Abstract
Recharge of aquifers by urban stormwater may trigger significant ecological changes that can be detrimental to the biodiversity and functioning of groundwater ecosystems. Here, the effects of aquifer recharge (AR) on three levels of parameters were investigated: dissolved organic carbon (DOC) quantity and quality, global biofilm characteristics, and diversity changes of bacterial communities. As DOC enrichment by AR can be mitigated by vadose zone (VZ) thickness, three AR sites with thin VZ (< 3 m) and three sites with thick VZ (> 10 m) were selected. For each AR site, clay beads were incubated over a 10-day-long rainy period through wells in recharged and non-recharged groundwaters. Total proteins, dehydrogenase, and hydrolytic activities were monitored from clay beads to assess biofilm development. Bacterial richness on beads was estimated by 16S rRNA-based metabarcoding. AR was found to significantly increase DOC and biodegradable DOC (BDOC) concentrations, biofilm development, and bacterial richness especially in sites with thin VZ. VZ thickness was inversely related to microbial growth indicators and bacterial richness in groundwater, through a control of DOC availability. The proportion of Bacteroidetes 16S rRNA gene reads was higher in recharged groundwater than in non-recharged groundwater, suggesting that this phylum could be used as an indicator of DOC enrichment associated with AR. Quantitative PCR assays for Bacteroides DNA confirmed these trends and showed an enrichment of this bacterial group in DOC-rich aquifer waters. The positive linear relationships between BDOC concentrations and biofilm variables highlighted a strong C-limitation of groundwater impacting bacterial species sorting and activity.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
In ecology, the species-energy theory (Wright 1983; Evans et al. 2005; Fuhrman 2009) is considered as a universal law stating that species richness correlates positively with energy availability. Smith (2007) reviewed 70 published studies in aquatic ecosystems and concluded that diversity-productivity patterns are similar between micro- and macro-organisms. Nonetheless, the shape of the diversity-productivity relationship was variable depending on the studied micro-organisms and environments (Smith 2007; Prosser et al. 2007; Bernstein et al. 2017). Although these relationships have been reported from many aquatic ecosystems including natural lakes, ponds, and oceans, few studies have investigated the influence of energy availability on the diversity of microbial communities in groundwater ecosystems (but see Hershey et al. 2018 for a deep karstic aquifer).
Groundwater ecosystems are among the most stable and energy-limited environments on Earth because of the lack of photosynthesis and low inputs of organic matter infiltrating from surface ecosystems (Gibert et al. 1994; Griebler and Lueders 2009). Nonetheless, connectivity and dissolved organic matter (DOM) transfers between surfaces and subterranean systems can be enhanced by aquifer recharge (AR) practices (e.g., Kolehmainen et al. 2007). In urban environments, these practices were developed to collect stormwater runoff to recharge aquifers by infiltration through porous media (Marsalek and Chocat 2002; Hunt et al. 2010). AR practices have been shown to increase DOM in groundwaters over the levels of those that were not influenced by recharge (Datry et al. 2004; Foulquier et al. 2010; Mermillod-Blondin et al. 2013). DOM is the main source of food for micro-organisms in groundwater ecosystems (Goldscheider et al. 2006), and its increase can lead to denser and more active microbial communities (Foulquier et al. 2011; Landa et al. 2013). For example, Mermillod-Blondin et al. (2013) showed that the growth and the esterase and dehydrogenase activities of biofilms were stimulated by DOM enrichment associated with aquifer recharge in an urban context. Nevertheless, the influence of DOM supplied by AR systems on microbial diversity is still poorly defined.
The aim of the present work was to evaluate the relative efficiencies of three levels of describers to detect the effect of stormwater inputs on groundwater: DOM quality, biofilm characteristics, and bacterial diversity. A field experiment was conducted during a rainy period in six AR sites that were divided into two groups: (1) those with a thin vadose zone (< 3 m) and (2) those with a thick vadose zone (> 10 m). The vadose zone plays a key role on DOM retention during its transfer from surface to groundwater (Pabich et al. 2001; Shen et al. 2015). Selection of sites with different vadose zone thicknesses should lead to significant differences in groundwater disturbance. Dissolved organic carbon (DOC) and biodegradable (BDOC) concentrations were measured together with the incubation of artificial substrates (i.e., clay beads) in recharged and non-recharged groundwaters. Clay beads were previously shown to be efficient artificial substrates that can allow reliable bacterial diversity analyses of groundwaters (Voisin et al. 2016; Mermillod-Blondin et al. 2019). Microbial productivity and activities of the biofilms on the clay beads could be monitored from biomass measurements (total proteins) and from esterase and dehydrogenase activities. Bacterial diversity was investigated through a next-generation sequencing (NGS) dataset of PCR-amplified 16S rRNA gene segments generated from biofilm DNA extracts. According to Datry et al. (2005), we expected that DOM concentrations would be higher in AR sites with a thin vadose zone in comparison with those with a thick vadose zone. Consequently, the hypothesis was that the influence of AR practices on microbial biomass, activity, and diversity should be highest in AR sites with the thinnest vadose zone (< 3 m). In addition, positive correlations between DOM concentrations over the six sites and bacterial richness of the clay bead biofilms developing in groundwaters should be observed.
Material and methods
Sites
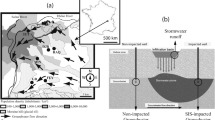
Aquifer recharge (AR) sites were located on the eastern aquifer of the Lyon metropolitan area, France (Fig. 1a). This aquifer (catchment area: 314 km2) is characterized by three corridors of highly permeable glacio-fluvial sediments (hydraulic conductivity 10−3–10−2 m/s) (Foulquier et al. 2009). The aquifer is artificially recharged with stormwater runoff at multiple AR sites to compensate for the reduced recharge caused by the sealing of urban surfaces. The AR sites of this study corresponded to retention/infiltration basins that collected stormwater from residential, commercial, and industrial areas.
Experimental design
Six AR sites (Fig. 1a, b) were selected: three with vadose zone thicknesses < 3 m (IUT, Grange Blanche (GB) and Minerve (MIN) basins) and three with vadose zone thicknesses > 10 m (Django Reinhardt (DJR), Raquin (RAQ), and Feyzin (FEY) basins). Each AR site is equipped with two wells: a well located outside the area of influence of the stormwater plume induced by infiltration which was used as a control well, and a well located in the immediate downstream vicinity of the infiltration basin used as a recharge well because it intersected the stormwater plume induced by the infiltration.
Clay beads (expanded clay aggregates, Botanic®, Saint-Julien en Genevois, France) of a diameter of 8 mm were incubated in these wells to trap micro-organisms. These clay beads favor biofilm developments from the groundwater microbiota (Voisin et al. 2016; Mermillod-Blondin et al. 2019) and enable a monitoring of parameters such as microbial productivity (total proteins) and esterase and dehydrogenase activities. Before incubation, clay beads were heated to 550 °C to eliminate organic matter traces and then autoclaved at 121 °C for 20 min. They were incubated in wells using small polypropylene bags (110 × 40 mm) with a mesh size of 3.4 mm, previously treated with ethanol (70%). A total of 30 clay beads were inserted per bag, and 3 bags were incubated per well after well purging with a submerged pump (see below). From each bag, clay beads were used to measure all microbial variables (total proteins, esterase and dehydrogenase activities, bacterial community structure, and qPCR assays on specific genes) in order to obtain mean biofilm parameters per well calculated from three values (corresponding to batches of clay beads from the three bags incubated in the same well). According to previous laboratory and field experiments (Voisin et al. 2016; Mermillod-Blondin et al. 2019), clay bead incubation lasted 10 days during a rainy period to obtain (1) a significant microbial colonization of clay beads submerged in groundwaters and (2) a time-integrated monitoring of micro-organisms in recharged groundwater during a rainy event. The experiment was performed between September 12 and September 22 in 2015. During this period, precipitations were of 26.6 mm on the 12, 34.6 mm on the 13, 1.6 mm on the 14, 49.6 mm on the 17, 2.8 mm on the 18, 3.2 mm on the 19, and 1 mm on the 20. For each AR site, groundwaters (recharged and non-recharged) were also collected from the same wells but at three dates over the clay bead incubation period (day 0, day 5, and day 10) to obtain mean values of DOC and BDOC per well during the incubation period of clay beads. Groundwaters were sampled in wells using a submerged pump (PP36 inox, SDEC, Reignac-sur-Indre, France). The first 50 pumped liters were used to rinse the sampling equipment. The following liter was collected for analyses of DOC and BDOC analyses. Samples were stored in pre-combusted glass bottles and brought back to the laboratory in an isotherm box at 4 °C.
Water physico-chemistry
DOC (dissolved organic carbon) was determined using a carbon analyzer (multi N/C® 3100, Analytik Jena, Jena, Germany) based on thermocatalytic oxidation (850 °C) of organic carbon and infrared detection of CO2, after removal of dissolved inorganic C with HCl. Biodegradable DOC (BDOC) was determined by the method of Servais et al. (1987, 1989) following Mermillod-Blondin et al. (2015). Briefly, water samples (100 mL) were filtered through a pre-washed 0.2-μm polycarbonate membrane and incubated with a bacterial inoculum at 20 °C for 30 days in the dark. DOC concentrations were measured from filtered (0.2 μm) water samples at the start and the end of the incubation period to determine the initial DOC concentration and the remaining DOC concentration after 30 days of incubation (representing the refractory dissolved organic carbon, RDOC), respectively. BDOC concentration was then calculated as the difference between initial DOC concentration and RDOC concentration.
Microbial analysis
Microbial biomass
Microbial biomasses on clay beads were assessed from Lowry’s total protein assay using the Sigma Protein Assay Kit P-5656 (Sigma Diagnostics, St Louis, MO, USA) according to Peterson (1977). For each incubated bag, five randomly sampled clay beads were treated with 4 mL of a 50% solution of Lowry’s reagent for 20 min. At the start of the reaction, the contact between proteins on beads and the reagent was improved by vortexing at 1000 rpm for 1 min. After the 20-min long reaction, 0.5 mL of Folin reagent was added, followed by a 30-min color reaction. The absorbance of the solution was then measured at 750 nm with a spectrophotometer (Aquamate, ThermoSpectronic, Cambridge, UK) and total protein content on beads was determined using a calibration curve performed with bovine serum albumin. Total proteins were expressed in μg of proteins per cm2 of clay bead, after subtracting values measured from three batches of five sterile clay beads.
Microbial activity
Hydrolytic activity of biofilms was estimated using the fluorescein diacetate (FDA) hydrolysis method (Fontvieille et al. 1992) following Voisin et al. (2016). For each incubated bag, five randomly sampled clay beads were incubated into 3 mL of a pH 7.6 phosphate buffer solution with 0.15 mL of 4.8 mM FDA solution. Incubation was performed at 15 °C for 3 h. The reaction was stopped by addition of 3 mL of acetone in the sample and storage at − 20 °C (Battin 1997). Fluorescein concentration was estimated by measuring the absorbance at 490 nm of the filtered supernatant (0.45 μm, HAWP, Millipore, Billerica, MA, USA). Then, the hydrolytic activity of biofilms was expressed in μmol of FDA hydrolyzed per hour and per cm2 of clay bead, after subtracting values measured from three batches of five sterile clay beads.
The 2-(p-iodophenyl)-3-(p-nitrophenyl)-5-phenyl tetrazolium chloride (INT) was used to measure dehydrogenase activity (respiratory activity) as modified from Houri-Davignon and Etcheber (1989) following Mermillod-Blondin et al. (2019). For each incubated bag, five randomly sampled clay beads were incubated in a 0.02% INT solution for 3 h at 15 °C and the supernatant was then filtered on a nylon membrane (0.22 μm, Millipore, Billerica, MA, USA) to retrieve floating microbial cells. Extraction of INT-formazan from clay beads and nylon membrane containing microbial cells was performed with 5 mL of methanol during 12 h. The INT-formazan concentration was determined by absorbance measurement at 480 nm. Then, the dehydrogenase activity of biofilms was expressed in μmol of INT-formazan produced per hour and per cm2 of clay bead, after subtracting values measured from three batches of five sterile clay beads.
16S rRNA gene bacterial community profiling
DNA extraction and qPCR assays
DNA extraction from clay bead biofilms was performed following Voisin et al. (2016). For each incubated bag, microbial cells were detached from 15 randomly selected clay beads by shaking at 2500 rpm for 2 min in 10 mL of 0.8% NaCl. Cells and aggregates were retrieved from the NaCl solution by a 0.22-μm filtration. DNA was extracted from filters using the FastDNA spin kit for soil (MP biomedicals). Then, the amount of extracted DNA was quantified spectrophotometrically using a Nanodrop ND1000 (Thermo Fisher Scientific, USA).
qPCR assays were performed on a Bio-Rad CFX96 real-time PCR instrument with Bio-Rad CFX Manager software, version 3.0 (Marnes-la-Coquette, France). Total Bacteroides DNA targets were PCR amplified according to Layton et al. (2006) using the Brilliant II SYBR Green low ROX qPCR master mix for TaqMan qPCR (Agilent, Vénissieux, France). The human-specific HF183 Bacteroides qPCR assay was performed according to Seurinck et al. (2005), and the assay for the 16S rRNA gene segment for total bacteria was performed according to Park and Crowley (2006) using primers 338F and 518R and using the Brilliant II SYBR green low ROX qPCR master mix for SYBR Green qPCR. Melting T° was 60 °C for all assays. Linearized plasmid DNA containing 16S rRNA genes from the targeted DNAs were run as standards using 10-fold dilutions of the plasmids. These plasmids were obtained from Marti et al. (2017). Presence of inhibitors was checked by spiking known amount of plasmid harboring int2 with 10 times dilution of sample DNA extracts (107 copies of plasmid per μl of DNA diluted solution). Number of cycles needed to have a significant signal was compared with wells where only plasmid harboring int2 was added to the qPCR mix. When a higher number of cycles was needed to observe a signal, a dilution by 5- or 10-fold was done and another run of tests was performed to confirm the absence of PCR inhibitions. Negative controls without template DNA were run in triplicate. Each assay was triplicated on distinct DNA extracts, and technical triplicates were performed.
V5-V6 analyses of 16S rRNA gene PCR products
Sequencing of the V5-V6 16S rRNA gene segments were performed at MrDNA (Shallowater, TX, USA) with the Illumina MiSeq technology. Primers 799F (5′-ACCMGGATTAGATACCCKG-3′) and 1193R (5′-CRTCCMCACCTTCCTC-3′) were used, and 20,000 reads (i.e., 2 × 300-bp paired-end reads) were generated per sample. Data processing was realized with the MOTHUR software version 1.35.1 (Schloss et al. (2009) following Voisin et al. (2018)). Briefly, raw sequences were filtered by discarding those having (1) read sizes < 200 bp and > 400 pb, (2) reads containing homopolymers > 8 bp, (3) more than one sequence mismatch to barcode DNA tags and more than two mismatches with the forward sequencing primer, (4) reads containing ambiguous bases, and (5) average sequencing quality score < 35 using a 50-bp sliding window (Schloss et al. 2011). After this filtering step, reads were aligned against the SILVA database release 123 (Quast et al. 2013) and reads that could not be reliably assigned to the V5-V6 region were discarded. A pre-cluster step was applied following Huse et al. (2010). Potential chimeric sequences were detected and removed using CHIMERA UCHIME (Edgar et al. 2011). Then, reads were clustered into operational taxonomic units (OTUs) by a clustering at 97% similarity (Schloss and Westcott 2011). OTUs were affiliated to taxonomic groups according to the SILVA database using an 80% minimum bootstrap cutoff (Wang 2007). DNA sequences classified as Archaea, Eukaryota, chloroplasts or mitochondria were discarded.
Data deposition
Sequence data are available at the European Nucleotide Archive (https://www.ebi.ac.uk/ena) under the project accession #PRJEB29925. Sample accession #ERS2912777 (SAMEA5128392) = run2. See Table S1 for barcode information.
Data analysis
The effects of artificial recharge (i.e., infiltration) and vadose zone thickness (VZT) on DOC and BDOC concentrations, biofilm characteristics (i.e., proteins, FDA, and INT assays), and diversity (number of OTUs, Shannon diversity index, and Pielou’s evenness index) were tested using repeated-measure analyses of variance (two-way RM-ANOVA) with VZT as fixed factor and recharge as repeated factor to take into account the non-independence of recharged and non-recharged wells in each site. Pearson’s correlations were computed to test the significance of the relations between DOM (i.e., DOC and BDOC) and clay bead biofilm variables (i.e., biomass, activities, OTU richness, and diversity indices). Most variables were logit transformed to satisfy the normality assumption verified by Shapiro’s test. Sphericity assumption for RM-ANOVA was verified using Mauchly’s sphericity test. As log-transformed qPCR datasets did not satisfy the assumption of normality, Kruskal-Wallis (KW) and Spearman correlation tests were applied on the qPCR datasets.
A non-metric multidimensional scaling (NMDS) was performed based on the Bray-Curtis dissimilarity distances. Differences in bacterial community structures among biofilms from recharged and non-recharged groundwaters in AR sites with thin and thick vadose zones (four groups taking into account 2 VZT conditions * 2 recharge conditions) were tested using permutational multivariate analysis of variance (PERMANOVA; Anderson 2001). Statistical tests were based on 999 permutations of the Bray-Curtis matrix and significance was accepted at p value < 0.05. All statistical tests were performed using the R software (R Development Core Team 2008). The ade4 and Vegan packages were used for nMDS and PERMANOVA analyses (Oksanen et al. 2007).
Results
DOM concentrations in groundwater of AR sites
DOC and BDOC concentrations in groundwater averaged 1.19 ± 0.55 mg L−1 and 0.56 ± 0.27 mg L−1, respectively. Both DOC and BDOC concentrations were significantly higher in recharged groundwaters than in non-recharged groundwaters (Fig. 2a, b; two-way RM-ANOVA, recharge effect, p < 0.05 for DOC and BDOC concentrations). DOC concentrations in recharged groundwater averaged 1.52 ± 0.54 mg L−1 whereas measured concentrations in non-recharged groundwater were 0.87 ± 0.33 mg L−1. The same pattern was observed for BDOC concentrations with higher values measured in recharged groundwater (0.69 ± 0.26 mg L−1) in comparison with concentrations from non-recharged groundwater (0.42 ± 0.22 mg L−1). The mean enrichment in DOM concentrations induced by groundwater recharge (recharged versus non-recharged groundwater) was more important in AR sites with thin vadose zones (mean enrichment of 0.89 mg L−1 and 0.37 mg L−1 for DOC and BDOC, respectively) than in AR sites with thick vadose zones (mean enrichment of 0.40 mg L−1 and 0.17 mg L−1 for DOC and BDOC, respectively). Consequently, DOC and BDOC concentrations in groundwaters were significantly higher in AR sites with a thin vadose zone than in AR sites with a thick vadose zone (two-way RM-ANOVA, VZT effect, p < 0.03).
a Concentrations of dissolved organic carbon (DOC) and b biodegradable dissolved organic carbon (BDOC) in non-recharged (NR, white) and recharged (R, gray) groundwater (GW). c Total proteins (i.e., proxy of biofilm biomass), d hydrolytic (FDA), e dehydrogenase (INT) activities, f OTU numbers, g Shannon diversity index, and h Pielou’s evenness index of biofilms developed on artificial substrates (clay beads) incubated in non-recharged (NR, white) and recharged (R, gray) groundwater (GW). The AR sites are used as replicates (n = 3) for the two conditions of vadose zone (VZ) thickness: thick VZ (> 10 m) on the left of the panel and thin VZ (< 3 m) on the right of the panel. Total proteins, hydrolytic and dehydrogenase activities were expressed per cm2 of artificial substrate (clay bead)
Influence of groundwater recharge on biofilm biomass and activities
Biofilm growth parameters were significantly influenced by recharge of groundwater with stormwater runoffs (two-way RM-ANOVA, recharge effect, p < 0.05). Total proteins measured on clay beads were significantly higher in biofilms developed in recharged groundwaters (mean total proteins: 7.34 ± 5.09 μg per cm2 of clay beads) than in biofilms recovered from non-recharged groundwaters (mean total proteins: 4.23 ± 1.79 μg per cm2 of clay beads) (Fig. 2c). The same trends were observed for hydrolytic and dehydrogenase activities measured on biofilms (Fig. 2d, e) although no significant statistical results were obtained due to the high variability among sites (two-way RM-ANOVA, recharge effect, 0.2 > p > 0.15 for hydrolytic activity and 0.1 > p > 0.05 for dehydrogenasic activity). On average, hydrolytic activity was more than 4-fold higher for biofilms developed in recharged groundwaters compared to those developed in non-recharged groundwaters (Fig. 2d). Groundwater recharge increased by more than 1.5-fold the dehydrogenase activity of biofilms over the activities obtained from non-recharged groundwaters (Fig. 2e). Quantitative PCR assays used to determine bacterial concentrations over the bead surfaces followed similar trends. High bacterial contents among the clay bead biofilms going from 2 × 106 to 1.88 × 107 16S rRNA gene copies per cm2 of clay beads incubated in non-recharged groundwater, and 2.35 × 106 to 6 × 108 copies per cm2 of clay beads incubated in recharged groundwater (Table S2). 16S rRNA qPCR DNA counts were significantly higher in clay bead biofilms from recharged wells with a thin vadose zone rather than those with a thick vadose zone (Kruskal-Wallis tests, p < 0.01). This VZT effect was confirmed with the other biofilm biomass and activity parameters which exhibited higher values in AR sites with thin vadose zones than in AR sites with thick vadose zones (two-way RM-ANOVA, VZT effect, p < 0.05 for the three variables). Although statistical analyses were close to significant level (two-way RM-ANOVA, “recharge x VZT” interaction, 0.1 > p > 0.05 for dehydrogenase activity), biomass and activities tended to be more stimulated by recharge in AR sites with vadose zone thicknesses < 3 m than in AR sites with vadose zone thicknesses > 10 m.
Influence of groundwater recharge on biofilm 16S rRNA gene diversity indices and bacterial community structure
The OTU numbers were estimated from 16S rRNA V5-V6 DNA segments generated from the clay bead biofilm DNA extracts (after a re-sampling step set at 32,000 sequences per sample, after the trimming-cleaning processes). These DNA reads were grouped into OTU when showing 3% or less divergence. Accordingly, the DNA extracts from recharged groundwaters were richer than biofilms from non-recharged ones (Fig. 2f, two-way RM-ANOVA, recharge effect, p < 0.05). Indeed, numbers of detected OTUs were 1224 ± 472 in recharged groundwaters and 870 ± 202 in non-recharged groundwaters (Fig. 2f). OTU richness also depended on VZT (two-way RM-ANOVA, VZT effect, p < 0.02). OTU increases were mainly observed in AR sites with vadose zone thicknesses < 3 m (mean increase of 607 OTUs). The other three sites with thicker vadose zones showed no clear changes in OTU numbers associated with AR (mean decrease of 102 OTUs). Shannon diversity indices computed from OTU numbers were higher in biofilms from recharged groundwaters than non-recharged ones (Fig. 2g; two-way RM-ANOVA, recharge effect, p < 0.05). Pielou’s evenness index indicated that these differences in OTU were associated with an increase in evenness among OTUs from biofilms of the recharged sites (Fig. 2h; two-way RM-ANOVA, recharge effect, p < 0.05). In addition, the NMDS ordination of OTUs from clay beads showed significant differences in bacterial community structure between biofilms developing in groundwater for the four tested conditions (2 recharge conditions * 2 VZT condition, Fig. 3, PERMANOVA, p < 0.001).
Core bacterial taxa inferred from aquifer biofilm 16S rRNA gene sequences
Only 130 OTUs which corresponded to 1.38% of the total number of OTUs (9435) were shared between the 36 clay bead biofilm samples. Nonetheless, these 130 OTUs represented 71.66% of all sequences. The largest groups of core OTUs were allocated to the following phyla: Proteobacteria (109 OTUs with 87% of sequences), followed by Bacteroidetes (13 OTUs with 11% of sequences), Actinobacteria (5 OTUs with 1.6% of sequences) and Nitrospirae (3 OTUs with only 0.05% of sequences). Proteobacteria were mainly represented by Gamma-Proteobacteria (28 OTUs with 44% of sequences), Beta-Proteobacteria (50 OTUs with 32% of sequences), and Alpha-Proteobacteria (27 OTUs with 10% of sequences).
Core clay bead biofilm communities of non-recharged groundwaters and recharged groundwaters were similar with Proteobacteria being predominant according to number of OTUs and read numbers (Fig. 4). Proteobacteria was followed by Bacteroidetes and Actinobacteria. Other phyla such as Nitrospirae and Elusimicrobia were found but in smaller proportions in both core communities. Chlorobi and Chloroflexi were only detected in core communities of recharged groundwaters, while Firmicutes were only found in core communities of non-recharged groundwaters. Except these rare phyla, the main difference between the core communities of recharged and non-recharged groundwaters was observed on the relative proportions of Bacteroidetes (Fig. 4): Bacteroidetes were represented more than 3-fold in the core community of recharged groundwaters (17.1% of sequences) than in core community of non-recharged groundwaters (4.9% of sequences). Quantitative PCR assays confirmed these trends (Table S2). Concentrations of total Bacteroides were found significantly higher by qPCR in clay bead biofilm DNA extracts from recharged wells than those recovered from non-recharged wells (Table S2) (for each VZT category, Kruskal-Wallis, p value < 0.01). Furthermore, biofilms from shallow aquifers were always found to have significantly higher total Bacteroides counts than those from aquifers with higher VZT (KW p value < 0.01). Total Bacteroides DNA could not be detected among most clay bead biofilms recovered from non-recharged wells from sites with a thick VZT except for two samples in the RAQ site (Table S2).
Relationships between DOM and biofilm biomass, activities, and diversity
Biofilm characteristics such as qPCR datasets, microbial biomass estimated from Lowry’s total proteins estimations or microbial activities estimated from FDA and INT were positively and significantly correlated with the concentrations of DOC and BDOC associated with stormwater infiltration (e.g., Table 1), with the exception of the relation between FDA activity and DOC that was not significant. Nevertheless, correlations between these three microbial parameters and dissolved organic matter were stronger with BDOC than with DOC. Number of bacterial cells among aquifer biofilms as estimated by qPCR were positively correlated with DOC and BDOC (Spearman’s rho factor > 0.6, p < 0.01). A positive and significant correlation was also detected between OTU number and BDOC (r = 0.79; p < 0.05) and between OTU number and DOC (r = 0.74, p < 0.05). When considering the main bacterial phyla, a positive correlation was observed between the number of OTUs belonging to Bacteroidetes and BDOC (r = 0.71; p < 0.01) and between Bacteroidetes and DOC (r = 0.67; p < 0.05) (Fig. 5). This correlation held true using total Bacteroides qPCR counts (Spearman’s rho factor > 0.7, p < 0.01) (according to Table S2). Furthermore, the total Bacteroides qPCR counts were found correlated with qPCR counts for the HF183 Bacteroides target which is specific for species associated with the human intestinal tract (Table S2) (Spearman’s rho factor > 0.7, p < 0.01). HF183 Bacteroides qPCR counts were also correlated with the DOC and BDOC concentrations (Spearman’s rho factor > 0.56, p < 0.01). HF183 were only detected in biofilms from AR sites with a thin vadose zone (all biofilm samples from recharged groundwater of GB site and one biofilm sample from recharged groundwater of IUT site, Table S2). The number of OTUs belonging to Alpha-proteobacteria and Beta-proteobacteria also significantly increased with DOC (r = 0.81; p < 0.01 for Alpha-proteobacteria and r = 0.71; p < 0.05 for Beta-proteobacteria) and BDOC concentrations (r = 0.79; p < 0.01 for Alpha-proteobacteria and r = 0.65; p < 0.05 for Beta-proteobacteria) (Fig. 5). In the dataset, one other phylum, Chlorobi, showed a significant and positive correlation between OTU numbers and BDOC concentrations (r = 0.66 and p < 0.05) (Fig. 5).
Linear regressions between BDOC concentrations and OTU number for selected phyla. a total number of bacterial OTUs bBacteroidetes, cChlorobi, dAlpha-proteobacteria and eBeta-proteobacteria. Pearson’s r factors are indicated with their respective p value. For Bacteroidetes (b), the relationship between richness and BDOC values remains significant (r = 0.7, p < 0.05) after suppression of the lowest and highest values of Bacteroidetes richness
Discussion
The influence of AR practices was very well illustrated by the DOC contents of groundwaters. The relationships between DOC and vadose zone thicknesses confirmed the hypothesis that DOM enrichment of groundwaters decreases with VZT (Pabich et al. 2001; Datry et al. 2006). Concentrations of total dissolved organic carbon (DOC) and biodegradable dissolved organic carbon (BDOC) were significantly higher in recharged than non-recharged groundwaters but mainly for AR sites with thin vadose zones (< 3 m). This effect was not significant for AR sites with a thick vadose zone (> 10 m). The influence of VZT on DOM reaching groundwater was thus linked to the DOM soil retention capacity. Biotic (microbial consumption of BDOC, Marschner and Kalbitz 2003; Li et al. 2013; Shen et al. 2015) and abiotic (sorption of DOM on clay particles, soil organic matter, or/and Fe and Al oxides/hydroxides, McKnight et al. 1992; Kalbitz et al. 2000; Saidy et al. 2013) soil components are likely involved in this VZT-related effect. Consequently, and according to several studies (e.g., Pabich et al. 2001; Shen et al. 2015), the thicker the VZ, the more efficient is its ability to limit groundwater contamination with DOM.
Based on groundwater samples from Cape Cod (USA), Pabich et al. (2001) determined a statistical relationship between VZT (in m) and DOC concentration (in mg L−1) in groundwater: DOC concentration = DOC concentration in surface water * e−0.7*VZT. By applying the same relationship using a mean DOC concentration of 5 mg L−1 in surface water (according to Voisin et al. 2018), we calculated DOC concentrations ranging between 0.9 and 3.05 mg L−1 for groundwaters influenced by recharge in sites with vadose zones < 3 m. Interestingly, the measured DOC concentrations at these sites were in the same range (i.e., 1.6–2.3 mg L−1). One would have expected that AR would have produced higher DOC concentrations than those expected from the model of Pabich et al. (2001) because of the stronger hydrological connectivity between the surface and the aquifer in these AR sites (Foulquier et al. 2011). Two mechanisms could explain the concordance between our measurements and simulations by the model of Pabich and colleagues: (1) soil and VZ of AR systems were very efficient in DOC retention (Zhang et al. 2012; Mermillod-Blondin et al. 2015) and (2) DOC from infiltrating waters was significantly diluted by groundwater (poor in DOC) in recharge zones of the aquifer (Foulquier et al. 2010). In some of our AR sites, two studies (Mermillod-Blondin et al. 2015; Voisin et al. 2018) tried to determine the respective influences of dilution and retention in organic matter dynamics by using chloride as conservative tracer to evaluate the role of dilution. They found that the dilution of surface water with groundwater could not explain the decrease of DOC concentrations from surface waters to the aquifer. Then, it is expected that abiotic and biotic retention of DOC during water infiltration through the soil and VZ was the main mechanism involved in DOC dynamics in AR sites. Nevertheless, this retention process was not efficient enough to limit significant DOC and BDOC enrichments of groundwater in AR sites with thin VZ. The concentration of organic carbon was thus a good indicator of the connectivity between surface and groundwater ecosystems.
As most groundwater ecosystems experience strong carbon limitation that severely limit growth of microbial communities (Bengtsson 1989; Kazumi and Capone 1994; Baker et al. 2000; Goldscheider et al. 2006), the biomass (total proteins), 16S rRNA gene counts, and activities of biofilm that grew on artificial substrates (clay beads) introduced in the aquifer were positively linked with DOC enrichment. The vadose zone thickness was also significantly related to the observed changes in microbial activities that were measured: artificial recharge significantly stimulated micro-organisms for AR sites with a thin vadose zone whereas it was not the case in AR sites with a thick vadose. To our knowledge, these results are the first to demonstrate that VZT influences microbial growth in groundwaters through its control over DOM availability in AR systems. Indeed, Datry et al. (2005) demonstrated the influence of VZT on DOC concentrations and groundwater invertebrates but they did not evaluate the response of the micro-organisms which are known to mediate energy fluxes from DOM towards invertebrates (Cooney and Simon 2009). Foulquier et al. (2011) estimated bacterial cell numbers (i.e., DAPI staining counts and Lowry’s total proteins assays) and microbial activities (i.e., esterase and dehydrogenase activities) on sediments collected from three MAR systems with a thin VZ but they did not consider systems with thick VZ.
The scarcity of studies on microbial biofilms from deep alluvial aquifers (i.e., when the vadose zone thickness exceeds 10 m) results from technical difficulties and costs associated with pumping or coring for sediment collection (Chapelle 2001). Thus, the use of artificial substrates (clay beads) appeared an efficient strategy to compare biofilm growth development and activities among sites of contrasted VZT including deep aquifers. The positive and significant correlations obtained between microbial datasets from biofilms developing on clay beads and DOC and/or BDOC concentrations (Table 1) confirmed the efficacy of this approach for monitoring trophic conditions in groundwater ecosystems (Williamson et al. 2012; Mermillod-Blondin et al. 2013; Voisin et al. 2016).
The influence of AR practices on the OTU richness of biofilms collected from artificial substrates (clay beads) was also related to the VZT. Indeed, OTU number was significantly higher in recharged groundwaters than in non-recharged ones for AR sites with thin VZ. In contrast, no significant effect of recharge was measured on OTU numbers in AR sites with thick VZ. These increasing OTU numbers were found to match the increase of 16S rRNA bacterial qPCR counts observed in recharged wells of AR sites with a thin vadose zone in comparison with recharged wells from AR sites with a thick vadose zone. In addition, Shannon diversity and Pielou’s evenness indices indicated that the higher OTU richness of biofilms from recharged wells in comparison with biofilms from non-recharged wells was not due to the addition of relatively rare or relatively abundant taxa in the community. It rather seemed to be associated with an increasing in numbers of less frequent OTU at the non-recharged sites, leading to a greater evenness in the datasets between the OTUs of the recharged sites. This confirms that bacterial community structures were significantly influenced by recharge (more OTUs and higher evenness in the bacterial community). NMDS ordination of OTUs and PERMANOVA test also highlighted that bacterial community structures of biofilms incubated in groundwaters were significantly different among the four tested conditions (2 VZT conditions * 2 recharge conditions). According to the observations of Hug et al. (2015) on groundwater microorganisms, the spatial distribution of the six AR sites (Fig. 1) would have led to high differences in bacterial community structures among these sites, reducing the ability to detect a significant influence of VZT and recharge conditions on bacterial communities using AR sites as replicates. As it was not the case, VZT and recharge could be considered as main factors structuring the bacterial communities of groundwater biofilms in the present study.
The V5-V6 16S rRNA gene-inferred richness indices were found coherent with changes in DOC availability in groundwaters associated with recharge, and positive significant relations were obtained between OTU numbers and BDOC concentrations. These trends were further confirmed by qPCR assays targeting Bacteroides species. Biofilms from shallow aquifers were found to have significantly higher total Bacteroides counts than those from aquifers with higher VZT. Furthermore, the application of a more specific PCR screening for Bacteroides species specialized for the human intestinal tract confirmed a strong relation between their counts in an aquifer biofilm and DOC/BDOC. Thus, the hypothesis of an increased richness of bacterial “species” (inferred from OTU numbers) with the availability of nutrient (i.e., BDOC being the biodegradable fraction of DOC for bacteria) in groundwater ecosystems is further supported by these datasets. However, DOC transfers were also found to point out probable transfers of allochtonous bacteria associated with organic matters such as those coming from human wastes.
We also showed that biomass (concentrations of total proteins), activities (fluorescein diacetate (FDA) hydrolysis, INT-formazan reduction), and 16S rRNA genetic diversity of biofilms growing on clay beads (in groundwaters) were all correlated positively with DOC and BDOC concentrations. These results are in line with the “more individuals” hypothesis (Srivastava and Lawton 1998) of the species–energy theory (Wright 1983; Cardinale et al. 2009) which indicates that resource availability positively influenced species richness by increasing population sizes and reducing the probability of extinction. This positive response of bacterial diversity with resource availability is more likely expected in ecosystems experiencing strong nutrient limitations (see Bienhold et al. 2012 for an example in Arctic deep-sea sediments). Given the oligotrophic conditions encountered in groundwater environments, it is thus probable that increased DOC and BDOC concentrations associated with AR practices would enable the maintenance of copiotrophic bacteria that were not adapted to live in the more oligotrophic areas of aquifers (e.g., Fierer et al. 2007; Lever et al. 2015).
These diversity changes were related to changes in the proportions of bacterial phyla among the core communities. The relative proportion of Bacteroidetes increased in the recharged groundwaters as observed from the OTU profiling and qPCR datasets. These observations are consistent with an increase of copiotrophic bacteria with increasing energy availability because members of the phylum Bacteroidetes are often abundant in nutrient-rich aquatic environments (e.g., Gómez-Pereira et al. 2010; Landa et al. 2013; Marti et al. 2017). Indeed, this phylum has been detected in most aquatic habitats (e.g., Alonso et al. 2007; Díez-Vives et al. 2014; Schauer et al. 2010) but bacteria of this group which are known to degrade polymeric organic matter (Cottrell and Kirchman 2000) are often associated with blooms of phytoplankton (e.g., Pinhassi et al. 2004; Teeling et al. 2012; Tada et al. 2011) and fecal contaminations (e. g. Marti et al. 2017; Bernardin-Souibgui et al. 2018). The latter origin was confirmed for some species of this phylum which are specific of the human intestinal tract (HF183 DNA target). The HF183 DNA target was found in the aquifer of AR sites with a shallow aquifer (but not for those with a thick vadose zone), suggesting a potential transfer from stormwater runoff to the aquifer. It is also to be noted that qPCR screenings for Bacteroides species appeared good indicators of the ability of a vadose zone at preventing bacterial contamination of the aquifer with surface bacteria. The total Bacteroides PCR screening could only detect this bacterial group among biofilm samples recovered from recharged wells of AR sites. In addition to the effects observed on Bacteroidetes, the diversities of most phyla in terms of OTU numbers showed positive linear relations with BDOC concentrations. The fact that dominant bacterial groups increased in diversity with nutrient (BDOC) availability in groundwater environment might have resulted from the harsh trophic conditions encountered by micro-organisms in subterranean ecosystems (Chapelle and Lovley 1990). Indeed, we measured relatively low BDOC concentrations (below 0.9 mg L−1) in groundwater samples. However, Li et al. (2013) showed that an increase in BDOC concentration from 0.5 to 1.1 mg L−1 was high enough to reduce 16S rRNA gene bacterial diversity of biofilms developing in sediment columns (under laboratory conditions). We can thus not exclude that a reduction of the diversity of some bacterial groups could occur due to BDOC enrichments of groundwater associated with AR practices if these enrichments lead to BDOC concentrations exceeding 1.1 mg L−1. Here, coupling measurements of DOC availability, biofilm variables, bacterial richness, and proportion of particular taxa among bacterial communities (e.g., all Bacteroides or particular species) appeared sufficient to infer the relative incidence of AR practices on aquifers. This approach based on functional indicators appears promising for water manager to evaluate groundwater ecosystem health. Moreover, the results of the present study clearly showed that the implementation of AR sites in porous aquifers would be recommended in areas with thick VZ (> 10 m) to prevent groundwater contamination.
References
Alonso C, Warnecke F, Amann R, Pernthaler J (2007) High local and global diversity of Flavobacteria in marine plankton. Environ Microbiol 9:1253–1266
Anderson MJ (2001) A new method for non-parametric multivariate analysis of variance. Austral Ecol 26:32–46
Baker MA, Valett HM, Dahm CN (2000) Organic carbon supply and metabolism in a shallow groundwater ecosystem. Ecology 81:3133–3148
Battin TJ (1997) Assessment of fluorescein diacetate hydrolysis as a measure of total esterase activity in natural stream sediment biofilms. Sci Total Environ 198:51–60
Bengtsson G (1989) Growth and metabolic flexibility in groundwater bacteria. Microb Ecol 18:235–248
Bernardin-Souibgui C, Barraud S, Bourgeois E, Aubin JB, Becouze-Lareure C, Wiest L, Marjolet L, Colinon C, Lipeme Kouyi G, Cournoyer B, Blaha D (2018) Incidence of hydrological, chemical, and physical constraints on bacterial pathogens, Nocardia cells, and fecal indicator bacteria trapped in an urban stormwater detention basin in Chassieu, France. Environ Sci Pollut Res 25:24860–24881
Bernstein HC, Brislawn C, Renslow RS, Dana K, Morton B, Lindemann SR, Song H-S, Atci E, Beyenal H, Fredrickson JK, Jansson JK, Morand JJ (2017) Trade-offs between microbiome diversity and productivity in a stratified microbial mat. ISME J 11:405–414
Bienhold C, Boetius A, Ramette A (2012) The energy–diversity relationship of complex bacterial communities in Arctic deep-sea sediments. ISME J 6:724–732
Cardinale BJ, Hillebrand H, Harpole WS, Gross K, Ptacnik R (2009) Separating the influence of resource "availability" from resource "imbalance" on productivity-diversity relationships. Ecol Lett 12:475–487
Chapelle FH (2001) Groundwater microbiology and geochemistry. Wiley Press, New York
Chapelle FH, Lovley DR (1990) rates of microbial metabolism in deep coastal plain aquifers. Appl Environ Microbiol 56:1865–1874
Cooney TJ, Simon KS (2009) Influence of dissolved organic matter and invertebrates on the function of microbial films in groundwater. Microb Ecol 58:599–610
Cottrell MT, Kirchman DL (2000) Community composition of marine bacterioplankton determined by 16S rRNA gene clone libraries and fluorescence in situ hybridization. Appl Environ Microbiol 66:5116–5122
Datry T, Malard F, Gibert J (2004) Dynamics of solutes and dissolved oxygen in shallow urban groundwater below a stormwater infiltration basin. Sci Total Environ 329:215–229
Datry T, Malard F, Gibert J (2005) Response of invertebrate assemblages to increased groundwater recharge rates in a phreatic aquifer. J North Am Benthol Soc 24:461–477
Datry T, Malard F, Gibert J (2006) Effect of artificial stormwater infiltration on urban groundwater ecosystems. In: Tellam JH, Rivett MO, Israfilov RG, Herringshaw LG (eds) Urban groundwater management and sustainability. Springer, Dordrecht, pp 331–345
Díez-Vives C, Gasol JM, Acinas SG (2014) Spatial and temporal variability among marine Bacteroidetes populations in the NW Mediterranean Sea. Syst Appl Microbiol 37:68–78
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200
Evans KL, Warren PH, Gaston KJ (2005) Species–energy relationships at the macroecological scale: a review of the mechanisms. Biol Rev 80:1–25
Fierer N, Bradford MA, Jackson RB (2007) Toward an ecological classification of soil bacteria. Ecology 88:1354–1364
Fontvieille DA, Outaguerouine A, Thevenot DR (1992) Fluorescein diacetate hydrolysis as a measure of microbial activity in aquatic systems: application to activated sludges. Environ Technol 13:531–540
Foulquier A, Malard F, Barraud S, Gibert J (2009) Thermal influence of urban groundwater recharge from stormwater infiltration basins. Hydrol Proced 23:1701–1713
Foulquier A, Malard F, Mermillod-Blondin F, Datry T, Simon L, Montuelle B, Gibert J (2010) Vertical change in dissolved organic carbon and oxygen at the water table region of an aquifer recharged with stormwater: biological uptake or mixing? Biogeochemistry 99:31–47
Foulquier A, Malard F, Mermillod-Blondin F, Montuelle B, Dolédec S, Volat B, Gibert J (2011) Surface water linkages regulate trophic interactions in a groundwater food web. Ecosystems 14:1339–1353
Fuhrman JA (2009) Microbial community structure and its functional implications. Nature 459:193–199
Gibert J, Danielopol DL, Stanford JA (1994) Groundwater ecology. Academic Press, San Diego
Goldscheider N, Hunkeler D, Rossi P (2006) Review: microbial biocenoses in pristine aquifers and an assessment of investigative methods. Hydrogeol J 14:926–941
Gómez-Pereira PR, Fuchs BM, Alonso C, Oliver MJ, Van Beusekom JE, Amann R (2010) Distinct flavobacterial communities in contrasting water masses of the North Atlantic Ocean. ISME J 4:472–487
Griebler C, Lueders T (2009) Microbial biodiversity in groundwater ecosystems. Freshw Biol 54:649–677
Hershey OS, Kallmeyer J, Wallace A, Barton MD, Barton HA (2018) High microbial diversity despite extremely low biomass in a deep karst aquifer. Front Microbiol 9 https://www.frontiersin.org/articles/10.3389/fmicb.2018.02823/full
Houri-Davignon J-C, Etcheber HC (1989) Measurement of actual electron transport system (ETS) activity in marine sediments by incubation with INT. Environ Technol 10:91–100
Hug LA, Thomas BC, Brown CT, Frischkorn KR, Williams KH, Tringe SG, Banfield JF (2015) Aquifer environment selects for microbial species cohorts in sediment and groundwater. ISME J 9:1846–1856
Hunt W, Traver R, Davis A, Emerson CH, Collins KA, Stagge JH (2010) Low impact development practices: designing to infiltrate in urban environments. In: Chang NB (Ed). Effects of urbanization on groundwater. American Society of Civil Engineers, 308–343
Huse SM, Welch DM, Morrison HG, Sogin ML (2010) Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ Microbiol 12:1889–1898
Kalbitz K, Solinger S, Park J-H, Michalzik B, Matzner E (2000) Controls on the dynamics of dissolved organic matter in soils: a review. Soil Sci 165:277–304
Kazumi J, Capone D (1994) Heterotrophic microbial activity in shallow aquifer sediments of Long Island, New York. Microb Ecol 28:19–37
Kolehmainen RE, Langwaldt JH, Puhakka JA (2007) Natural organic matter (NOM) removal and structural changes in the bacterial community during artificial groundwater recharge with humic lake water. Water Res 41:2715–2725
Landa M, Cottrell MT, Kirchman DL, Blain S, Obernosterer I (2013) Changes in bacterial diversity in response to dissolved organic matter supply in a continuous culture experiment. Aquat Microb Ecol 69:157–168
Layton A, McKay L, Williams D, Garrett V, Gentry R, Sayler G (2006) Development of Bacteroides 16S rRNA gene TaqMan-based real-time PCR assays for estimation of total, human, and bovine fecal pollution in water. Appl Environ Microbiol 72:4214–4224
Lever MA, Rogers KL, Lloyd KG, Overmann J, Schink B, Thauer RK, Hoehler TM, Jørgensen BB (2015) Life under extreme energy limitation: a synthesis of laboratory and field-based investigations. FEMS Microbiol Rev 39:688–728
Li D, Alidina M, Ouf M, Sharp JO, Saikaly P, Drewes JE (2013) Microbial community evolution during simulated managed aquifer recharge in response to different biodegradable dissolved organic carbon (BDOC) concentrations. Water Res 47:2421–2430
Marsalek J, Chocat B (2002) International report: Stormwater management. Water Sci Technol 46:1–17
Marschner B, Kalbitz K (2003) Controls of bioavailability and biodegradability of dissolved organic matter in soils. Geoderma 113:211–235
Marti R, Ribun S, Aubin J-B, Colinon C, Petit S, Marjolet L, Gourmelon M, Schmitt L, Breil P, Cottet M, Cournoyer B (2017) Human-driven microbiological contamination of benthic and hyporheic sediments of an intermittent peri-urban river assessed from MST and 16S rRNA genetic structure analyses. Front Microbiol 8:19
McKnight DM, Bencaia KE, Zeiiweger GW, Aiken GR, Feder GL, Thorn KA (1992) Sorption of dissolved organic carbon by hydrous aluminum and iron oxides occurring at the confluence of deer creek with the Snake River, Summit County, Colorado. Environ Sci Technol 26:1388–1396
Mermillod-Blondin F, Foulquier A, Maazouzi C, Navel S, Negrutiu Y, Vienney A, Simon L, Marmonier P (2013) Ecological assessment of groundwater trophic status by using artificial substrates to monitor biofilm growth and activity. Ecol Indic 25:230–238
Mermillod-Blondin F, Simon L, Maazouzi C, Foulquier A, Delolme C, Marmonier P (2015) Dynamics of dissolved organic carbon (DOC) through stormwater basins designed for groundwater recharge in urban area: assessment of retention efficiency. Water Res 81:27–37
Mermillod-Blondin F, Voisin J, Marjolet L, Marmonier P, Cournoyer B (2019) Clay beads as artificial trapping matrices for monitoring bacterial distribution among urban stormwater infiltration systems and their connected aquifers. Env Monitor Assess 191:58
Oksanen J, Kindt R, Legendre P, O'Hara BV (2007) The vegan package. Community ecology package, R package version 1
Pabich WJ, Valiela I, Hemond HF (2001) Relationship between DOC concentration and vadose zone thickness and depth below water table in groundwater of Cape Cod, USA. Biogeochemistry 55:247–268
Park J-W, Crowley DE (2006) Dynamic changes in nahAc gene copy numbers during degradation of naphthalene in PAH-contaminated soils. Appl Microbiol Biotechnol 72:1322–1329
Peterson GL (1977) A simplification of the protein assay method of Lowry et al. which is more generally applicable. Anal Biochem 83:346–356
Pinhassi J, Sala MM, Havskum H, Peters F, Guadayol O, Malits A, Marrasé C (2004) Changes in bacterioplankton composition under different phytoplankton regimens. Appl Environ Microbiol 70:6753–6766
Prosser JI, Bohannan BJ, Curtis TP, Ellis RJ, Firestone MK, Freckleton RP, Green JL, Green LE, Killham K, Lennon JJ, Osborn AM, Solan M, van der Gast CJ, Young JPW (2007) The role of ecological theory in microbial ecology. Nat Rev Microbiol 5:384–392
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596
R Development Core Team (2008) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Saidy AR, Smernik RJ, Baldock JA, Kaiser K, Sanderman J (2013) The sorption of organic carbon onto differing clay minerals in the presence and absence of hydrous iron oxide. Geoderma 209–210:15–21
Schauer R, Bienhold C, Ramette A, Harder J (2010) Bacterial diversity and biogeography in deep-sea surface sediments of the South Atlantic Ocean. ISME J 4:159–170
Schloss PD, Westcott SL (2011) Assessing and improving methods used in operational taxonomic unit-based approaches for 16S rRNA gene sequence analysis. Appl Environ Microbiol 77:3219–3226
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF (2009) Introducing mothur: open-source, platform-independent, community supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541
Schloss PD, Gevers D, Westcott SL (2011) Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS One 6:e27310
Servais P, Billen G, Hascoët MC (1987) Determination of the biodegradable fraction of dissolved organic matter in waters. Water Res 21:445–450
Servais P, Anzil A, Ventresque C (1989) Simple method for determination of biodegradable dissolved organic carbon in water. Appl Environ Microbiol 55:2732–2734
Seurinck S, Defoirdt T, Verstraete W, Siciliano SD (2005) Detection and quantification of the human-specific HF183 Bacteroides 16S rRNA genetic marker with real-time PCR for assessment of human faecal pollution in freshwater. Environ Microbiol 7:249–259
Shen Y, Chapelle FH, Strom EW, Benner R (2015) Origins and bioavailability of dissolved organic matter in groundwater. Biogeochemistry 122:61–78
Smith VH (2007) Microbial diversity-productivity relationships in aquatic ecosystems. FEMS Microbiol Ecol 62:181–186
Srivastava DS, Lawton JH (1998) Why more productive sites have more species: an experimental test of theory using tree-hole communities. Am Nat 152:510–529
Tada Y, Taniguchi A, Nagao I, Miki T, Uematsu M, Tsuda A, Hamasaki K (2011) Differing growth responses of major phylogenetic groups of marine bacteria to natural phytoplankton blooms in the Western North Pacific Ocean. Appl Environ Microbiol 77:4055–4065
Teeling H, Fuchs BM, Becher D, Klockow C, Gardebrecht A, Bennke CM, Kassabgy M, Huang S, Mann AJ, Waldmann J, Weber M, Klindworth A, Otto A, Lange J, Bernhardt J, Reinsch C, Hecker M, Peplies J, Bockelmann FD, Callies U, Gerdts G, Wichels A, Wiltshire KH, Glockner FO, Schweder T, Amann R (2012) Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science 336:608–611
Voisin J, Cournoyer B, Mermillod-Blondin F (2016) Assessment of artificial substrates for evaluating groundwater microbial quality. Ecol Indic 71:577–586
Voisin J, Cournoyer B, Vienney A, Mermillod-Blondin F (2018) Aquifer recharge with stormwater runoff in urban areas: influence of vadose zone thickness on nutrient and bacterial transfers from the surface of infiltration basins to groundwater. Sci Total Environ 637:1496–1507
Wang Q (2007) Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267
Williamson WM, Close ME, Leonard MM, Webber JB, Lin S (2012) Groundwater biofilm dynamics grown in situ along a nutrient gradient. Ground Water 50:690–703
Wright DH (1983) Species–energy theory: an extension of species-area theory. Oikos 41:496–506
Zhang X, Zhao X, Zhang M (2012) Functional diversity changes of microbial communities along a soil aquifer for reclaimed water recharge. FEMS Microbiol Ecol 80:9–18
Acknowledgments
The authors thank Léa Dantony, Mathilde Pigneret, and Felix Vallier for their support and advice during field and laboratory work and Pierre Marmonier for editing an earlier draft of this manuscript. This work was performed within the framework of the EUR H2O’Lyon (ANR-17-EURE-0018) of Université de Lyon (UdL), within the program “Investissements d'Avenir” operated by the French National Research Agency (ANR).
Funding
The Auvergne-Rhône-Alpes region for Jérémy Voisin’s Ph.D provided grant. This work was supported by l’Agence Nationale de la Recherche [ANR-16-CE32–0006 FROG], Lyon Metropole and Agence de l’Eau Rhône-Méditerranée-Corse within the framework of the experimental observatory for urban hydrology (OTHU, http://www.graie.org/othu) and Rhône Basin LTER (ZABR), and the French national research program for environmental and occupational health of Anses under the terms of project “Iouqmer” EST 2016/1/120.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Responsible editor: Diane Purchase
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Voisin, J., Cournoyer, B., Marjolet, L. et al. Ecological assessment of groundwater ecosystems disturbed by recharge systems using organic matter quality, biofilm characteristics, and bacterial diversity. Environ Sci Pollut Res 27, 3295–3308 (2020). https://doi.org/10.1007/s11356-019-06971-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-019-06971-5