Abstract
Riparian forests are critically endangered many anthropogenic pressures and natural hazards. The importance of riparian zones has been acknowledged by European Directives, involving multi-scale monitoring. The use of this very-high-resolution and hyperspatial imagery in a multi-temporal approach is an emerging topic. The trend is reinforced by the recent and rapid growth of the use of the unmanned aerial system (UAS), which has prompted the development of innovative methodology. Our study proposes a methodological framework to explore how a set of multi-temporal images acquired during a vegetative period can differentiate some of the deciduous riparian forest species and their health conditions. More specifically, the developed approach intends to identify, through a process of variable selection, which variables derived from UAS imagery and which scale of image analysis are the most relevant to our objectives.
The methodological framework is applied to two study sites to describe the riparian forest through two fundamental characteristics: the species composition and the health condition. These characteristics were selected not only because of their use as proxies for the riparian zone ecological integrity but also because of their use for river management.
The comparison of various scales of image analysis identified the smallest object-based image analysis (OBIA) objects (ca. 1 m2) as the most relevant scale. Variables derived from spectral information (bands ratios) were identified as the most appropriate, followed by variables related to the vertical structure of the forest. Classification results show good overall accuracies for the species composition of the riparian forest (five classes, 79.5 and 84.1 % for site 1 and site 2). The classification scenario regarding the health condition of the black alders of the site 1 performed the best (90.6 %).
The quality of the classification models developed with a UAS-based, cost-effective, and semi-automatic approach competes successfully with those developed using more expensive imagery, such as multi-spectral and hyperspectral airborne imagery. The high overall accuracy results obtained by the classification of the diseased alders open the door to applications dedicated to monitoring of the health conditions of riparian forest. Our methodological framework will allow UAS users to manage large imagery metric datasets derived from those dense time series.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Despite their relatively low area coverage, riparian forests are central landscape features that provide several ecosystem services: stream bank stabilization, reduction of fine sediment inputs and nutrient contamination, aquatic and terrestrial habitat improvement, and recreational and educational opportunities (Guo et al. 2011; Vidal-Abarca Gutiérrez and Suárez Alonso 2013; Clerici et al. 2014). Nevertheless, riparian forests are critically endangered in European countries by human pressures, such as livestock grazing, land use pressures, canalizations and wastewater, and also by natural hazards, such as the black alder (Alnus glutinosa) disease caused by Phytophthora alni (Cech 1998; Gibbs et al. 2003; De Merlier et al. 2005). The importance of riparian zones has been acknowledged by European directives (notably the Habitats Directive and the Water Framework Directive), involving multi-scale monitoring (local to network scales). Because of the typical linear and narrow shape of the riparian zones, field-based monitoring involves sampling, high labor costs, and time-consuming travels (Debruxelles et al. 2009; Myers 1989). The continuous improvement of the spatial resolution of remote sensing data combined with more powerful computer capacity and new geomatic procedures to extract information make the remote sensing approach more competitive (Alber and Piégay 2011; Carbonneau and Piégay 2012; Johansen et al. 2010; Michez et al. 2013; Roux et al. 2014). The use of this very-high-resolution (VHR) imagery in a multi-temporal approach is an emerging topic (Ardila et al. 2012; Champion 2012; Lasaponara et al. 2014). The trend is reinforced by the recent and rapid growth of the use of the unmanned aerial system (UAS), which has prompted the development of innovative methodology.
The use of the UAS in environmental science has become more common since the late 2000s. UAS imagery can be an effective alternative for cost-effectively describing riparian zones at the local scale (Dunford et al. 2009; Husson et al. 2014). In the ecological scope, the scientific community is very enthusiastic about the use of the UAS, proclaiming the “dawn of drone ecology” (Koh and Wich 2012) and that the UAS “will revolutionize spatial ecology” (Anderson and Gaston 2013). The two most important characteristics of UAS imagery are said to be the very high spatial and temporal resolution, allowing description of events occurring at a very local scale in a finite time window (e.g., the flowering of or the attack of a pathogen on a given tree species). Because of its very high spatial resolution (<0.1-m ground sampling distance (GSD)), UAS imagery is regularly characterized as hyperspatial imagery (Carbonneau and Piégay 2012; Greenberg et al. 2005; Laliberte et al. 2007; Strecha et al. 2012). Many studies have taken advantage of these two characteristics, for a broad range of environmental applications, such as landslide mapping (Lucieer et al. 2014), forest fire mapping (de Dios et al. 2011; Merino et al. 2012; Urbahs et al. 2013), precision farming (Bendig et al. 2013), wildlife census (Lisein et al. 2013a; Vermeulen et al. 2013), tree and forest characterization (Lisein et al. 2013b; Zarco-Tejada et al. 2014), forest biodiversity assessment (Getzin et al. 2012), and forest species composition (Dunford et al. 2009; Gini et al. 2014). Recent studies have demonstrated the potential of UAS imagery to finely describe the forest canopy (Dandois and Ellis 2010, 2013). When a good-quality digital terrain model is available, UAS-derived photogrammetric point clouds (>10 points/m2) can provide a canopy height model (CHM) with a quality comparable to light detection and ranging (LiDAR) CHM but with significant cost differences (Lisein et al. 2013b). References on classification of forest species by use of UAS imagery are still rare in the literature and include only the single-date approach (Dunford et al. 2009; Gini et al. 2014). However, useful pioneering studies are available on vegetation mapping projects based on UAS imagery (Knoth et al. 2013; Zaman et al. 2011).
Classification of forest species using remote-sensing data is relatively well documented. Successful classifications have been carried out using multi-spectral aerial/satellite imagery (Immitzer et al. 2012; Ke et al. 2010; Key et al. 2001; Korpela et al. 2011; Lucas et al. 2008; Sesnie et al. 2008) and hyperspectral data (Dalponte et al. 2014; 2012). The scale of analysis varies from tree crown pixel or object (object-based image analysis) to the crown itself. When available, 3D data (ALS LiDAR, SRTM DEM) can improve the classification accuracy (Dalponte et al. 2014; Ke et al. 2010; Sesnie et al. 2008). Forest species classification has also been studied through the use of multi-temporal remote sensing dataset which allows to highlight phenological differences between forest species (Key et al. 2001; Hill et al. 2010; Zhu and Liu 2014). These multi-temporal studies were intended to identify discriminant phenological stages (and related date) in order to schedule future imagery acquisition at lower cost within the identified time window. These studies identified the late growing season as the most relevant for these purposes. As UAS imagery allows acquisition of cost-effective dense time series of very-high-resolution imagery, new approaches can be considered.
Few studies have taken advantage of the temporal resolution of UAS imagery and the subsequent possibility of redundancy. Interesting case studies can be found, but they are outside the proper ecological scope. In precision farming, Bendig et al. (2013) computed UAS-derived digital surface model time series to measure the growth rate of experimental field crops while Torres-Sánchez et al. (2014) used UAS orthophotos to differentiate weeds in wheat fields in the early season. Multi-temporal data from UAS imagery have also been used to evaluate landslide and rockslide evolution (Lucieer et al. 2014; Ragg and Fey 2013). Most of these previous contributions used the temporal resolution of UAS imagery to fit the acquisition period to the occurrence of the events to be mapped, because UAS allow the final user to perform (at least to really get involved in) the survey. Subsequently, a methodological gap remains to be filled in terms of approaches that utilize dense (UAS) time series for a unified and multi-temporal classification purpose.
We propose a methodological framework to explore multi-temporal hyperspatial UAS imagery acquired during a single vegetative period in order to characterize riparian forest species and health condition. More specifically, we intend to identify which type of variables derived from hyperspatial imagery and which scale of image analysis are the most relevant to reach our objectives. Another major ambition is to highlight the best time window and the configuration of off-the-shelf camera.
The methodological framework is applied to describe the riparian forest through two fundamental characteristics: the species composition and the health condition. These characteristics were selected because of their applicability as proxies for ecological integrity of the riparian zones (Innis et al. 2000; Naiman et al. 2005) and for their usefulness in targeting management strategies (Debruxelles et al. 2009).
Methods
Study sites
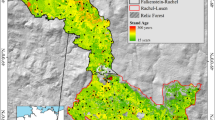
Both study sites are riparian forests located in two different landscapes in Wallonia, southern Belgium (Fig. 1) in the Ardennes eco-region. To ensure the riparian character of the studied forest, forest patches located along the river bank (<20 m from the wetted channel) were selected.
Riparian forests of site 1 (Vielsalm, Salm River) are located in an agricultural landscape (pastures). They are highly pressured by farm activities and remain mostly in a managed forest strip along the riverbanks (<10 m). The population of black alders in site 1 is known to be infested by P. alni (Di Prinzio et al. 2013a). Riparian forests of site 2 (Felenne—Houille River) are located in a relatively undisturbed forest landscape. These strips are mainly unmanaged and have a regular presence of standing dead woods.
In terms of species composition, riparian forests of both sites are dominated by A. glutinosa with non-native coniferous stands (Picea abies and Larix decidua) in site 2. A. glutinosa is the typical overstory tree species in the riparian zones of Wallonia (Claessens et al. 2010; Debruxelles et al. 2009).
Acquisition of UAS imagery
Our workflow is based on a time series of 25 UAS flights (Table 1) allowing production of 18 and 7 orthophotos in site 1 and site 2, respectively, using a Gatewing X100 (wingspan 100 cm, weight 2 kg, cruising speed 80 km/h, flight height 100 to 750 m). Two pre-calibrated Ricoh GR3 cameras (10-megapixel sensor, focal length 6 mm) were used. One camera was adapted for near-infrared acquisition (Fig. 2). The UAS can only carry one camera per flight. The flight duration never exceeded 45 min, and the overlap between images was set to 75 %.
Idealized filter profiles (lines) and channel sensitivity (surface) for the RGB (top) camera and the RGNIR modified camera (bottom). The RGNIR camera was modified by removing the internal hot-mirror filter and adding a blue-light-blocking filter (i.e., a yellow long-pass filter). Adapted from Nijland et al. (2014)
Field staff performed the maximum number of flight surveys during approximately 2 h around solar noon. The number of flights per day was directly influenced by the meteorological conditions and logistical constraints (i.e., period of air space restrictions, UAS equipment failures, etc.). These circumstances resulted in a varying number of flights per day (Table 1). The weather conditions induced varying lighting conditions within and between the orthophotos. Although orthophotos which are the closest to solar noon should theoretically present the best quality to reach our objectives, the varying lighting conditions seriously complicated the selection of a single orthophoto per flight date. With the willingness to build a computer-based approach, all available orthophotos were therefore used to avoid an operator-mediated selection.
Because one of the main objectives of the study was the characterization of riparian forest health condition, the aerial data survey was planned for the late growing season, which is the best time window (Gibbs et al. 2003) for catching the leaf symptoms of the most common riparian forest pathogen in Wallonia: P. alni on the black alder (Di Prinzio et al. 2013b; Pintos Varela et al. 2010). This time window also matches well with the time windows found in other multi-temporal remote sensing studies for differentiating forest species (Key et al. 2001; Hill et al. 2010; Zhu and Liu 2014).
General workflow
We propose an innovative and replicable multi-platform workflow to handle large and complex datasets induced by the spatial and temporal resolution of the UAS time series in a computer-based perspective to accurately map riparian forest species and health conditions (see summary in Fig. 3).
Photogrammetric process
General photogrammetric workflow
Agisoft Photoscan 1.0 Professional, which is now relatively common in the UAS community, was used to perform photogrammetric surveys with proven efficiency (Dandois and Ellis 2013; 2010; Sona et al. 2014). Every flight dataset was processed by following the workflow described in Fig. 4.
Our photogrammetric workflow was based on the work of Lisein et al. (2013b) in the Photoscan environment. On the basis of the information of the inertial measurement unit (IMU), the GPS positions recorded by the UAS, and a set of ground control points (GCPs), an initial aerotriangulation, is performed (number 1 in Fig. 4). The georeferencing process is based on a set of particular points (road crossings, roofs, pedestrian crossings, etc.) that are used as GCPs in Photoscan. For site 1, the GCPs have been measured with a real-time kinematic (RTK) GPS (Leica 200+). For site 2 (low mobile network coverage), the GCP planimetric coordinates were defined on the regional orthophoto coverage (0.25-m GSD), and the altimetric positions were extracted from LiDAR digital terrain model (DTM). Bare ground areas of the low-resolution 3D model (number 2 in Fig. 4) are aligned on a LiDAR DTM within CloudCompare (http://www.danielgm.net/cc/), by use of the iterative closest-point matching strategy. The matching process results in a global transformation matrix (number 3 in Fig. 4) of six parameters (translation and rotation), which is used to perform a rigid transformation of the GCP coordinates. The registered GCP positions are used to finely georeference the initial aerotriangulation (number 4 in Fig. 4) and compute a high-resolution dense matching (number 5 in Fig. 4).
Orthophoto generation
The photogrammetric workflow (Fig. 4) allows generation of orthophotos for every flight dataset. The orthophotos were resampled to 0.1-m GSD. The orthophoto generation process used the “mosaic” blending mode with the color correction option enabled in Photoscan 1.0. Table 1 presents the list of the orthophotos, with the reconstruction error provided by Photoscan.
Canopy height model computation
For each site, we computed a single CHM (0.1-m GSD) by the combination of a LiDAR DTM and a photogrammetric digital surface model (DSM) (DSM—DTM). The two “hybrid” CHMs have been used as the sole 3D reference for the segmentation/classification process, under the condition that the surface remained unchanged during the time window of the surveys (<6 months).
In each site, a reference flight date was selected to compute the two site reference DSM (see Table 1). The flights were selected manually on the basis of the visual quality of the individual raw images (ground visibility, luminosity, and sharpness). For each site, a new photogrammetric project (Table 2) was conducted with all images of the selected date, by following the same photogrammetric workflow (Fig. 4).
Reference dataset
A georeferenced database of 1186 deciduous trees (434 in site 1, 752 in site 2) was compiled. The position of the stem was measured with a field-map system (http://www.fieldmap.cz), initially positioned using a DGPS (GNSS Leica GPS1200+) on site 1 and with a post-processed SXBlue II GPS (http://sxbluegps.com) on site 2.
All overstory trees were mapped and identified (species level) within the two study sites. Because site 1 has been previously identified as infested by P. alni, the health condition of the black alders was estimated by a skilled operator according to two classes: asymptomatic tree and tree with P. alni foliar symptoms (defoliation and/or foliage yellowing). The field survey was conducted in August 2012 in site 1, at the time the symptoms of the P. alni disease were the most distinctive (Gibbs et al. 2003; Di Prinzio et al. 2013b). In site 2, the field survey was conducted in October 2012 and was only dedicated to identifying species.
Crowns of georeferenced trees were delineated by the use of the generated orthophotos and the two site-reference CHMs by the operator who performed the field survey. Respectively, A. glutinosa represent 67 and 48 % of the trees in sites 1 and 2 (see details in Table 3).
Multi-resolution image segmentation
The segmentation was performed by the use of eCognition developer, with a multi-resolution image segmentation algorithm (Blaschke 2010). This region-growing segmentation algorithm starts from individual pixels that are merged to most-similar adjacent regions. The size and the shape of the resulting objects are defined by a scale parameter (related to the object size), shape, and compactness criterion (object homogeneity). To define which object size is the most relevant, we tested six different scale parameters: 5, 10, 15, 30, 45, and 60 (Fig. 5). The mean size of the different objects of homogeneity criteria (shape and compactness) was equally balanced and set to 0.5.
The segmentation process is performed on selected layers to which a pre-defined weight has been assigned. To identify an object that can be considered as constant during the time window of the multi-temporal dataset, the segmentation process used in this study was based on the two canopy height models.
We extracted from each of the image objects a set of metrics (Table 4) at eight scales of analysis. In addition to the six OBIA scales, we tested one pixel-based scale (4 pixels square objects) and the manually delineated tree crown scale. To exploit the whole time series in a single automated classification process on the basis of a random forest machine-learning algorithm, various spectral, texture, and vertical structure metrics were derived from the UAS imagery.
We choose five gray level co-occurrence matrix (GLCM) derivatives based on the work of Haralick et al. (1973) and computed with eCognition: entropy, standard deviation, correlation, mean, and contrast (see Table 4). Those derivatives were chosen, most notably, because of their proven efficiency in similar approaches (Laliberte and Rango 2009; Stumpf and Kerle 2011). Those GLCM derivatives are typically computed through symmetric matrices for pixels neighboring directly at 0°, 45°, 90°, or 135°. In our case, we select an omnidirectional approach, in which the sums of all four directions of GLCM derivatives are calculated before texture calculation.
We test several simple metrics (see Table 4 for details), derived from the spectral layers’ mean itself and from different band ratios. Our approach was based on normalized band ratios (such as NDVI or GNDVI) and individual band ratios, such as the green ratio vegetation index.
On the basis of the two canopy height models, we compute three metrics (mean, SD, and skewness) that describe the vertical structure of a riparian forest, considered as constant during the time series (see Table 4).
Classification process
Generation of sample database
Objects with at least 75 % of relative overlay with delineated tree crowns were selected to compile the tree crowns’ object database (Table 5). This threshold is relatively restrictive regarding to other comparable studies (60 % for Ma et al. (2015) and even 50 % Stumpf and Kerle (2011)). This choice was justified because of the relatively high similarity between classes compared to these two studies (land cover classes for Ma et al. (2015) and “landslide” vs “other objects” for Stumpf and Kerle (2011)).
We test two classification scenarios for the riparian forest species and health condition (Table 6). For each of the sites, we established a classification with five classes. The first four classes group the spatial units of each of the four most common species from the field survey, whereas the fifth class (“other riparian species”) regroups the spatial units corresponding to the other species of the field survey. The purpose of this selection was to ensure a sufficient number of objects at every scale of image analysis. Because the black alders of the riparian forest of site 1 were known to be highly contaminated by P. alni (De Merlier et al. 2005; Di Prinzio et al. 2013b),. a focus was placed on the distinction between black alders with and without foliage symptoms.
Selection of variables by the use of random forest
Random forests’ machine-learning algorithms have proved their ability to handle very complex and high-dimensional remote-sensing datasets (Cutler et al. 2007; Lawrence et al. 2006; Stumpf and Kerle 2011; Watts et al. 2009). Most of the previously cited studies used the initial implementation of the random forest algorithm (Breiman 2001) in the R software. Moreover, different studies have provided some improvement to this first package, notably to use the random forest algorithm for variable selection (Diaz-Uriarte 2007; Genuer et al. 2010). To identify which variable is the most relevant to differentiate species or tree health conditions, we use an implementation of the random forest algorithm by Genuer et al. (2010) and implemented within the R package “VSURF” ( http://cran.r-project.org/ ). This process of variable selection was developed to handle high-dimensional data (number of variables exceeding number of observations) by following three steps (Genuer et al. 2010). The first step (“thresholding step”) runs a set of classic random forest processes (ntree parameter 2000) and eliminates irrelevant variables from the dataset through the study of variable importance (Gini index), computed during the random forest processes (see Breiman (2001) for details). The second step (“interpretation step”) selects the best model through successive calls of random forest processes, starting with the random forest build utilizing only the most important variable and ending with all variables. The last step (“prediction step”) refines the selected variables by eliminating redundancy in the set of previously selected variables. The first model is based only on the most important variable, and other variables are added to the model in a stepwise manner. To remain in the final model, a variable must have a bigger impact on the model accuracy than a noise variable (variables left out by interpretation). This last concern is tuned into VSURF through the “number of mean jump (nmj)” parameter, which was set to five with a trial and error approach (default value 1).
Classification and accuracy assessment
Once the selected variables have been identified, the original sample database is randomly resampled (50 %) to create training and evaluation sets. The training set is used to run a new RF process (ntree parameter 2000). The model is then applied to the evaluation set for the accuracy assessment. The entire classification and accuracy process has been repeated 50 times to compute mean ± SD values of accuracies
Results
Riparian forest species composition
The classification process was run on the entire sample database, classifying riparian tree crown objects into five classes (Table 6). The accuracy of the classification varies with the scale of analysis (Table 7), suggesting that the overall accuracy is improving while the size of the analyzed objects is decreasing. Pixel-based or close to pixel OBIA objects (scale 5) perform the best (approximately 80 % of overall accuracies).
The study of the selected variables (Table 7) at various scales reveals that the data derived from flights in the late growing season (site 1) or in partially leaf-off conditions (site 2) are the most appropriate. Phenological differences are enhanced during the late season, especially in the riparian forest foliage. In very late survey dates, various states of phenology can be found from fully photosynthetically active foliage in coniferous stands to completely “leaf-off” deciduous riparian trees.
Compared to selected variables of the site 1, variables derived from the modified RGNIR camera are more frequent in selected variables of site 2 (Table 7). The variables derived from the unmodified RGB camera (regardless the scale of analysis) are far more used on site 1 (Fig. 6).
Results presented in Fig. 7 show that most of the selected variables are derived from the spectral information. The results also highlight the advantage of adding additional information to the classical spectral information from 3D layers or texture information. The 3D information, from derivatives of the CHM, is used for the finest scales of analysis, whereas the texture information, from GLCM derivatives, is used at coarser scale of analysis.
Scale of analysis and variable type frequency in selected variables for the species composition classification approach (site 1). Height: variables derived from a canopy height model. Texture: GLCM derivative variable. Spectral: variables derived from the spectral layers. Spectral variables are the most used. Texture variables are used at coarse scales, whereas 3D variables are used at fine scales
The study of the confusion matrices for the best performing classification models (species composition approach) for sites 1 and 2 (Tables 8 and 9) reveal contrasted individual classification errors.
Classification of alder health conditions
The variables used to classify the health condition of the black alders differ from those used for the species composition classification in two important ways. The health condition classification presents better overall accuracy value (>90 %) and requires fewer variables to construct the optimal classification models (12 vs 9 for site 1) (Table 10).
The classification of the health condition of A. glutinosa in site 1 is based more on variables derived from NIR imagery, which allow healthy alders to be distinguished from those that presented defoliation symptoms during the field survey.
Discussion
Riparian forest species and health condition
Overall accuracies of our tree species classifications reached 79.5 % (site 1) and 84.1 % (site 2), which are comparable with those raised by recent researchers who used much more expensive hyperspectral imagery (79 %, Dalponte et al. 2014) or multi-spectral imagery (83 %, Waser et al. 2011). High overall accuracy results were obtained for the classification of the health condition of the typical overstory tree species of Walloon riparian forest (A. glutinosa, 90.6 %).
The study of individual classification errors (see Table 8 and Table 9) for the best performing species classification models can be analyzed through the phenological traits of the tree species. In site 2 (Table 9), Acer pseudoplatanus and A. glutinosa present similar and relatively high classification error. These higher classification errors can be explained by high intraspecific phenological variations for Acer species (reported by Hill et al. 2010) and by variation in the health condition of A. glutinosa (recorded presence of P. alni). P. abies or Quercus species present very low classification errors in site 2. The phenological characteristics of P. abies are quite obvious because this species forms distinct even-aged and evergreen stands along site 2. In site 1, the highest classification error is related to the class of Fraxinus excelsior and can be explained by the presence of the highly pathogenic Chalaria fraxinea (recorded during field survey).
Operational recommendations
Time window
The most appropriate time windows to perform the flight surveys were found to be the late season of vegetation, when phenological differences of riparian forest trees are the most enhanced. These results are in line with previous studies such as Key et al. (2001) and Hill et al. (2010), who used aerial multi-spectral imagery, and Zhu and Liu (2014), who used satellite imagery.
Number of surveys
Respectively, 6 and 5 days of surveys were required to achieve the best classification results for sites 1 and 2, whereas 6 days of flight surveys were required to assess the health condition of black alders of site 1. These results highlight the importance of the multi-temporal character of the dataset. However, the health condition of black alders in site 1 can be evaluated with a sufficient accuracy (81 %) using two variables derived from a single flight survey with the modified RGNIR camera.
Scale of analysis
The relation between the scale of the analysis and the accuracy of the classification shows evidence of higher global accuracies for the classification scenarios performed with smaller objects (from segmented objects <1 m2 to 4 pixel objects, 0.04 m2). The trend is more obvious for the “species composition” scenario.
This result highlights the importance of the tree crown object segmentation process. The closer the size of the segmented object is to the tree crown scale, the more accurate the segmentation process must be to avoid objects overlapping several individual trees. The oversegmentation induced by the reduction of the OBIA scale parameters limits this overlapping. However, tree crown delineation is a research topic in itself, and the multi-resolution segmentation process was, in our approach, intentionally simple, without scale-related fine-tuning. Fine-tuning of the segmentation process in relation to the scale of image analysis would especially improve the performance of coarser scales of image analysis but would turn the approach into a more time-consuming and less computer-mediated perspective.
Besides the quality of segmented objects, larger objects present higher spectral heterogeneity. This fact combined to a lower number of training objects (due to the fixed sampling ratio) induces the decrease of the overall accuracy as the size of the objects increases. These findings are in line with recent publication of Ma et al. (2015) who also used UAS imagery. The lower overall accuracy values of the manually delineated tree crown scale (despite a lower mean area value than the OBIA 60 scale objects) can be further explained by the fact that a major part of delineated individuals in the reference dataset is actually clump of trees. In this case, a single individual polygon can regroup up to ten crowns, with shadow and blurry effect between and around them, inducing even higher spatial heterogeneity in these objects (compared to larger “OBIA 60” objects).
Relevant type of variable
The analysis of the selected variables also confirms the value of the use of 3D information (derived from a canopy height model) at fine scale and texture information at a coarser scale. The selection of the “mean height” at fine scale can be explained by height differences between “healthy” and “unhealthy” riparian trees. At the local scale, the dense matching process can fail to reconstruct the canopy of trees with extreme defoliation symptom, leading to an underestimation of the height. The low efficiency of texture metrics (and thus the high performance of spectral metrics) can also be explained by the orthophoto generation process. Because of the very high complexity of the surface of the canopy, tree crowns on orthophotos can present a blurry aspect. Considering that the best results were achieved at finer scale (<1-m2 objects), we do not recommend the use of texture metrics, which are time-consuming and were unused by classifiers at best performing scales.
Use of modified RGNIR off-the-shelf camera
The site 1 being less contrasted in terms of forest type and presenting the “densest” time series, it can be used as a model for comparing the performance of the modified RGNIR camera. The results (see Fig. 6) highlight the higher performance of the RGB camera, which can be explained by the spectral overlap between the spectral bands of the modified RGNIR camera (see Fig. 2).
Compared to site 1, the more intense use of RGNIR imagery in site 2 can be explained by the forest species composition. As the appropriate time windows to perform the flight surveys were found to be the late season of vegetation, the simultaneous presence in site 2 of photosynthetically active (evergreen coniferous stands) and non-active species (riparian leaf-off deciduous trees) induces higher contrast in NIR. This fact can also explain the selection of the blue band capture in the late season of vegetation in site 2, because the soil (and subsequently leaf-off trees) has a specific response within the blue wavelength.
Conclusions
The quality of the classification models developed with a UAS-based, cost-effective, and semi-automatic approach competes successfully with those developed using more expensive imagery, such as multi-spectral and hyperspectral airborne imagery. Our study uses the temporal resolution of the UAS to compile a redundant dataset of UAS imagery to highlight riparian forest phenological differences. This innovative approach is the first forest case study that takes advantage of both spatial and temporal resolution of UAS imagery to successfully describe subtle features, such as individual tree species or health condition.
The accuracy of the classification of the diseased alders opens the door to applications dedicated to monitoring of the health conditions of riparian forests. Our approach is directly applicable in other context, notably to characterize other emerging dieback diseases that also induce defoliation symptoms (e.g., C. fraxinea on F. excelsior in Europe).
Regarding the forest characterization, most of the previous research on time series imagery aimed to identify a specific time window when the classification would be the most reliable. The final operational application was subsequently to schedule imagery acquisition at a lower cost within the identified time window. With very low operational costs induced by UAS imagery, applications can integrate dense time series imagery on an operational basis. Our methodological framework will provide UAS users with a solution to manage large imagery metric datasets derived from those dense time series. In temperate climate areas, our results confirm the relevance of imagery acquired in the late season of vegetation. Our results also highlight the importance of time series to characterize riparian forest species while the health condition of A.glutinosa can be assessed through a single flight survey with satisfactory accuracy.
Future work should focus on the use of denser multi-temporal datasets, covering several entire vegetation seasons linked with the climatic conditions. This effort will limit the influence of early phenological disturbances and allow identification of more general phenological traits to develop better classification models.
References
Alber, A., & Piégay, H. (2011). Spatial disaggregation and aggregation procedures for characterizing fluvial features at the network-scale: application to the Rhône basin (France). Geomorphology, 125, 343–360. doi:10.1016/j.geomorph.2010.09.009.
Anderson, K., & Gaston, K. J. (2013). Lightweight unmanned aerial vehicles will revolutionize spatial ecology. Frontiers in Ecology and the Environment, 11, 138–146. doi:10.1890/120150.
Ardila, J. P., Bijker, W., Tolpekin, V. A., & Stein, A. (2012). Multi-temporal change detection of urban trees using localized region-based active contours in VHR images. Remote Sensing of Environment, 124, 413–426. doi:10.1016/j.rse.2012.05.027.
Bendig, J., Bolten, A., & Bareth, G. (2013). UAV-based imaging for multi-temporal, very high resolution crop surface models to monitor crop growth variability. Monitoring des Pflanzenwachstums mit Hilfe multi-temporaler und hoch auflösender Oberflächenmodelle von Getreidebeständen auf Basis von Bildern aus UAV-Befliegungen. Photogrammetrie Fernerkundung Geoinformation, 2013, 551–562. doi:10.1127/1432-8364/2013/0200.
Birth, G. S., & McVey, G. R. (1968). Measuring the color of growing turf with a reflectance spectrophotometer. Agronomy Journal, 60(6), 640–643.
Blaschke, T. (2010). Object based image analysis for remote sensing. ISPRS Journal of Photogrammetry and Remote Sensing, 65, 2–16. doi:10.1016/j.isprsjprs.2009.06.004.
Breiman, L. (2001). Random forests. Machine Learning, 45, 5–32.
Carbonneau, P., & Piégay, H. (2012). Fluvial remote sensing for science and management. Chichester: Wiley.
Cech, T. L. (1998). Phytophthora decline of alder (Alnus spp.) in Europe. Journal of Arboriculture, 24, 339–342.
Champion, N. (2012). Automatic cloud detection from multi-temporal satellite images: towards the use of Pléiades time series. ISPRS - The International Archives of the Photogrammetry, Remote Sensing and Spatial Information Sciences., XXXIX-B3, 559–564. doi:10.5194/isprsarchives-XXXIX-B3-559-2012.
Claessens, H., Oosterbaan, A., Savill, P., & Rondeux, J. (2010). A review of the characteristics of black alder (Alnus glutinosa (L.) Gaertn.) and their implications for silvicultural practices. Forestry, cpp038
Clerici, N., Paracchini, M.L., Maes, J. (2014). Land-cover change dynamics and insights into ecosystem services in European stream riparian zones. Ecohydrology & Hydrobiology, 14(2), 107–120.
Cutler, D. R., Edwards, T. C., Beard, K. H., Cutler, A., Hess, K. T., Gibson, J., & Lawler, J. J. (2007). Random forests for classification in ecology. Ecology, 88, 2783–2792. doi:10.1890/07-0539.1.
Dalponte, M., Bruzzone, L., & Gianelle, D. (2012). Tree species classification in the Southern Alps based on the fusion of very high geometrical resolution multispectral/hyperspectral images and LiDAR data. Remote Sensing of Environment, 123, 258–270. doi:10.1016/j.rse.2012.03.013.
Dalponte, M., Ørka, H. O., Ene, L. T., Gobakken, T., & Næsset, E. (2014). Tree crown delineation and tree species classification in boreal forests using hyperspectral and ALS data. Remote Sensing of Environment, 140, 306–317. doi:10.1016/j.rse.2013.09.006.
Dandois, J. P., & Ellis, E. C. (2010). Remote sensing of vegetation structure using computer vision. Remote Sensing, 2, 1157–1176. doi:10.3390/rs2041157.
Dandois, J. P., & Ellis, E. C. (2013). High spatial resolution three-dimensional mapping of vegetation spectral dynamics using computer vision. Remote Sensing of Environment, 136, 259–276. doi:10.1016/j.rse.2013.04.005.
De Dios, J. R. M., Merino, L., Caballero, F., & Ollero, A. (2011). Automatic forest-fire measuring using ground stations and unmanned aerial systems. Sensors, 11, 6328–6353. doi:10.3390/s110606328.
De Merlier, D., Chandelier, A., Debruxelles, N., Noldus, M., Laurent, F., Dufays, E., Claessens, H., & Cavelier, M. (2005). Characterization of alder Phytophthora isolates from Wallonia and development of SCAR primers for their specific detection. Journal of Phytopathology, 153, 99–107. doi:10.1111/j.1439-0434.2005.00936.x.
Debruxelles, N., Claessens, H., Lejeune, P., & Rondeux, J. (2009). Design of a watercourse and riparian strip monitoring system for environmental management. Environmental Monitoring and Assessment, 156, 435–450.
Di Prinzio, J., Chandelier, A., Henrotay, F., Claessens, H. (2013a). La maladie de l’aulne en Wallonie: évolution depuis son émergence. Forêt Wallonne, (124).
Di Prinzio, J., Henrotay, F., Claessens, H. (2013b). Le point sur l’évolution de la maladie de l’aulne en Région wallonne. Silva Belgica, 120(3).
Diaz-Uriarte, R. (2007). GeneSrF and varSelRF: a web-based tool and R package for gene selection and classification using random forest. BMC Bioinformatics, 8, 328. doi:10.1186/1471-2105-8-328.
Dunford, R., Michel, K., Gagnage, M., Piégay, H., & Trémelo, M.-L. (2009). Potential and constraints of Unmanned Aerial Vehicle technology for the characterization of Mediterranean riparian forest. International Journal of Remote Sensing, 30, 4915–4935. doi:10.1080/01431160903023025.
Genuer, R., Poggi, J.-M., & Tuleau-Malot, C. (2010). Variable selection using random forests. Pattern Recognition Letters, 31, 2225–2236.
Getzin, S., Wiegand, K., & Schöning, I. (2012). Assessing biodiversity in forests using very high-resolution images and unmanned aerial vehicles. Methods in Ecology and Evolution, 3, 397–404. doi:10.1111/j.2041-210X.2011.00158.x.
Gibbs, J., van Dijk, C., Webber, J. (2003). Phytophthora disease of alder in Europe, Forestry Commission. Edinburgh, UK: Forestry Commission: Forestry Commission Bulletin 126
Gini, R., Passoni, D., Pinto, L., & Sona, G. (2014). Use of unmanned aerial systems for multispectral survey and tree classification: a test in a park area of northern Italy. European Journal of Remote Sensing, 47, 251–269. doi:10.5721/EuJRS20144716.
Greenberg, J. A., Dobrowski, S. Z., & Ustin, S. L. (2005). Shadow allometry: estimating tree structural parameters using hyperspatial image analysis. Remote Sensing of Environment, 97, 15–25.
Guo, E., Sun, R., & Chen, L. (2011). Main ecological service functions in riparian vegetation buffer zone: research progress and prospects. Chinese Journal of Ecology, 30, 1830–1837.
Haralick, R. M., Shanmugam, K., & Dinstein, I. (1973). Textural features for image classification. IEEE Transactions on Systems, Man, and Cybernetics, SMC-3, 610–621. doi:10.1109/TSMC.1973.4309314.
Hill, R. A., Wilson, A. k., George, M., & Hinsley, S. a. (2010). Mapping tree species in temperate deciduous woodland using time-series multi-spectral data. Applied Vegetation Science, 13, 86–99. doi:10.1111/j.1654-109X.2009.01053.x.
Husson, E., Lindgren, F., Ecke, F. (2014). Assessing biomass and metal contents in riparian vegetation along a pollution gradient using an unmanned aircraft system. Water Air and Soil Pollution 225(6), 1–14. doi:10.1007/s11270-014-1957-2
Immitzer, M., Atzberger, C., & Koukal, T. (2012). Tree species classification with random forest using very high spatial resolution 8-band worldView-2 satellite data. Remote Sensing, 4, 2661–2693. doi:10.3390/rs4092661.
Innis, S. A., Naiman, R. J., & Elliott, S. R. (2000). Indicators and assessment methods for measuring the ecological integrity of semi-aquatic terrestrial environments. Hydrobiologia, 422–423, 111–131. doi:10.1023/A:1017033226325.
Jensen, J. R. (2006). Remote sensing of the environment: an earth resource perspective (2nd ed.). Upper Saddle River: Prentice Hall. http://www.pearsonhighered.com/educator/product/Remote-Sensing-of-the-Environment-An-Earth-Resource-Perspective/9780131889507.page
Johansen, K., Arroyo, L. A., Armston, J., Phinn, S., & Witte, C. (2010). Mapping riparian condition indicators in a sub-tropical savanna environment from discrete return LiDAR data using object-based image analysis. Ecological Indicators, 10, 796–807. doi:10.1016/j.ecolind.2010.01.001.
Ke, Y., Quackenbush, L. J., & Im, J. (2010). Synergistic use of QuickBird multispectral imagery and LIDAR data for object-based forest species classification. Remote Sensing of Environment, 114, 1141–1154. doi:10.1016/j.rse.2010.01.002.
Key, T., Warner, T. A., McGraw, J. B., & Fajvan, M. A. (2001). A Comparison of multispectral and multi-temporal information in high spatial resolution imagery for classification of individual tree species in a temperate hardwood forest. Remote Sensing of Environment, 75, 100–112. doi:10.1016/S0034-4257(00)00159-0.
Knoth, C., Klein, B., Prinz, T., & Kleinebecker, T. (2013). Unmanned aerial vehicles as innovative remote sensing platforms for high-resolution infrared imagery to support restoration monitoring in cut-over bogs. Applied Vegetation Science. doi:10.1111/avsc.12024. n/a–n/a.
Koh, L. P., & Wich, S. A. (2012). Dawn of drone ecology: low-cost autonomous aerial vehicles for conservation. Tropical Conservation Science, 5, 121–132.
Korpela, I., Heikkinen, V., Honkavaara, E., Rohrbach, F., & Tokola, T. (2011). Variation and directional anisotropy of reflectance at the crown scale—implications for tree species classification in digital aerial images. Remote Sensing of Environment, 115, 2062–2074. doi:10.1016/j.rse.2011.04.008.
Laliberte, A. S., & Rango, A. (2009). Texture and scale in object-based analysis of subdecimeter resolution unmanned aerial vehicle (UAV) IMAGERY. IEEE Transactions on Geoscience and Remote Sensing, 47, 761–770. doi:10.1109/TGRS.2008.2009355.
Laliberte, A.S., Winters, C., Rango, A. (2007). Acquisition, orthorectification, and classification of hyperspatial UAV imagery. In: Fourth Annual Symposium: Research Insights in Semiarid Scosystems, RISE, University of Arizona, Tucson. http://www.tucson.ars.ag.gov/rise/2007/Posters/19SlaughterPoster.pdf. Acessed 30 Nov 2015.
Lasaponara, R., Masini, N., Sabia, C. (2014). Multi-temporal analysis of Pleiades data for study of archaeological crop marks. In EGU General Assembly Conference Abstracts (vol. 16, p. 7534).
Lawrence, R. L., Wood, S. D., & Sheley, R. L. (2006). Mapping invasive plants using hyperspectral imagery and Breiman Cutler classifications (randomForest). Remote Sensing of Environment, 100, 356–362. doi:10.1016/j.rse.2005.10.014.
Lisein, J., Linchant, J., Lejeune, P., Bouché, P., & Vermeulen, C. (2013a). Aerial surveys using an unmanned aerial system (UAS): comparison of different methods for estimating the surface area of sampling strips. Tropical Conservation Science, 6, 506–520.
Lisein, J., Pierrot-Deseilligny, M., Bonnet, S., & Lejeune, P. (2013b). A photogrammetric workflow for the creation of a forest canopy height model from small unmanned aerial system imagery. Forests, 4, 922–944. doi:10.3390/f4040922.
Lucas, R., Bunting, P., Paterson, M., & Chisholm, L. (2008). Classification of Australian forest communities using aerial photography, CASI and HyMap data. Remote Sensing of Environment, 112, 2088–2103. doi:10.1016/j.rse.2007.10.011.
Lucieer, A., Jong, S. M., & Turner, D. (2014). Mapping landslide displacements using Structure from Motion (SfM) and image correlation of multi-temporal UAV photography. Progress in Physical Geography, 38, 97–116. doi:10.1177/0309133313515293.
Ma, L., Cheng, L., Li, M., Liu, Y., & Ma, X. (2015). Training set size, scale, and features in Geographic Object-Based Image Analysis of very high resolution unmanned aerial vehicle imagery. ISPRS Journal of Photogrammetry and Remote Sensing, 102, 14–27.
Merino, L., Caballero, F., Martínez-De-Dios, J. R., Maza, I., & Ollero, A. (2012). An unmanned aircraft system for automatic forest fire monitoring and measurement. Journal of Intelligent and Robotic Systems: Theory and Applications, 65, 533–548. doi:10.1007/s10846-011-9560-x.
Michez, A., Piégay, H., Toromanoff, F., Brogna, D., Bonnet, S., Lejeune, P., & Claessens, H. (2013). LiDAR derived ecological integrity indicators for riparian zones: application to the Houille river in Southern Belgium/Northern France. Ecological Indicators, 34, 627–640. doi:10.1016/j.ecolind.2013.06.024.
Motohka, T., Nasahara, K. N., Oguma, H., & Tsuchida, S. (2010). Applicability of green-red vegetation index for remote sensing of vegetation Phenology. Remote Sensing, 2, 2369–2387. doi:10.3390/rs2102369.
Myers, L. (1989). Riparian area management. Inventory and monitoring of riparian areas. Bureau of Land Management. BLM/YA/PT-89/022+ 1737, Service Center, CO.
Naiman, R., Décamps, H., McClain, M. E. (2005). Riparia: ecology, conservation, and management of streamside communities. Elsevier Academic Press, London.
Nijland, W., de Jong, R., de Jong, S. M., Wulder, M. A., Bater, C. W., & Coops, N. C. (2014). Monitoring plant condition and phenology using infrared sensitive consumer grade digital cameras. Agricultural and Forest Meteorology, 184, 98–106. doi:10.1016/j.agrformet.2013.09.007.
Pintos Varela, C., Rial Martínez, C., Mansilla Vázquez, J. P., & Aguín Casal, O. (2010). First report of Phytophthora rot on Alders caused by Phytophthora alni subsp. alni in Spain. Plant Disease, 94, 273. doi:10.1094/PDIS-94-2-0273A.
Ragg, H., & Fey, C. (2013). UAs in the mountains: monitoring mountain rockslides using multi-temporal point clouds. GIM International, 27, 29–31.
Roux, C., Alber, A., Bertrand, M., Vaudor, L., & Piégay, H. (2014). “FluvialCorridor”: a new ArcGIS toolbox package for multiscale riverscape exploration. Geomorphology. doi:10.1016/j.geomorph.2014.04.018.
Sesnie, S. E., Gessler, P. E., Finegan, B., & Thessler, S. (2008). Integrating Landsat TM and SRTM-DEM derived variables with decision trees for habitat classification and change detection in complex neotropical environments. Remote Sensing of Environment, 112, 2145–2159. doi:10.1016/j.rse.2007.08.025.
Sona, G., Pinto, L., Pagliari, D., Passoni, D., & Gini, R. (2014). Experimental analysis of different software packages for orientation and digital surface modelling from UAV images. Earth Science Informatics, 7, 97–107. doi:10.1007/s12145-013-0142-2.
Sripada, R. P., Heiniger, R. W., White, J. G., & Meijer, A. D. (2006). Aerial color infrared photography for determining early in-season nitrogen requirements in corn. Agronomy Journal, 98, 968. doi:10.2134/agronj2005.0200.
Strecha, C., Fletcher, A., Lechner, A., Erskine, P., & Fua, P. (2012). Developing species specific vegetation maps using multi-spectral hyperspatial imagery from unmanned aerial vehicles. ISPRS Annals of the Photogrammetry, Remote Sensing and Spatial Information Sciences, 3, 311–316.
Stumpf, A., & Kerle, N. (2011). Object-oriented mapping of landslides using Random Forests. Remote Sensing of Environment, 115, 2564–2577. doi:10.1016/j.rse.2011.05.013.
Torres-Sánchez, J., Peña, J. M., de Castro, A. I., & López-Granados, F. (2014). Multi-temporal mapping of the vegetation fraction in early-season wheat fields using images from UAV. Computers and Electronics in Agriculture, 103, 104–113. doi:10.1016/j.compag.2014.02.009.
Urbahs, A., Petuhova, J., Urbaha, M., Carjova, K., Andrejeva, D. (2013). Monitoring of Forest (Fire) using micro class unmanned aerial vehicles. In: Transport Means 2013: Proceedings of the 17th International Conference, Lithuania, Kaunas, 24–25 October, 2013. Kaunas: Technologija, 2013, pp 61–65.
Vermeulen, C., Lejeune, P., Lisein, J., Sawadogo, P., Bouché, P. (2013). Unmanned Aerial Survey of Elephants. PloS one, 8(2), 1–7. doi:10.1371/journal.pone.0054700
Vidal-Abarca Gutiérrez, M. R., & Suárez Alonso, M. L. (2013). Which are, what is their status and what can we expect from ecosystem services provided by Spanish rivers and riparian areas? Biodiversity and Conservation, 22, 2469–2503. doi:10.1007/s10531-013-0532-2.
Waser, L. T., Ginzler, C., Kuechler, M., Baltsavias, E., & Hurni, L. (2011). Semi-automatic classification of tree species in different forest ecosystems by spectral and geometric variables derived from Airborne Digital Sensor (ADS40) and RC30 data. Remote Sensing of Environment, 115, 76–85. doi:10.1016/j.rse.2010.08.006.
Watts, J. D., Lawrence, R. L., Miller, P. R., & Montagne, C. (2009). Monitoring of cropland practices for carbon sequestration purposes in north central Montana by Landsat remote sensing. Remote Sensing of Environment, 113, 1843–1852. doi:10.1016/j.rse.2009.04.015.
Zaman, B., Jensen, A.M., McKee, M. (2011). Use of high-resolution multispectral imagery acquired with an autonomous unmanned aerial vehicle to quantify the spread of an invasive wetlands species. Presented at the International Geoscience and Remote Sensing Symposium (IGARSS), pp. 803–806. doi:10.1109/IGARSS.2011.6049252
Zarco-Tejada, P. J., Diaz-Varela, R., Angileri, V., & Loudjani, P. (2014). Tree height quantification using very high resolution imagery acquired from an unmanned aerial vehicle (UAV) and automatic 3D photo-reconstruction methods. European Journal of Agronomy, 55, 89–99. doi:10.1016/j.eja.2014.01.004.
Zhu, X., & Liu, D. (2014). Accurate mapping of forest types using dense seasonal Landsat time-series. ISPRS Journal of Photogrammetry and Remote Sensing, 96, 1–11. doi:10.1016/j.isprsjprs.2014.06.012.
Acknowledgments
The authors would like to acknowledge the Walloon public service (Walloon Non-Navigable Watercourses department) that provided financial support for the conduct of the research. Authors warmly thank Frédéric Henrotay, Jennifer Di Prinzio, and Benoit Mackels for their field work to acquire the georeferenced tree database. Authors address sincere thanks to Cédric Geerts and Alain Monseur who performed most of the UAS flight surveys. Authors also wish to thank Yves Brostaux for his helpful assistance in the understanding of the random forest algorithms.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Michez, A., Piégay, H., Lisein, J. et al. Classification of riparian forest species and health condition using multi-temporal and hyperspatial imagery from unmanned aerial system. Environ Monit Assess 188, 146 (2016). https://doi.org/10.1007/s10661-015-4996-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10661-015-4996-2