrification enzyme activity
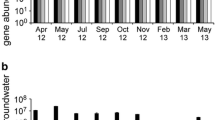
Bacterial densities were greater in the sediment compartment than in groundwater in three cases out of five (Fig. 2). Furthermore the DEA responses of the free-living and attached communities were markedly different between the two compartments (groundwater and sediment; Fig. 3). No DEA was detected (detection threshold = 0.32 mg N–N2O m−3 d−1) in the groundwater, while DEA ranged from 2 to 1,389 mg N–N2O m−3 d−1 in the sediment (Fig. 3). Note that during incubation for DEA measurements, total bacterial numbers (= sample volume or mass × BD), ranged from 0.4 × 109 to 3.7 × 109 cells and were similar for groundwater and sediment assays. The DEA values recorded were significantly higher in IHD wells (p9 and p17) than in OHD wells (p10, p11 and p26) (Mann-Whitney, P < 0.05) (Fig. 3).
Free-living and attached bacterial community composition
The BCC of the total communities of the two compartments were compared based on DGGE profiles. DGGE profiles (BCC) were significantly differentiated between sediment-attached and free-living communities (Monte-Carlo, P < 0.001) (Fig. 4a). Regarding the IHD/OHD groups of samples, no clear clustering pattern was observed for either free-living or sediment attached samples sharing less than 30% of bands.
DGGE fingerprints of 16S rRNA in (a) natural bacterial communities and (b) cultivable denitrifying bacterial communities selected by enrichment culture from the alluvial aquifer. On the left gel pictures and on the right UPGMA dendrograms constructed from presence-absence matrices from groundwater (GW) and sediment-attached (SED) bacteria of the five wells (p9, p10, p11, p17, and p26) and of the river (Gar)
As assessed by culture enrichment coupled to 16S rRNA genes DGGE, the BCC of the cultivable denitrifying fraction of the communities was found to be significantly different (Monte-Carlo, P < 0.001) between free-living and attached communities (Fig. 4b). Some bands of cultivable denitrifiers were shared by sediment and groundwater communities (similarities 19%). Within the sediment cluster, average similarities were lower than 35% and did not reveal any grouping pattern between IHD or OHD samples. The most intense bands (14 out of 58) were excised and sequenced. Sequenced bands are operational taxonomic units (OTU). All 16S rDNA gene partial sequences proved to be affiliated with members of Proteobacteria (β and γ subclasses). Out of the 14 sequenced bands, 11 were present in both groundwater and sediment compartments and were clustered with bacteria like, e.g., Azoarcus sp. (OTU 9 AY816205), Pseudomonas spp. (OTU 6 AY816202 and OTU 13 AY816207), Dechloromonas spp. (OTU 8 AY816204 and 5 AY816201) and Hydrogenophaga sp. (OTU 3 AY816199) (Fig. 5).
Phylogenetic tree for the partial bacterial 16S rDNA sequences recovered after excision of dominant DGGE bands from samples elaborated with denitrifying enrichment culture medium. Tree was obtained using neighbor-joining method with Methanobacterium formicicum as outgroup. Bootstrap values are based on 1,000 runs and are shown where >50%. Similar trees were obtained using maximum-likelihood and maximum-parsimony analyses
Discussion
In this study we intended to combine structural and functional descriptors of free-living and sediment attached bacterial communities. This would permit to study the relationship between biodiversity and functioning for denitrifying bacterial communities in a river–groundwater interface.
The DNA fingerprinting approach used to assess the BCC, PCR-DGGE based on 16S rRNA genes, has been recently introduced in microbial ecology (Muyzer et al., 1997). Only dominant taxa (>1% of the relative abundance) are detected (Casamayor et al., 2000). Each band detected in the lane of the gel would correspond to one bacterial taxon. Co-migration of DNA fragments from different taxa to the same position in the lane of the gel can however happen while separate bands can occur for one taxon (Muyzer & Smalla, 1998). PCR-DGGE is a powerful tool to compare the BCC of different samples, β–diversity sensu Forney et al. (2004) despite imprecision caused by dominant population selection and bias caused by the inaccuracy of the assumption that one band correspond to one bacterial taxon. This technique has been successfully applied to monitor bacterial communities from various environments, e.g., soils (Enwall et al., 2005), river biofilms (Lyautey et al., 2005), sediments (An et al., 2004), plankton (Casamayor et al., 2000) and groundwaters (Röling et al., 2000). On the other hand, regarding the reliability of the obtained BCC, one of the critical point is the sample size with respect to sediment spatial heterogeneity. In soils, 1 g proved to be a good trade off to cover soil heterogeneity (Ranjard et al., 2003). The amount of sandy sediment analyzed in the present study accounted for these recommendations and was in the range of the sample size (0.25–10 g) employed in previous studies on aquifer sediment (Röling et al., 2000; Islam et al., 2004). The amount of metagenomic DNA used as PCR template is also critical for BCC analysis of a natural assemblage (Lyautey et al., 2005). As comparable amounts of metagenome (20 ng) were analyzed here for free-living and attached fractions, differences between the two bacterial communities were unlikely to be due to representativeness bias.
As the functional descriptor used, DEA measures the potential capacity of denitrifying communities under optimal conditions (Groffman et al., 1992). Previous studies comparing simultaneous in situ denitrification and DEA measurements concluded that DEA was 18- to 34-fold higher than in situ denitrification, whereas both methods described similar spatial and temporal patterns of activity (Clément et al., 2002; Well et al., 2003). DEA values measured in the present study in respectively IHD and OHD zones were consistent with previously measured in situ patterns of denitrification activity (Sánchez-Pérez et al., 2003a, b; Baker & Vervier, 2004). Many factors such as denitrification or other microbial metabolisms, plant assimilation and dilution account for changes in the alluvial groundwater nitrate concentrations. In the studied riparian zone, however, the variation of nitrate concentrations calculated by means of a two-end-member mixing model based on chloride concentrations can be considered as a geochemical signature of denitrification (Sánchez-Pérez et al., 2003b). The calculated variations of nitrate concentrations indicated a strong elimination of nitrate in IHD zones. This was consistent with measured DEA indicating that both approaches lead to the same result. It can be assumed that the measured DEA satisfactorily relate to the functional response (denitrification) of the studied communities.
As hypothesized, DEA measurements indicated important denitrifying capability in the sediment compartment but not in the groundwater. In accordance, the BCC of sediment-attached and groundwater free-living assemblages differed. Differences between particle attached and free-living bacteria assemblages within the same bulk have been reported for aquifers (Lehman et al., 2002; Besemer et al., 2005) but also for other environments such as, e.g., marine snow (Grossart et al., 2006), estuary (Crump et al., 1999), marine or lake sediments (Friedrich et al., 1999; Ahn et al., 2006). The growth and succession of free living bacterial populations colonizing the surface of particles result in the formation of biofilms (Watnick & Kolter, 2000; Wimpenny et al., 2000). In these biofilms, new environmental conditions (niches and resources) are generated irrespective of conditions that prevail in the water (Jackson, 2003). For instance, the microzones of oxygen depletion generated by the heterotrophic activity in the biofilm may favor anoxic metabolic activity such as denitrification (Paerl & Pinckney, 1996). The facilitation of chemical microgradient formation within sediment-bacteria aggregates, as in biofilms, explains why attached and free-living bacteria communities in the aquifer exhibited different functional composition and denitrification capability. In the studied aquifer, the free-living bacterial populations likely provide the bacterial species that form the sediment-attached assemblage and selection during the attachment process and later growth on the substrate may thus explain the differentiation that was observed between free-living and attached bacterial assemblages. Moreover, in the aquatic fraction, denitrifying bacteria might represent a lower percentages of the total community compared to sediment total community. Since environmental conditions (e.g., higher dissolved oxygen concentration) in this compartment are not favorable for their development, except in one well (p17), this explains the undetectable DEA in the water compartment.
In the natural assemblages studied, enrichment culture coupled to molecular fingerprinting indicated that different communities of cultivable denitrifying bacteria were present in the groundwater free-living and sediment-attached fractions, in agreement with the differences observed for the total BCC. The enrichment culture step was designed to target the functional community of denitrifiers. This approach has been successfully used to target heterotrophic bacteria in a photoautotrophic assemblage (Jonkers & Abed, 2003a, b; Rafidinarivo et al., 2007). In environmental bacteriology, main bias related to the use of culture is the selection of the so-called cultivable fraction of the community (Barer & Harwood, 1999; Colwell & Grimes, 2000). A part of the diversity, estimated to be higher than 90% in some environments, is not detected by such culture and remains cryptic supporting the interest of molecular approaches (Amann et al., 1995). Culture based communities approaches are thus considered to give a poor estimate of the α–diversity sensu Forney et al. (2004). The 14 OTUs (out of 58 detected bands) that were identified in the present study should be considered as a limited set of the bacterial taxa responsible for the denitrification in the studied zone. All were affiliated to a single group of Bacteria: the Proteobacteria. Taxa exhibiting low genetic distance with Azoarcus and Pseudomonas genus were found that have previously been recorded in several aquifers (Röling et al., 2000; Cho et al., 2003; An et al., 2004; Cavalca et al., 2004). By selecting a fraction of the cultivable community that grow in the laboratory conditions, enrichment culture step might have favored the selection of the same subset of strains in different assemblages. This approach may thus lessen the differences that actually occur between two assemblages, i.e., the β-diversity sensu Forney et al. (2004). Conversely, when differences are evidenced between two assemblages by enrichment culture techniques, reliable conclusions on the β–diversity of the two assemblages can be drawn. With 11 of the 14 OTUs common to sediment and groundwater compartments, it can be suspected that the enrichment culture step may have lessened the differences between the two compartments. Although free-living and sediment-attached assemblages showed similar groups of bacteria, the structure of the communities was different. The groundwater and sediments proved to harbor significantly different cultivable denitrifying communities in relation with the DEA values.
Our study confirms that DEA differs between sediment and groundwater habitat during the low flow period in the unconfined aquifer of the River Garonne. Moreover differences in DEA would be explained by differences in the BCC of sediment-attached and free-living assemblages in relation with the habitat conditions. To date, none of the existing models to predict denitrification in riparian zones take into account the biomass and/or the diversity of denitrifying bacteria (McClain et al., 2003). Here we suggest that the structuring of communities as biofilm aggregate likely facilitates denitrification expression and/or denitrifying strains’ selection. Modelling biofilm development and associated bacterial community structuring and functioning would thus be a way to include community dynamics in biogeochemical models of the aquifers.
A new challenge in functional ecology is to describe how biodiversity dynamics, ecosystem processes and abiotic factors interact (Loreau et al., 2001). This question is equally relevant for microbial taxa (Schimel, 1995). By means of manipulation of the diversity, the contribution of bacteria species diversity to ecosystem functioning was revealed in artificial assemblages (Bell et al., 2005). In natural assemblages from soil, denitrification rates were shown to be related to the putative BCC (Holtan-Hartwig et al., 2000; Cavigelli & Robertson, 2001). The present study would support these conclusions in aquatic assemblages. Monitoring the dynamics of BCC and denitrification expression during a colonization experiment would allow in-depth investigation on the role of communities structuring as biofilm in the denitrification expression in such systems.
According to the hot spots hot moments’ hypothesis, variable hydrological flow paths generate localized activity events that are not permanent (McClain et al., 2003). This study was realized during a low water hydrological period, representative of 40% of time during a year. As defined in this study, IHD and OHD zones may vary with respect to, e.g., the river discharge, precipitation events. This would also affect attached communities maturation and denitrification capability. Further investigations would be necessary to assess the functional significance of free living and sediment attached communities over the entire hydrological cycle in the studied riparian zone.
Conclusions
This study confirmed differences between free-living and attached BCC, and showed that they had different functional significance in the studied aquifer. Both BCC and denitrification can be linked to the structuring of an attached community assemblage. Sediment-attached aggregates form new and original habitats where, e.g., anoxic metabolisms are favored. This conclusion would need to be tested in in situ colonization experiments in order to show that attached bacterial communities result from selection of populations originating from the free-living community. In the aquifer, the interplay between the groundwater liquid phase, which is responsible for transport, and the sedimentary solid phase, which is responsible for biological transformation, would thus explain the occurrence of activity hotspots and the link between diversity and activity.
References
Amann, R. I., W. Ludwig & K. H. Schleifer, 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiological Reviews 59: 143–169.
Ahn, T. S., O. S. Kim, K. S. Joh, L. P. Spiglazov, V. V. Drucker & S.-H. Hong, 2006. Community analysis of aggregated bacteria in southern Lake Baikal. Hydrobiologia 568: 5–8.
An, Y.-J., Y.-H. Joo, I.-Y. Hong, H.-W. Ryu & K.-S. Cho, 2004. Microbial characterization of toluene-degrading denitrifying consortia obtained from terrestrial and marine ecosystems. Applied Microbiology and Biotechnology 65: 611–619.
APHA, 1992. Standard methods for the examination of water and wastewater. American Public Health Association 18th Edition, Washington DC.
Baker, M. A. & P. Vervier, 2004. Hydrological variability, organic matter supply and denitrification in the Garonne River ecosystem. Freshwater Biology 49: 181–190.
Barer, M. R. & C. R. Harwood, 1999. Bacterial viability and culturability. Advances in Microbial Physiology 41: 93–137.
Bekins, B. A., E. M. Godsy & E. Warren, 1999. Distribution of microbial physiologic types in an aquifer contaminated by crude oil. Microbial Ecology 37: 263–275.
Bell, T., J. A. Newman, B. W. Silverman, S. L. Turner & A. K. Lilley, 2005. The contribution of species richness and composition to bacterial services. Nature 436: 1157–1160.
Besemer, K., M. M. Moeseneder, J. M. Arrieta, G. J. Herndl & P. Peduzzi, 2005. Complexity of bacterial communities in a river-floodplain system (Danube, Austria). Applied and Environmental Microbiology 71: 609–620.
Brettar, I., J. M. Sánchez-Pérez & M. Trémolières, 2002. Nitrate elimination by denitrification in hardwood forest soils of the Upper Rhine floodplain—correlation with redox potential and organic matter. Hydrobiologia 469: 11–21.
Brinson, M. M., H. D. Bradshaw & E. S. Kane, 1984. Nutrient assimilative capacity of an alluvial floodplain swamp. Journal of Applied Ecology 21: 1041–1057.
Brüsch, W. & B. Nilsson, 1993. Nitrate transformation and water movement in a wetland area. Hydrobiologia 251: 103–111.
Burgin, A. J. & S. Hamilton, 2007. Have we overemphasized the role of denitrification in aquatic ecosystems? A review of nitrate removal pathways. Frontiers in Ecology and the Environment 5: 89–96.
Burt, T. P., L. S. Matchett, K. W. T. Goulding, C. P. Webster & N. E. Haycock, 1999. Denitrification in riparian buffer zones: the role of floodplain hydrology. Hydrological Processes 13: 1451–1463.
Casamayor, E. O., H. Schäfer, L. Bañeras, C. Pedrós-Alió & G. Muyzer, 2000. Identification of and spatio-temporal differences between microbial assemblages from two neighboring sulphurous lakes: comparison by microscopy and denaturing gradient gel electrophoresis. Applied and Environmental Microbiology 66: 499–508.
Cavalca, L., E. Dell’Amico & V. Andreoni, 2004. Intrinsic bioremediability of an aromatic hydrocarbon-polluted groundwater: diversity of bacterial population and toluene monooxygenase genes. Applied Microbiology and Biotechnology 64: 576–587.
Cavigelli, M. A. & G. P. Robertson, 2001. Role of denitrifier diversity in rates of nitrous oxide consumption in a terrestrial ecosystem. Soil Biology & Biochemistry 33: 297–310.
Cho, H.-B., J.-K. Lee & Y.-K. Choi, 2003. The genetic diversity analysis of the bacterial community in groundwater by denaturing gradient gel electrophoresis (DGGE). The Journal of Microbiology 41: 327–334.
Clément, J.-C., G. Pinay & P. Marmonier, 2002. Seasonal dynamics of denitrification along topohydrosequences in three different riparian wetlands. Journal of Environmental Quality 31: 1025–1037.
Colwell, R. R., & D. J. Grimes, 2000. Non culturable microorganisms in the environment. ASM Press, Washington DC.
Crump, B. C., E. V. Armbrust & J. A. Baross, 1999. Phylogenetic analysis of particle-attached and free-living bacterial communities in the Columbia River, its estuary, and the adjacent coastal ocean. Applied and Environmental Microbiology 65: 3192–3204.
Duff, J. H., A. P. Jackman, F. J. Triska, R. W. Sheibley & R. J. Avanzino, 2007. Nitrate retention in riparian groundwater at natural and elevated nitrate levels in North Central Minnesota. Journal of Environmental Quality 36: 343–353.
Enwall K., L. Philippot & S. Hallin, 2005. Activity and composition of the denitrifying bacterial community respond differently to long-term fertilization. Applied and Environmental Microbiology 71: 8335–8343.
Forney, L. J., X. Zhou & C. J. Brown, 2004. Molecular microbial ecology: land of the one-eye king. Current Opinion in Microbiology 7: 210–220.
Friedrich, U., M. Schallenberg & C. Holliger, 1999. Pelagic bacteria-particle interactions and community-specific growth rates in four lakes along a trophic gradient. Microbial Ecology 37: 49–61.
Garabétian, F., M. Petit & P. Lavandier, 1999. Does storage affect epifluorescence microscopic counts of total bacteria in freshwater samples? Comptes Rendus de l’Academie des Sciences. Paris 322: 779–784.
Groffman, P. M., A. J. Gold & R. C. Simmons, 1992. Nitrate dynamics in riparian forests: Microbial studies. Journal of Environmental Quality 21: 666–671.
Grossart, H. P., T. Kiørboe, K. W. Tang, M. Allgaier, E. M. Yam & H. Ploug, 2006. Interactions between marine snow and heterotrophic bacteria: aggregate formation and microbial dynamics. Aquatic Microbial Ecology 42: 19–26.
Harvey, R. W., R. L. Smith & L. George, 1984. Effect of organic contamination upon microbial distributions and heterotrophic uptake in a Cape Cod, Mass., aquifer. Applied and Environmental Microbiology 48: 1197–1202.
Haycock, N. E. & T. P. Burt, 1993. Role of floodplain sediments in reducing the nitrate concentration of subsurface run-off: a case study in the Cotswolds, UK. Hydrological Processes 7: 287–295.
Hill, A. R., K. J. Devito, S. Campagnolo & K. Sanmugadas, 2000. Subsurface denitrification in a forest riparian zone: interactions between hydrology and supplies of nitrate and organic carbon. Biogeochemistry 51: 193–223.
Hinkle, S. R., J. H. Duff, F. J. Triska, A. Laenen, E. B. Gates, K. E. Bencala, D. A. Wentz & S. R. Silva, 2001. Linking hyporheic flow and nitrogen cycling near the Willamette River—a large river in Oregon, USA. Journal of Hydrology 244: 157–180.
Holtan-Hartwig, L., P. Dörsch & L. R. Bakken, 2000. Comparison of denitrifying communities in organic soils: kinetics of NO3 − and N2O reduction. Soil Biology and Biochemistry 32: 833–843.
Islam, F. S., A. G. Gault, C. Boothman, D. A. Polya, J. M. Charnock, D. Chatterjee & J. R. Lloyd, 2004. Role of metal-reducing bacteria in arsenic release from Bengal delta sediments. Nature 430: 68–71.
Jackson, C. R., 2003. Changes in community properties during microbial succession. Oikos 101: 444–448.
Jackson, M. L., 1985. Soil chemical analysis—advanced course, 2nd ed. 11th printing published by the author, Madison, WI (Revised from 1956 edition).
Jonkers, H. M. & R. M. M. Abed, 2003a. Identification of aerobic heterotrophic bacteria from the photic zone of a hypersaline microbial mat. Aquatic Microbial Ecology 30: 127–133.
Jonkers, H. M., R. Ludwig, R. De Wit, O. Pringault, G. Muyzer, H. Niemann, N. Finke & D. De Beer, 2003b. Structural and functional analysis of a microbial mat ecosystem from a unique permanent hypersaline inland lake: ‘La Salada de Chiprana’ (NE Spain). FEMS Microbiology Ecology 44: 175–189.
Kropf, S., H. Heuer, M. Grüning & K. Smalla, 2004. Significance test for comparing complex microbial community fingerprints using pairwise similarity measures. Journal of Microbiological Methods 57: 187–195.
Lambs, L., 2000. Correlation of conductivity and stable isotope 18O for the assessment of water origin in river system. Chemical Geology 164: 161–170.
Lehman, R. M., F. F. Roberto, D. Earley, D. F. Bruhn, S. E. Brink, S. P. O’Connell, M. E. Delwiche & F. S. Colwell, 2001. Attached and unattached bacterial communities in a 120-Meter corehole in an acidic, crystalline rock aquifer. Applied and Environmental Microbiology 67: 2095–2106.
Lehman, R. M., & S. P. O’Connell, 2002. Comparison of extracellular enzyme activities and community composition of attached and free-living bacteria in porous medium columns. Applied and Environmental Microbiology 68: 1569–1575.
Loreau, M., S. Naeem, P. Inchausti, J. Bengtsson, J. P. Grime, A. Hector, D. U. Hooper, M. A. Huston, D. Raffaelli, B. Schmid, D. Tilman & D. A. Wardle, 2001. Biodiversity and ecosystem functioning: current knowledge and future challenges. Science 294: 804–808.
Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Förster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. König, T. Liss, R. Lüßmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode & K.-H. Schleifer, 2004. ARB: a software environment for sequence data. Nucleic Acids Research 32: 1363–1371.
Lyautey, E., B. Lacoste, L. Ten-Hage, J. L. Rols & F. Garabétian, 2005. Analysis of bacterial diversity in river biofilms using 16S rDNA PCR-DGGE: methodological settings and fingerprints interpretation. Water Research 39: 380–388.
Maître, V., A. C. Cosandey, E. Desagher & A. Parriaux, 2002. Effectiveness of groundwater nitrate removal in a river riparian area: the importance of hydrogeological conditions. Journal of Hydrology 278: 76–93.
McClain, M. E., E. W. Boyer, C. L. Dent, S. E. Gergel, N. B. Grimm, P. M. Groffman, S. C. Hart, J. W. Harvey, C. A. Johanston, E. Mayorga, W. H. McDowell & G. Pinay, 2003. Biogeochemical hot spots and hot moments at the interface of terrestrial ad aquatic ecosystems. Ecosystems 6: 301–312.
McMahon, P. B. & J. K. Böhlke, 1996. Denitrification and mixing in a stream-aquifer system: effects on nitrate loading to surface water. Journal of Hydrology 186: 105–128.
Mosier, A. R., J. W. Doran & J. R. Freney, 2002. Managing soil denitrification. Journal of Soil and Water Conservation 57: 505–513.
Mouné, S., P. Caumette, R. Matheron & J. C. Willison, 2003. Molecular sequence analysis of prokaryotic diversity in the anoxic sediments underlying cyanobacterial mats of two hypersaline ponds in Mediterranean salterns. FEMS Microbiology Ecology 44: 117–130.
Muyzer, G., T. Brinkhoff, U. Nübel, C. Santegoeds, H. Schäfer & C. Wawer, 1997. Denaturing gradient gel electrophoresis (DGGE) in microbial ecology. In Akkermans, A. D. L., J. D. van Elsas, F. J. De Bruijn (eds), Molecular Microbial Ecology Manual. Kluwer Academics Publishers. Dordrecht, The Netherlands: 1–27.
Muyzer, G. & K. Smalla, 1998. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie van Leeuwenhoek, 73: 127–141.
Paerl, H. W. & J. L. Pinckney, 1996. A mini-review of microbial consortia: their roles in aquatic production and biogeochemical cycling. Microbial Ecology 31: 225–247.
Peterjohn, W. T. & D. L. Correll, 1984. Nutrient dynamics in an agricultural watershed: observations on the role of a riparian forest. Ecology 65: 1466–1475.
Pinay, G., C. Ruffinoni, S. Wondzell & F. Gazelle, 1998. Change in groundwater nitrate concentration in a large river floodplain: denitrification, uptake or mixing? Journal of North American Benthological Society 17: 179–189.
Ranjard, L., D. P. H. Lejon, C. Mougel, L. Schehrer, D. Merdinoglu & R. Chaussod, 2003. Sampling strategy in molecular microbial ecology: influence of soil sample size on DNA fingerprinting analysis of fungal and bacterial communities. Environmental Microbiology 5: 1111–1120.
Rafidinarivo, H. F., S. Fujimoto, K. Watanabe, K. Kitazato & N. Kobayashi, 2007. Topographic effects of coastal seas on the composition of the culturable bacterial communities in marine sediments. Hydrobiologia 583: 205–212.
Röling, W. F. M., B. M. van Breukelen, M. Braster, M. T. Goeltom, J. Groen & H. W. van Verseveld, 2000. Analysis of microbial communities in a landfill leachate polluted aquifer using a new method for anaerobic physiological profiling and 16S rDNA based fingerprinting. Microbial Ecology 40: 177–188.
Ruffinoni, C., 1994. Rôle des ripisylves dans la réduction des pollutions diffuses en milieu fluvial. Thesis Université Paul Sabatier, Toulouse: 63.
Sabater, S., A. Butturini, J. C. Clément, T. Burt, D. Dowrick, M. Hefting, V. Maître, G. Pinay, C. Postolache, M. Rzepecki & F. Sabater, 2003. Nitrogen removal by riparian buffers along a European climatic gradient: patterns and factors of variation. Ecosystems 6: 20–30.
Sánchez-Pérez, J. M. & T. Trémolières, 2003. Change in groundwater chemistry as a consequence of suppression of floods: the case of the Rhine floodplain. Journal of Hydrology 270: 89–104.
Sánchez-Pérez, J. M., C. Bouey, S. Sauvage, S.Teissier, I. Antiguedad & P. Vervier, 2003a. A standardised method for measuring in situ denitrification in shallow aquifers: numerical validation and measurements in riparian wetlands. Hydrology and Earth System Sciences 7: 87–96.
Sánchez-Pérez, J. M., P. Vervier, F. Garabétian, S. Sauvage, M. Loubet, J. L. Rols, T. Bariac & P. Weng, 2003b. Nitrogen dynamics in the shallow groundwater of a riparian wetland zone of the Garonne, Southwestern France: Nitrate inputs, bacterial densities, organic matter supply and denitrification measurements. Hydrology and Earth System Sciences 7: 97–107.
Schimel, J., 1995, Ecosystem consequences of microbial diversity and community structure. In F. S. Chapin III, C. Korner (eds), Arctic and alpine biodiversity: patterns, causes and ecosystem consequences. Springer Verlag, New York, NY: 234–259.
Soriano S. & N. Walker, 1968. Isolation of ammonia-oxidizing autotrophic bacteria. Journal of Applied Bacteriology 31: 413–497.
Takatert, N., J. M. Sánchez-Pérez & M. Trémolières, 1999. Spatial and temporal variations of nutrient concentration in the groundwater of a floodplain: effect of hydrology, vegetation and substrate. Hydrological Processes 13: 1511–1526.
Tiedje, J. M., 1994. Denitrifiers. Methods of soil analysis, part 2: Microbiological and biochemical properties, Soil Science Society of America Journal. Madison. WI: 245–267.
Van Cleemput, O., P. Boeckx, P. E. Lindgren, & K. Tonderski, 2007. Denitrification in wetlands. In Bothe, H., S. J. Ferguson, W. E. Newton (eds), Biology of the nitrogen cycle. Elsevier, Amsterdam, The Netherlands: 359–367.
Watnick, P. & R. Kolter, 2000. Biofilm, City of Microbes. Journal of Bacteriology 182: 2675–2679.
Well, R., J. Augustin, K. Meyerc & D. D. Myrold, 2003. Comparison of field and laboratory measurement of denitrification and N2O production in the saturated zone of hydromorphic soils. Soil Biology & Biochemistry 35: 783–799.
Weng, P., J. M. Sánchez-Pérez, S. Sauvage, P. Vervier & F. Giraud, 2003. Assessment of the quantitative and qualitative buffer function of an alluvial wetland: hydrological modelling of a large floodplain (Garonne River, France). Hydrological Processes 17: 2375–2392.
Wimpenny, J., W. Manz & U. Szewzyk, 2000. Heterogeneity in biofilms. FEMS Microbiology Reviews 24: 661–671.
Yoshinari, T. & R. Knowles, 1976. Acetylene inhibition of nitrous oxide reduction by denitrifying bacteria. Biochemical and Biophysical Research Communications 69: 705–710.
Acknowledgements
This work was funded by GIS ECOBAG (Groupement d’Intérêt Scientifique – Ecologie et Economie du Bassin Adour Garonne), A. Iribar was supported by FEDER (Fonds Européen de Développement Régional). We are grateful to C. Mur and D. Dalger for water analysis and F. Julien for field assistance. We also thank the two anonymous reviewers for constructive comments on a previous version of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Handling editor: J. Padisak
Rights and permissions
About this article
Cite this article
Iribar, A., Sánchez-Pérez, J.M., Lyautey, E. et al. Differentiated free-living and sediment-attached bacterial community structure inside and outside denitrification hotspots in the river–groundwater interface. Hydrobiologia 598, 109–121 (2008). https://doi.org/10.1007/s10750-007-9143-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10750-007-9143-9