Abstract
Mapping tree species at the single-tree level is an active field of research linking ecology and remote sensing. However, the discrimination of tree species requires the selection of the relevant spectral features derived from imagery. We can extract an extensive number of image parameters even from images with a low spectral resolution, such as Red-Green-Blue (RGB) or near-infrared (NIR) images. Hence, identifying the most relevant image parameters for tree species discrimination is still an issue. We generated 42 parameters from very high resolution images acquired by Unmanned Aerial Vehicles (UAV), such as chromatic coordinates, spectral indices, texture measures and a canopy height model (CHM). The aim of this study was to compare the relevance of these components for classifying savannah tree species. We obtained very high (5 cm) pixel resolution RGB-NIR imagery with a delta-wing UAV in a thorn bush savannah landscape in central Namibia in April 2016. Simultaneously, we gathered ground truth data on the location of 478 individual trees and large shrubs belonging to 16 species. We then used a Random Forest classifier on single and combined thematic sets of image data, e.g. RGB, NIR, texture and in combination with CHM. The best average overall accuracy was 0.77 and the best Cohen´s Kappa value was 0.63 for a combination of RGB imagery and the CHM. Our results are comparable to other studies using hyperspectral data and LiDAR information. We further found that the abundance of the tree species is crucial for successful mapping, with only species with a high abundance being classified satisfactorily. Diverse ecosystems such as savannahs could therefore be a challenge for future tree mapping projects. Nevertheless, this study indicates that UAV-borne RGB imagery seems promising for detailed mapping of tree species.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
Introduction
Classifying and mapping individual trees is increasingly applied in forestry, urban management and nature conservation. According to a review by Fassnacht et al. (2016), since the year 2000 and particularly from 2010 onwards, there has been a dramatic increase in the number of studies that compare the suitability of different datasets, classifiers, and sensor platforms. However, most studies on the classification of tree species use expensive technology to capture data, e.g. hyperspectral or LiDAR sensors, with only a few studies applying relatively cheap solutions such as UAVs carrying consumer-grade cameras that provide Red-Green-Blue (RGB) or Near-Infrared (NIR) imagery. Furthermore, most studies to date have focussed on temperate or boreal forest ecosystems while Savannah ecosystems, which are relatively rich in tree species, remain understudied.
This chapter evaluates the suitability of image parameters derived from low-cost, UAV-borne, consumer-grade cameras for classifying tree species in a savannah ecosystem. In particular, we aim to test (a) whether savannah tree species can be discriminated successfully with very high resolution UAV imagery, (b) whether RGB or NIR spectral indices perform better, and (c) if a canopy height model can significantly improve the classification. Finally, we discuss the role of a species abundance for it´s potential to be accurately mapped.
Background
Mapping the distribution of tree species using remote sensing means producing a vector or raster layer that contains the information on locations of tree species either at the stand-level or single-stem level. These maps or data sets are valuable in nature conservation, particularly in a biodiversity monitoring context. However, until recently, the most commonly used image data for mapping tree species were from hyperspectral and LiDAR sensors (Fassnacht et al. 2016) which are costly, difficult to preprocess, and require expert knowledge in their analysis. The recent advent of drones, also called Unmanned Aerial Vehicles (UAVs), provides new tools and the opportunity to obtain more spatial detail for tree species mapping, and the use of UAVs for mapping tree species is becoming increasingly popular (Singh et al. 2015; Lisein et al. 2015). UAVs have several advantages over satellite or airborne data. They are extremely flexible in usage, can be scheduled in a very short time interval (e.g. daily or weekly), are easily carried to diverse locations and, unlike satellites, are not limited by clouded skies. The main drawbacks are the limited spatial coverage and a restricted carrying capacity which renders UAVs unsuitable for heavy hyperspectral or LiDAR sensors and thus being restricted to consumer-grade cameras or small multispectral cameras. Only a few studies have evaluated RGB spectral indices for species discrimination, e.g. Rasmussen et al. (2013), who tested the potential of RGB indices for site-specific weed management, Dvořák et al. (2015) used RGB imagery for invasive plant detection, and Rasmussen et al. (2016) studied the performance of RGB indices to measure barley biomass. Hence, the question remains open as to whether very high spatial resolution imagery taken by a standard UAV can successfully discriminate tree species. If so, nature conservation could make use of a very flexible image acquisition platform for monitoring small areas, i.e. covering several square kilometres with a small number of flights. Furthermore, the question how consumer-grade cameras with RGB or a NIR-filter perform in such a task needs to be addressed. Is NIR really necessary or are observations in RGB sufficient?
Most of the studies reviewed by Fassnacht et al. (2016) had two things in common: they used hyperspectral imagery in combination with LiDAR data and were undertaken in temperate or boreal forests. Bunting and Lucas (2006) and Lucas et al. (2008) established the use of CASI and HYMAP hyperspectral data for discriminating tree species in open woodlands and forests in Queensland, Australia, confirming that differences in the mean spectra from crown objects increased the accuracy of discrimination. However, only a few studies set out to classify savannah tree species in southern Africa (Naidoo et al. 2012; Cho et al. 2012; Colgan et al. 2012). These studies classified between six and 15 tree species. Cho et al. (2015) also tested the suitability of very high resolution satellite imagery for this purpose, but only used three out of ten dominant canopy species. It seems that the abundance of a tree species also contributes to its capability for being mapped precisely. We are of the opinion that this issue has not been sufficiently highlighted in the literature (but see comments in Fassnacht et al. 2016).
Study Area
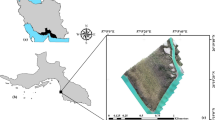
The study was part of the Biodiversity Observatory S05 of the BIOTA Africa project (www.biota-africa.org), which is a cross-country biodiversity monitoring project with a standardized monitoring approach performing monitoring in southern, western, and northern Africa (Jürgens et al. 2012). The observatory is located on the cattle farm Erichsfelde (coordinates: 16.935° E 21.597° S) in central Namibia. The Biodiversity Observatory spans 1 km2 and is divided into 100 ha from which 20 were selected in the year 2001 for permanent annual monitoring of vegetation and animal diversity. The vegetation monitoring was undertaken within plots of 20 × 50 m, which were situated at the mid-point of a selected hectare. The vegetation consisted of typical Thornbush savanna sensu Giess (1998), dominated by Acacia mellifera subsp. detinens and Boscia albitrunca. Other Acacia species also occurred, in particular A. hebeclada subsp. hebeclada, A. tortilis, A. reficiens and A. karroo. The vegetation structure is a typical semi-closed bushland, with a low grass layer, and shrubs of up to 4 m high (Fig. 1). These are interspersed by few trees, between 4 and 8 m in height. Erichsfelde is a private cattle farm of about 13,000 ha that is used extensively for meat production (cattle grazing). Game species, including Oryx and Kudu are also present. The Observatory is not excluded from regular land-use.
Landscape perspective of the thorn bush savannah vegetation on the private cattle-farm Erichsfelde. The tree layer consists mainly Acacia mellifera subsp. detinens with one larger Acacia tortilis in the back. Small shrubs and a dense grass layer leaving some open soil patches characterize the landscape. Picture taken 05. April 2016, by L. Naftal
Methods
UAV Imagery Acquisition
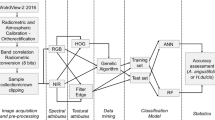
On 21.03.2016, we acquired an image mosaic with 5 cm ground resolution for the whole Biodiversity Observatory S05, i.e. approximately 1 km2 (Fig. 2). We covered the area in two flights with an eBee 3 drone (costs ca. 30.000€, SenseFly 2015, Cheseaux-Lausanne, Switzerland). The settings for the flight missions were 70% longitudinal and 60% lateral overlap, with a flying height of 115 m above take-off point. Each image had a width of 160 m and a length of 120 m. The first flight was conducted using a modified Canon S110 where the blue filter was replaced by a NIR filter, recording at 850 nm. This camera also recorded a green band (550 nm) and a red band (625 nm). The second flight was performed using a regular RGB camera (Canon S110), recording at 450 nm (blue), 520 nm (green) and 660 nm (red) (SenseFly 2014). The flights to collect the images took about 30 min each and took place on the same day between 10h45 and 12h00. The first flight (NIR) yielded 411 single images; the second flight (RGB) 358 images. We then mosaicked the image sets into two single orthomosaics using the PiX4D software. We did not use ground control points for the mosaicking procedure, so we needed to adjust the imagery by shifting the NIR image manually by five pixels on the x-axis and three pixels on the y-axis to ensure proper matching with RGB imagery. We used QGIS v.2.16 (QGIS Development Team 2016) to shift the image.
Ground Truth Data
After image processing, subsections of the orthomosaics corresponding to each of 20 permanent monitoring plots were taken to the field. Within each plot, all trees and shrubs in the image were compared to each tree and shrub in the vegetation plot. Then, for each single-stem individual, an outline for each individual was drawn onto the image. This was necessary as the crowns of species were commonly overlapping where multiple stemmed individuals occurred. In total, 16 species were recorded and only living individuals were considered. However, we excluded seven of these species from further analysis because they occurred with an abundance of less than ten individuals within the samples (Table 1). We then converted our paper drawings into polygons per species in QGIS based on the RGB orthomosaic. Although several software solutions exist that promise to segment tree crowns from point clouds and RGB imagery, such as the TIDA algorithm (Culvenor 2002), these algorithms are difficult to handle and come with their own error. As the number of polygons required was small, we preferred the manual approach of delineating tree crowns. In total, we drew 478 polygons to build the species dataset. The abundance of the observed species is described in Table 1. Note that this includes only living individuals.
RGB and NIR Spectral Indices
As the aim of this study was to identify the most suitable predictors for species discrimination, we derived an extensive set of parameters from both the RGB and NIR imagery. Based on the RGB imagery, we calculated the chromatic coordinates (Woebbecke et al. 1995; Meyer and Neto 2008) as well as the excessive Red (exR) and Green (exG) indices (Table 2). Recent studies had found these to be the most suitable to discriminate crop species (Woebbecke et al. 1995; Meyer and Neto 2008) or to predict vegetation parameters based solely on RGB imagery (Zhang et al. 2010; Schirrmann et al. 2016; Vergara-Díaz et al. 2016). In addition, we calculated the normalised green-red difference index (NGRDI) and the exG–exR parameter. A recent, simplified version of the excessive greenness index (Rasmussen et al. 2016) was also calculated, in this study called exG2.
To exploit the additional NIR information, we calculated slope and distance based vegetation indices (Silleos et al. 2006). Slope based vegetation indices make use of the difference in the slope of the red and NIR channel; the famous Normalized Difference Vegetation Index (NDVI, Tucker 1979) belongs here. The SAVI is a modified NDVI which adjusts for potential effects of bare soil (Huete 1988). Thiam’s vegetation index improves on the NDVI by multiplying the absolute NIR and Red band values with their square root (Thiam 1998). Distance based vegetation indices make use of the concept of the so-called “soil-line” (Silleos et al. 2006). The distances refer to the distance of samples in the two dimensional red-NIR spectral space to the soil line, that describes the lower boundary of pixels in this space, usually aligning across a clearly visible axis. To determine the soil line parameters required for the calculation of the distance based vegetation indices, a set of n = 100 bare soil pixels were selected, stratified by the hectare grid of the Biodiversity Observatory, and the NIR and red values were extracted. Based on these values, a linear regression (R2 = 0.89, p < 0.001) was used to estimate the intercept and the slope of the soil line. The linear regression parameters intercept (alpha = −227.29) and slope (beta=1.877) were used to calculate the Perpendicular Vegetation Index III (Qi et al. 1994; Silleos et al. 2006). All indices were calculated and image manipulations were performed with the open source software SAGA-GIS (Conrad et al. 2015). For all individual tree crown polygons, we calculated values for the mean and standard deviations using the zonal statistics tool in SAGA-GIS.
Image Texture
Richards (2013) suggested that the texture of an image can be described as smooth, rough or repetitive in terms of the spatial arrangement of grey values. In terms of canopy cover this would describe whether tree crowns consist of repeating patterns of shadow and greenness or whether the canopy is closed and thus equal in colour. Often texture measures will improve remote sensing classifiers (Krefis et al. 2011). As our main interest was to discriminate between tree species canopies, the greenness (exG) of the canopy seemed to be a good parameter for a texture analysis. We used the Orfeo Toolbox v.5.6.1 (McInerney and Kempeneers 2015), a free open source software for remote sensing image analysis, to calculate eight different types of simple image texture measures. Haralick’s grey level occurrence matrix (GLCM, Haralick et al. 1973), which is a standard for describing image texture, was the basis for calculating all of the texture measures. We choose a constant window size of 5×5 pixels and an offset of 1 for x and y. The number of grey levels was set to 16. We then loaded the calculated image texture measures into SAGA-GIS and extracted the texture as average and standard deviation for each individual tree crown canopy polygon.
Canopy Height Model
We generated a canopy height model (CHM) based on the overlapping single image tiles. We used the software Postflight Terra 3D Vs4.0.104 (SenseFly 2015) to generate dense point clouds as *.las-files. Then we imported the *.las point cloud files into the LAStools software (Isenburg 2016) to generate buffered tiles. Ground points representing bare ground were identified visually. Based on these ground points, we calculated the height (height normalization) for all non-ground points of all tiles. These tiles were then mosaicked in SAGA-GIS using a b-spline interpolation with feathering to create a seamless normalized Digital Surface Model (nDSM). This nDSM describes the maximum heights of the point cloud. Next, we generated a Digital Terrain Model (DTM), that describes the minimum heights of the point cloud. Finally, the CHM was generated by subtracting the DTM from the DSM, which gave values in the range of −0.11 to 1.89 m. The lower range was adjusted to zero. Average canopy height and its standard deviation were extracted for each canopy polygon.
Random Forest Classification
The Random Forest algorithm (Breiman Breimanx) is now a common standard non-parametric classifier with high performance as was found by many comparative studies in a remote sensing context (Pal 2005; Duro et al. 2012; Qian et al. 2014). Random Forest makes use of the concept of classification and regression trees (CART) but combines them with ensemble modelling and bagging. Random Forest is a non-parametric classifier that creates thousands of single decision trees and averages their results. Each decision tree is a subsample of the whole dataset. The split for each tree node is determined by the Gini criterion, which measures the entropy of the dataset. The best split is that parameter value that leads to the largest decrease in the Gini criterion. When the classifier is applied to the test dataset, the final class label is then based on the majority vote of all constructed decision trees (Immitzer et al. 2012).
A Random Forest classifier was used to predict species labels, with this achieved by first dividing the dataset into training and testing polygons, with an 80:20 ratio per class. To establish if any single set of parameters were sufficient alone, the dataset was split into a RGB, a NIR, a texture and a complete dataset (ALL). For quantifying the importance of the CHM, we added these values to each parameter dataset. Before classification, all parameters with Pearson correlations higher than 0.75 were deleted to ensure that multicollinearity issues would not be an issue. Only two texture parameters where omitted because of multicollinearity, the Cluster Prominence and Haralick’s correlation. The latter was correlated with “correlation (corrL)” and the first with “inverse distance moment (IDM)”. Finally, in order to test the effect of species abundance on the classification results, the species data were divided into three datasets, considering (a) all species with an abundance larger than 10, (b) frequent species with an abundance larger than 30 and (c) infrequent species with abundances between 30 and 10 individuals (Table 2). These species subsets were all tested in combination with all parameter subsets, leading to a final number of 24 single datasets.
For each single dataset, a classifier was produced and its accuracy was verified using the test dataset. For accuracy assessment, confusion matrices were generated and the following accuracy measures were derived: Overall accuracy (OA), confidence limits for OA based on cross-validation, and Cohen´s Kappa which takes class imbalance into account (Kuhn and Johnson 2013). As a null-model for the overall accuracy, we calculated the No-Information Rate (Kuhn and Johnson 2013), which is simply defined as the proportion of the largest class expressed as a percentage. A one-sided test of equal proportions was then conducted to provide a p-value for the null-model.
The relevance of the single predictors was assessed by calculating their variable importance. Variable importance describes the relationship between each parameter and the outcome of the classification or regression procedure. It is measured as the loss in performance when the respective parameter is not considered. Variable importance was measured for all parameters in the three species subsets in order to identify consistently important predictors across all predictors considered.
Random Forests were run with 5000 trials. The parameter mtry was set to 1/3 of the number of variables considered. The parameter mtry describes the number of parameters that are included in each single decision tree. In addition, a repeated cross validation was implemented using a tenfold cross validation with five repetitions to be able to achieve standard errors and confidence intervals for the overall accuracy. Classification was performed in the free and open source software R (R Core Team 2016) using the packages caret (Kuhn et al. 2016), randomForest (Liaw and Wiener 2002) and e1071 (Meyer et al. 2015).
Results
Only frequent species constantly exhibited significant p-values (Table 3, Fig. 3) meaning that OA was higher than the respective null-model. When using all species or only infrequent species, this was not the case. The species subsets “All” and “Infrequent” had always low Kappa and OA values except for infrequent species with the ALL and ALL+CHM (Table 3).
Average overall accuracy with confidence limits based on tenfold cross-validation with five repetitions. The stars denote the overall accuracy derived by a null-model. Stars within confidence limits signify models that were not significantly better than the null-model and thus do not provide credible results. Note the very high range of confidence limits for the infrequent species data set (Spinfreq)
The highest OA and Kappa values were obtained for the combined RGB and CHM dataset for the frequent species, with an OA value of 0.77 and a Kappa value of 0.63. Globally, the “ALL” model was ranked second by Kappa for frequent species. However, “ALL” is much more complex (42 parameters) than RGB+CHM (16 parameters). Thus, the simpler RGB solution can be regarded as much more informative and easier to reproduce as fewer parameters have to be derived from the imagery (Table 3). The lower quality of the infrequent species dataset was also evident by the large confidence intervals (Fig. 3). These are much smaller in the case for ALL and frequent species. This effect was attributed to the small number of samples within the infrequent species (72 samples spread across five classes, see Table 1). Models that used texture (TEXT) alone or a combination of texture and CHM (TEXTCHM) were never significant in any of the species sets (Table 3). Best results for texture models were found for the frequent species with an OA of 0.70 and a Kappa value of 0.49.
The inclusion of the CHM led to an increase in model quality for 7 out of 12 image parameter pairs (Table 3). The largest increase in the Kappa (0.22) was found for the RGB – RGB+CHM pair in the frequent species dataset. However, the second largest change was a decrease of 0.13 for the NIR – NIR+CHM in the infrequent species dataset (Table 3). Except for these two values, the average increase in Kappa was zero. Hence, we did not find that the CHM contributed additional information.
In the variable importance analysis (Fig. 4), none of the NIR-derived image parameters occurred in the top ten parameters. The RGB indices exG2, exR, exG and NGRDI occurred in the All and Frequent species subsets. Highest importance values were achieved by the RGB indices exG2 and chrR both with 32% and 20% for Frequent and All species subsets respectively. In the case of the infrequent species, two texture measures appear in the ten most important variables. However, their variable importance did not reach values higher than five percent, rendering all parameters for the infrequent species redundant.
Variable importance of the ten most important image parameters for the three species subsets, i.e. all species (a), frequent species (b) and infrequent species (c). For full names of image parameters see Table 2
In summary, the frequent species dataset together with RGB+CHM image parameters provided the highest accuracy in the discrimination of tree species, used the lowest number of predictors and provided the smallest confidence intervals.
Discussion
We evaluated the relevance of different parameters derived from very high resolution RGB-NIR imagery for the discrimination of savannah tree species. We could confirm the commonly found pattern that information based on the visual part of the spectrum is important for discriminating tree species (Fassnacht et al. 2016). In particular, we found that RGB-based spectral indices (Meyer and Neto 2008; Rasmussen et al. 2016) and simple chromatic coordinates (Woebbecke et al. 1995) in combination with a canopy height model (CHM) achieved the best results. By contrast, the performance of the NIR image parameters was weak and deteriorated when combined with the CHM. Our overall accuracy of 77% (on average, maximum was 0.83) is comparable to the results of the recent review of Fassnacht (see Fig. 3 in Fassnacht et al. (2016)) who analysed 129 case studies on tree species mapping.
Most case studies have used a combination of hyperspectral and/or LiDAR for tree species mapping and have typically achieved overall accuracies of between 75 and 90%. Of these, three were carried out in southern Africa, all in Kruger National Park, and all used hyperspectral image data and height information derived from LiDAR sensors. Naidoo et al. (2012) achieved 82% with four hyperspectral indices (including NDVI) and height information; whilst Cho et al. (2012) also used hyperspectral data but resampled them to seven World View 2 multispectral bands and combined them with LiDAR based height information. They achieved OA values of between 63 and 81%. Colgan et al. (2012) used LiDAR-based height information and BRDF corrected reflectance values for the VIS-NIR region. The bidirectional reflectance distribution function (BRDF) is a function describing the change in reflectance values due to view angle and sun position during image assessment. The BRDF correction improved the hyperspectral information and thus led to OAs of between 70 and 78%. Hence, our UAV based tree species discrimination approach, requiring only a ≈100$ RGB camera, performed equally well, when compared to the technically more sophisticated, and also much more expensive, hyperspectral and LiDAR sensors.
VIS-NIR imagery is by far the most commonly used data source for generating spectral indices. Pure RGB based spectral information has been used less often in remote sensing studies that have focused on the discrimination and mapping species. In their review on tree species mapping, Fassnacht et al. (2016) also state that the VIS region (350–650 nm) contains the most often selected features for tree species mapping, without mentioning the relation to RGB. A recent study comparing spectral indices derived from RGB and NIR camera images to multispectral imagery (Rasmussen et al. 2016) showed that cheaper RGB / NIR cameras are equal in performance for mapping barley biomass in agricultural fields. Another study (Fischer et al. 2012) compared an NDVI calculated from high spectral resolution field spectrometer (i.e. ASD Field Spec 3) with an NDVI derived from a modified Olympus consumer-grade camera for mapping the spatial variability of NDVI in biotic soil crusts; they found strong correlations with R2 of 0.91.
In this study, we found that RGB bands were an important predictor but the least important was texture. This is surprising as we thought that texture would have a high potential for describing crown properties related to shadow patterning or variation in greenness. Fassnacht et al. (2016) list several studies that applied texture to improve tree species classification by 10–15%. However, using texture also creates problems that make its use seem clumsy and time consuming. First of all, the idea of texture is a multiscale problem. The relevant scale (i.e., the size of the window in which a texture value is calculated) has to be identified empirically. Therefore, different window sizes have to be compared with these usually being 3×3, 5×5, 7×7, 9×9 and so on. When UAV data volumes are large (>1 GB), this quickly becomes unwieldy. Also, with very high resolution images, larger window sizes are needed but these slow processing times. Other options such as the offset and the number of grey levels considered, provide further opportunities to optimize the results; yet leading to a seemingly endless endeavour in finding the right parameter settings. Second, different species might require different window sizes. This seems logical but is difficult to realize technically. Third, the large number of available texture measures makes it difficult to select those that are optimal or most appropriate. This is complicated by the typically high correlation between the different texture measures. In our study, all parameters were kept stable (i.e., a window size of 5×5, offset of 1×1 and 16 grey levels). Not testing different settings might explain the poor performance of the texture parameters. We also used only a small fraction of the available texture measures. Other texture measures related to tree crown shape and size could have been considered (Fassnacht et al. 2016). The Orfeo Toolbox provides around 40 different texture measures in total. Hence, texture measures derived from UAV imagery require more studies on selecting and optimising the best measures and optimal window sizes for tree species discrimination or tree crown analyses.
Tree species mapping through remote sensing data can become an efficient tool in biodiversity monitoring. However, the nature of biodiversity is that communities under study almost always consist of common and rare species (Magurran and McGill 2011). The occurrence of rare tree species (rare equal to less than ten individuals overall) severely affected the potential to classify the whole tree species pool. Out of 16 species, seven species were considered as too rare to be used in the classification. Another five infrequent species, i.e. with less than 30 tree crowns for training and testing, led to poor classification results due to the small amount of training data. However, the infrequent species were impossible to classify correctly, as can be seen from the non-significant or low quality models (Table 2, Fig. 3). Thus out of 16 species, only the four frequent species could be classified to acceptable levels of accuracy. The pattern that only frequent species can be mapped with sufficient accuracy is confirmed by many other studies (Naidoo et al. 2012; Immitzer et al. 2012; Cho et al. 2012; Baldeck et al. 2015). The review of Fassnacht et al. (2016) reports the number of species that were classified in the analysed studies ranging from two to seventeen with an average of five or six. This finding has important implications for future biodiversity monitoring that should be based on tree species mapping leading to a complete census. Mapping rare tree and shrub species becomes a challenge when too few individuals can be found for training and testing a classifier. Thus, in future studies, more emphasis should be put on high quality ground truth data gathering an equal number of ground truth tree locations per species. In the study of Colgan et al. (2012), three species made up 30% of the landscape while the category “other” also had 20% of all occurring tree crowns. Thus, rare species can make a large fraction of tree crowns in a savannah but are represented by a small number of individuals per species. Common trees however, bear different challenges. For example, a high genus – species ratio (i.e. where many species of the same genus occur as in the genus Acacia or Combretum) means these species are sometimes lumped together into a single tree category at genus level (Naidoo et al. 2012). The species abundances in (semi-)natural ecosystems are much more complex than in temperate forests and will require special considerations for an operative tree species mapping based on remote sensing imagery.
Although our study was successful in discriminating selected savanna tree species with a UAV-borne RGB camera, the limitations of UAVs in comparison to airborne or satellite-borne sensors requires discussion. In our case, the largest obstacle was the mismatch in the co-registration of NIR and RGB imagery, which had to be corrected manually. Better results could be achieved when using multispectral cameras or even lightweight hyperspectral cameras. The spatial mismatch could have been the reason why the averaged tree crown parameters were worse for NIR than for RGB. Digitization of the tree crowns was also undertaken manually using only the RGB imagery. Hence, it is possible that the NIR imagery parameters contained a higher shadow fraction or parts of neighbouring tree crowns. Although manual digitization seems straightforward, it is also error prone and could be avoided by using specifically designed algorithms or software packages, e.g. TIDA (Culvenor, 2002), JSEG (Kang et al. 2016) or ITCsegment (Dalponte and Coomes 2016). Other serious problems connected to light and shadowing effects that can occur when using UAV imagery are discussed by Rasmussen et al. (2016). In our study, the different flight directions during the drone overflight affected the brightness pattern. Rasmussen et al. (2016) also mentioned that BRDF effects affect the outcome of a study when not taken into consideration. These issues, co-registration and changing light conditions (including BRDF effects), seem to compromise the utility of UAV imagery. Ground control points should be essential for proper image co-registration, however, often these require expensive differential-GPS equipment. Further improvement of technical equipment or standardized procedures for UAV image acquisition should bring remedies in the future.
Conclusions
In this study, we evaluated the relevance of RGB and NIR image products, derived from UAV images, for discriminating tree species in a Namibian savannah. We found that data acquired in the NIR wavelength region only were not sufficient or even necessary, although this conclusion might have been incorrectly drawn because of co-registration problems between the NIR and RGB imagery. Permanently marked, well-surveyed ground control points therefore need to be planned for future image acquisition campaigns. Nevertheless, the OAs achieved with RGB data and CHM were comparable to other studies that used more expensive hyperspectral data and LiDAR instruments. This indicated that UAVs have a high potential for future tree species mapping tasks if areas less than 1 km2 are to be monitored. However, the number of species that can be mapped or discriminated seems independent of the sensor type. The assumption is that hyperspectral data theoretically can outperform RGB-NIR data when a large number of species are present. However, for the process of training a classifier, such as a Random Forest, the number of training polygons needs to be at least 30 in order to achieve sufficient and acceptable accuracies. This seems not feasible when rare species (i.e., less than ten individuals per square kilometre) are present. Hence, a significant future challenge is the task of mapping species with low abundances.
Practical Application for Nature Conservation
This chapter dealt with the application of UAV-borne consumer grade cameras for discriminating savannah tree species and has several important messages for practical applications in nature conservation. Firstly, the delta-wing UAV that we employed, the eBee system (SenseFly 2015), is capable of capturing an area of 1 km2 during a single flight when the desired resolution is a 5 cm pixel size or greater. Smaller pixel sizes, e.g. 2 cm, can only be achieved in several flights (four to five). However, this this also doubles the disk space required for storing the imagery. Affordable quadcopter systems cannot usually cover 1 km2 in a single flight. Secondly, we showed that tree species discrimination based solely on RGB + Canopy Height is possible, suggesting that a second flight with a NIR camera is potentially unnecessary. However, we need more studies comparing RGB based spectral indices to NIR based spectral indices in order to see whether RGB can replace NIR indices in the future. Finally, we found that ground truthing should take the abundance or frequency of the species into consideration. We suggest using a minimum of 50 individuals per species for training purposes in order to be successfully mapped. Species from the same genus, e.g. the different Acacia species in our study, often share similar spectral properties and thus are very difficult to distinguish. One alternative is to map these at the genus level, if it is not the users demand to produce species specific map. In conclusion, we have shown that using UAVs to map the individual stems of tree species could be a cheap and very flexible tool for nature conservation in the near future.
References
Baldeck, C.A., Asner, G.P., Martin, R.E., et al.: Operational tree species mapping in a diverse tropical forest with airborne imaging spectroscopy. PLoS One. 10, e0118403 (2015). doi:10.1371/journal.pone.0118403
Baret, F., Guyot, G., Major, D.: TSAVI: a vegetation index which minimizes soil brightness effects on LAI and APAR estimation. In: 12th Canadian Symposium on Remote Sensing and IGARSS’90, p. 4, Vancouver, Canada, 10–14 July 1989 (1989)
Breiman, L.: Random forests. Mach. Learn. 45, 5–32 (2001). doi:10.1023/A:1010933404324
Bunting, P., Lucas, R.: The delineation of tree crowns in Australian mixed species forests using hyperspectral Compact Airborne Spectrographic Imager (CASI) data. Remote Sens. Environ. 101, 230–248 (2006). doi:10.1016/j.rse.2005.12.015
Cho, M.A., Mathieu, R., Asner, G.P., et al.: Mapping tree species composition in South African savannas using an integrated airborne spectral and LiDAR system. Remote Sens. Environ. 125, 214–226 (2012). doi:10.1016/j.rse.2012.07.010
Cho, M.A., Malahlela, O., Ramoelo, A.: Assessing the utility WorldView-2 imagery for tree species mapping in South African subtropical humid forest and the conservation implications: Dukuduku forest patch as case study. Int. J. Appl. Earth Obs. Geoinf. 38, 349–357 (2015). doi:10.1016/j.jag.2015.01.015
Colgan, M.S., Baldeck, C.A., Féret, J.-B., Asner, G.P.: Mapping savanna tree species at ecosystem scales using support vector machine classification and BRDF correction on airborne hyperspectral and LiDAR data. Remote Sens. 4, 3462–3480 (2012). doi:10.3390/rs4113462
Conrad, O., Bechtel, B., Bock, M., et al.: System for automated geoscientific analyses (SAGA) v. 2.1.4. Geosci. Model Dev. 8, 1991–2007 (2015). doi:10.5194/gmd-8-1991-2015
Culvenor, D.S.: TIDA: an algorithm for the delineation of tree crowns in high spatial resolution remotely sensed imagery. Comput. Geosci. 28, 33–44 (2002). doi:10.1016/S0098-3004(00)00110-2
Dalponte, M., Coomes, D.A.: Tree-centric mapping of forest carbon density from airborne laser scanning and hyperspectral data. Methods Ecol. Evol. 7, 1236–1245 (2016). doi:10.1111/2041-210X.12575
Duro, D.C., Franklin, S.E., Dubé, M.G.: A comparison of pixel-based and object-based image analysis with selected machine learning algorithms for the classification of agricultural landscapes using SPOT-5 HRG imagery. Remote Sens. Environ. 118, 259–272 (2012). doi:10.1016/j.rse.2011.11.020
Dvořák, P.J., Müllerová J., Bartaloš, T., Brůna J.: Unmanned aerial vehicles for alien plant species detection and monitoring. ISPRS – international archives of the photogrammetry, remote sensing and spatial information sciences XL-1/W4: 83–90 (2015). doi:10.5194/isprsarchives-XL-1-W4-83-2015
Fassnacht, F.E., Latifi, H., Stereńczak, K., et al.: Review of studies on tree species classification from remotely sensed data. Remote Sens. Environ. 186, 64–87 (2016). doi:10.1016/j.rse.2016.08.013
Fischer, T., Veste, M., Eisele, A., et al.: Small scale spatial heterogeneity of Normalized Difference Vegetation Indices (NDVIs) and hot spots of photosynthesis in biological soil crusts. Flora – Morphol. Distrib. Funct. Ecol. Plants. 207, 159–167 (2012). doi:10.1016/j.flora.2012.01.001
Giess, W.: A preliminary vegetation map of Namibia. Dinteria. 4, 1–112 (1998)
Haralick, R.M., Shanmugam, K., Dinstein, I.: Textural features for image classification. IEEE Trans. Syst. Man Cybern. 3, 610–621 (1973)
Huete, A.R.: A soil-adjusted vegetation index (SAVI). Remote Sens. Environ. 25, 295–309 (1988). doi:10.1016/0034-4257(88)90106-X
Immitzer, M., Atzberger, C., Koukal, T.: Tree species classification with random forest using very high spatial resolution 8-Band WorldView-2 satellite data. Remote Sens. 4, 2661–2693 (2012). doi:10.3390/rs4092661
Isenburg, M.: LAStools, efficient LiDAR processing software. rapidlasso GmbH (2016)
Jürgens, N., Schmiedel, U., Haarmeyer, D.H., et al.: The BIOTA biodiversity observatories in Africa—a standardized framework for large-scale environmental monitoring. Environ. Monit. Assess. 184, 655–678 (2012). doi:10.1007/s10661-011-1993-y
Kang, J., Wang, L., Chen, F., Niu, Z.: Identifying tree crown areas in undulating eucalyptus plantations using JSEG multi-scale segmentation and unmanned aerial vehicle near-infrared imagery. Int. J. Remote Sens. 38, 1–17 (2016). doi:10.1080/01431161.2016.1253900
Klaassen, E.S., Kwembeya, E.G.: A checklist of Namibian indigenous and naturalised plants. 2013. Occasional Contributions No 5, National Botanical Research Institute, Windhoek, Namibia (2013).
Krefis, A.C., Schwarz, N.G., Nkrumah, B., et al.: Spatial analysis of land cover determinants of malaria incidence in the Ashanti Region, Ghana. PLoS One. 6, e17905 (2011). doi:10.1371/journal.pone.0017905
Kuhn, M., Johnson, K.: Applied Predictive Modeling. Springer, New York (2013)
Kuhn, M.K., Weston, S., Williams, A., et al.: Caret: Classification and Regression Training. R package version 6.0-70. https://CRAN.Rproject.org/package=caret (2016)
Kyalangalilwa, B., Boatwright, J.S., Daru, B.H., et al.: Phylogenetic position and revised classification of Acacia s.l. (Fabaceae: Mimosoideae) in Africa, including new combinations in Vachellia and Senegalia. Bot. J. Linn. Soc. 172, 500–523 (2013). doi:10.1111/boj.12047
Liaw, A., Wiener, M.: Classification and regression by random Forest. R News. 2, 18–22 (2002)
Lisein, J., Michez, A., Claessens, H., Lejeune, P.: Discrimination of deciduous tree species from time series of unmanned aerial system imagery. PLoS One. 10, e0141006 (2015). doi:10.1371/journal.pone.0141006
Lucas, R., Bunting, P., Paterson, M., Chisholm, L.: Classification of Australian forest communities using aerial photography, CASI and HyMap data. Remote Sens. Environ. 112, 2088–2103 (2008). doi:10.1016/j.rse.2007.10.011
Magurran, A.E., McGill, B.J.: Biological Diversity: Frontiers in Measurement and Assessment, 1st edn. Oxford University Press, Oxford/New York (2011).
McInerney, D., Kempeneers, P.: Orfeo toolbox. In: Open Source Geospatial Tools, pp. 199–217. Springer International Publishing, Basel (2015).
Meyer, G.E., Neto, J.C.: Verification of color vegetation indices for automated crop imaging applications. Comput. Electron. Agric. 63, 282–293 (2008). doi:10.1016/j.compag.2008.03.009
Meyer, D., Dimitriadou, E., Hornik, K., et al.: e1071: Misc Functions of the Department of Statistics, Probability Theory Group (Formerly: E1071), TU Wien. R package version 1.6-7 https://CRAN.R-project.org/package=e1071 (2015)
Naidoo, L., Cho, M.A., Mathieu, R., Asner, G.: Classification of savanna tree species, in the Greater Kruger National Park region, by integrating hyperspectral and LiDAR data in a Random Forest data mining environment. ISPRS J. Photogramm. Remote Sens. 69, 167–179 (2012). doi:10.1016/j.isprsjprs.2012.03.005
Pal, M.: Random forest classifier for remote sensing classification. Int. J. Remote Sens. 26, 217–222 (2005). doi:10.1080/01431160412331269698
Perry, C.R., Lautenschlager, L.F.: Functional equivalence of spectral vegetation indices. Remote Sens. Environ. 14, 169–182 (1984). doi:10.1016/0034-4257(84)90013-0
QGIS Development team. QGIS Geographic Information System. Open Source Geospatial Foundation (2016)
Qi, J., Chehbouni, A., Huete, A.R., Kerr, Y.H., Sorooshian, S.: A modified soil adjusted vegetation index. Remote Sens. Environ. 48, 119–126 (1994). doi:10.1016/0034-4257(94)90134-1
Qian, Y., Zhou, W., Yan, J., et al.: Comparing machine learning classifiers for object-based land cover classification using very high resolution imagery. Remote Sens. 7, 153–168 (2014). doi:10.3390/rs70100153
R Core Team: R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna (2016)
Rasmussen, J., Nielsen, J., Garcia-Ruiz, F., Christensen, S., Streibig, J.C.: Potential uses of small unmanned aircraft systems (UAS) in weed research. Weed Res. 53, 242–248 (2013). doi:10.1111/wre.12026
Rasmussen, J., Ntakos, G., Nielsen, J., et al.: Are vegetation indices derived from consumer-grade cameras mounted on UAVs sufficiently reliable for assessing experimental plots? Eur. J. Agron. 74, 75–92 (2016). doi:10.1016/j.eja.2015.11.026
Richards, J.A.: Remote Sensing Digital Image Analysis: An Introduction, 5th edn. Springer, Berlin (2013)
Schirrmann, M., Giebel, A., Gleiniger, F., et al.: Monitoring agronomic parameters of winter wheat crops with low-cost UAV imagery. Remote Sens. 8, 706 (2016). doi:10.3390/rs8090706
SenseFly. User Manual: S110 RGB/NIR /RE camera. SenseFly Ltd., Lausanne, Switzerland (2014)
SenseFly. eBee Sensefly: Extended User MANUAL eBee and eBee Ag. Revision 17, June 2015. SenseFly Ltd., Lausanne, Switzerland (2015)
Silleos, N.G., Alexandridis, T.K., Gitas, I.Z., Perakis, K.: Vegetation indices: advances made in biomass estimation and vegetation monitoring in the Last 30 years. Geocarto Int. 21, 21–28 (2006). doi:10.1080/10106040608542399
Singh, M., Evans, D., Tan, B.S., Nin, C.S.: Mapping and characterizing selected canopy tree species at the Angkor World Heritage Site in Cambodia using aerial data. PLoS One. 10, e0121558 (2015). doi:10.1371/journal.pone.0121558
Thiam A.K.: Geographic Information Systems and Remote Sensing Methods for Assessing and Monitoring Land Degradation in the Sahel Region: The Case of Southern Mauritania (1998)
Tucker, C.J.: Red and photographic infrared linear combinations for monitoring vegetation. Remote Sens. Environ. 8, 127–150 (1979)
Vergara-Díaz, O., Zaman-Allah, M.A., Masuka, B., et al.: A novel remote sensing approach for prediction of maize yield under different conditions of nitrogen fertilization. Front. Plant Sci. (2016). doi:10.3389/fpls.2016.00666
Woebbecke, D.M., Meyer, G.E., Von Bargen, K., Mortensen, D.A.: Color Indices for Weed Identification Under Various Soil, Residue, and Lighting Conditions. Trans. ASAE. 38, 259–269 (1995). doi:10.13031/2013.27838
Zhang, F., Zaman, Q.U., Percival, D.C., Schumann, A.W.: Detecting bare spots in wild blueberry fields using digital color photography. Appl. Eng. Agric. 26, 723–728 (2010)
Acknowledgments
Our gratitude to the Pommersche Farmereigesellschaft and their staff for allowing us to work on the farm Erichsfelde. The work was financially supported by the SASSCAL initiative, with funding by the German Federal Ministry of Education and Research; BMBF Funding Nr: 01LG1201M.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this chapter
Cite this chapter
Oldeland, J., Große-Stoltenberg, A., Naftal, L., Strohbach, B.J. (2017). The Potential of UAV Derived Image Features for Discriminating Savannah Tree Species. In: Díaz-Delgado, R., Lucas, R., Hurford, C. (eds) The Roles of Remote Sensing in Nature Conservation. Springer, Cham. https://doi.org/10.1007/978-3-319-64332-8_10
Download citation
DOI: https://doi.org/10.1007/978-3-319-64332-8_10
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-64330-4
Online ISBN: 978-3-319-64332-8
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)