Abstract
Tumor metastasis is the process by which tumor cells disseminate from tumors and enter nearby and distant microenvironments for new colonization. Bif-1 (BAX-interacting factor 1), which has a BAR domain and an SH3 domain, has been reported to be involved in cell growth, apoptosis and autophagy. However, the influence of Bif-1 on metastasis has been less studied. To understand the role of Bif-1 in metastasis, we studied the expression levels of Bif-1 in human HCC specimens using immunohistochemistry, a tissue microarray and quantitative PCR. The function of Bif-1 was assessed in migration and translocation assays and the pulmonary metastatic animal model. The relationship between Bif-1 and the Rho family was determined using immunoblot analyses and chromatin immunoprecipitation. The results showed that the expression of Bif-1 was higher in hepatocellular carcinoma (HCC) than matched adjacent non-tumor liver tissues. Increased Bif-1 expression was associated with tumor size and the intercellular spread and metastasis of HCC. Analysis of the relationship between Bif-1 expression and patients’ clinical characteristics revealed that patients with higher levels of Bif-1 had shorter disease-free and overall survival rates. Knockdown of Bif-1 with RNAi suppressed the migration of HCC cells and pulmonary metastasis and decreased the expression of Cdc42, a member of the Rho family. Bif-1 localized to the cytosol and nucleus and interacted with the promoter transcription region of Cdc42, which may regulate Cdc42 expression. Our results demonstrate a novel role of Bif-1 in HCC, in which Bif-1 promotes cell metastasis by regulating Cdc42 expression and activity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Why tumor cells undergo metastasis has puzzled clinical and scientific researchers for many years, and tumor metastasis is still a leading cause of patient mortality. Metastasis occurs in two stages: cancer cells disseminate from the primary tumor site to the microenvironment of distal tissues, and new clones are formed in a remote site [1, 2]. In this process, metastatic tumor cells are regulated by the following factors: (1) Migration-related chemotactic factors; (2) cell surface receptors related to the chemotactic factors; (3) activation of cytoskeletal proteins and their regulatory systems, including the Rho family and its regulatory factors; (4) epithelial-mesenchymal transition (EMT).
The endophilin family has five members in two categories (endophilin A and B). Bif-1 (endophilin B1 or SH3GLB1) belongs to the endophilin B group. The N-terminal region contains a BAR (Bin/Amphiphysin/Rvs) domain, and the C-terminal region has an SH3 (Src homology 3) domain. This protein was originally identified as a BAX-binding protein that alters the conformation of BAX to stimulate apoptosis [3]. Bif-1 interacts with the cytoplasmic membrane through helical regions in the BAR domain. Wang et al. [4] found that Bif-1 binds to Beclin 1 via UVRAG and forms a complex that regulates autophagy. In response to nutritional deprivation, Bif-1 forms autophagosomes with LC3 and Atg5 and enriches Atg9 in these vesicles, regulating autophagosome formation [5]. Bif-1 regulates the activation of PI3K and Vps34 by interacting with UVRAG and Beclin 1 [4]. Knockdown of Bif-1 in HeLa cell lines and mouse embryonic fibroblasts significantly inhibited the activity of PI3K in the formation of the autophagosome, delayed EGFR endocytosis and degradation, and maintained the sustained activation of Erk1/2, increasing the chemotaxis of breast cancer cells in response to EGF or serum [6]. However, human HCC cells were not shown to respond to stimulation with EGF in our chemotaxis assay, suggesting that Bif-1 may regulate the migration of HCC via other mechanisms [7].
Increasing evidence has shown that Bif-1 is present in various tumors, such as colorectal cancer [8], gallbladder cancer [9], pancreatic cancer [10], breast cancer [6, 11], bladder cancer [9], prostate cancer [12], stomach cancer [13], HCC [14] and Merkel cell tumors [15]. Bif-1 has different structures and functions in different tissues. HCC is one of the most lethal malignancies. We found that Bif-1 expression was increased in human HCC samples. Given that Bif-1 is activated through the BAR domain and is tethered to the membrane by the SH3 domain in conjunction with receptor tyrosine kinase phosphorylation, we speculated that Bif-1 may be involved in the progression and development of HCC.
In this study, we found that the expression of Bif-1 increased in HCC and was significantly associated with tumor size and HCC intercellular spread and remote metastasis in a large number of clinical samples. Inhibition of Bif-1 reduced cell metastasis and invasion in vitro and in vivo. Bif-1-mediated migration was associated with the expression and activity of the Cdc42 protein. Therefore, our results provide new evidence indicating that Bif-1 promotes the metastasis of HCC via Cdc42.
Methods and materials
Cell lines and culture
The HepG2 HCC cell line was obtained from the Shanghai Cell Bank, LM3 cells were obtained from the Zhongshan Hospital, Fudan University, Medical University, and the HCC cell line CSQT2 was derived from an HCC portal vein tumor thrombus in our laboratory [16]. Lentiviral pCSII-H1-PGK-puro-WPRE-shRNA-Bif-1 and its control viral vector were prepared by Hanbio Technology (Shanghai, China) and used to generate the stable Bif-1 knockdown cell line LM3-shBif-1 and the control cell line LM3-shCON. Cells were routinely cultured in DMEM (Invitrogen) supplemented with 10 % fetal bovine serum (FBS; Life Technologies) in a humidified incubator containing 5 % CO2 at 37 °C.
Patients, samples, and Immunohistochemistry
HCC samples from tumor patients were obtained during resection. Written consent was obtained before the operation. The protocol for this study was approved by the Ethics Committee of Eastern Hepatobiliary Surgery Hospital. Fifty pairs of HCC samples (with tumor and the adjacent non-tumor tissues) were used to measure Bif-1 expression. Sixty-nine pairs of tumor samples were used for tissue arrays that were analyzed to determine Bif-1 expression using immunohistochemistry. Twenty-three HCC samples were assessed to determine the relationship between Bif-1 (Novus Biologicals, #NBP2-24733) and Cdc42 (Santa Cruz, sc-8401). For immunohistochemistry, slides were deparaffinized in xylene, rehydrated in an alcohol series, and then processed for antigen retrieval. Samples were blocked with 1 % BSA for 30 min at 37 °C and then incubated with primary antibodies against Bif-1 (1:50) at 4 °C overnight. Sections were subsequently incubated with horseradish peroxidase (HRP)-labeled anti-mouse immunoglobulin for 30 min, followed by incubation with freshly prepared 3, 3ˈ-diaminobenzidine (DAB) solution at room temperature. Finally, the sections were counterstained with hematoxylin and observed under an Olympus fluorescence microscope. To determine the levels of Bif-1 in the tissue arrays, three individuals without prior knowledge of the clinical data determined the Bif-1 scores based on a 12-point system, as previously reported [17]. The median scores were chosen as the cut-off for high and low Bif-1 expression levels, which were used to analyze overall survival using the log-rank test, as described previously.
Real-time PCR
Total RNA was extracted from tumor and the adjacent non-tumor tissues using TRIzol solution (Invitrogen), and cDNA was synthesized with M-MLV Reverse Transcriptase (Promega). Gene primers were synthesized by Sangon Biotech (Shanghai) and are as follows: Bif-1: Forward: 5′-CGCTGTCTGAATGACTTTGT-3′, Reverse: 5′-CCTTTCTGCTGCCACTACAC-3′; 18S: Forward: 5′-CGGCTACCACATCCAAGGAA-3′, Reverse: 5′-GCTGGAATTACCGCGGCT-3′; Cdc42: Forward: 5′-GGCGATGCTGTTGGTAA -3′, Reverse: 5′-GCGGTCGTAATCTGTCATAATCCT-3′. Real-time PCR was performed using SYBR Green PCR Master Mix (TaKaRa Biotechnology Corporation) and carried out in an ABI-7300 system (Applied Biosystems).
Proliferation assay using cell count kit 8
Cells were harvested after incubation for 24, 48, or 72 h, and proliferation was detected with a CCK8 kit according to the instructions (Dojindo Laboratories).
Briefly, stable cell lines (1 × 105/ml) were seeded into 96-well plates, and growth was detected at the indicated time. For measurement, a mixture of 10 μl reagent and 90 μl DMEM was added to each well after the wells were rinsed with PBS once for the removal of dead cells. The reaction was performed at 37°C for 1 h, and cell viabilities were measured at 450 nm with the BioTek Gen5 system (BioTek, USA). The data are expressed as the mean ± SD.
Western blot
Total cell lysates were separated by SDS-PAGE and then transferred to nitrocellulose membranes. The membranes were blocked with 5 % milk in TBS for 1 h and probed with a primary antibody at 4 °C overnight. The membranes were then incubated with secondary antibodies for 1 h at room temperature. After washing with TBST, the membranes were scanned with an Odyssey system. The primary antibodies used in this study were anti-Bif-1 (Novus Biologicals, #NBP2-24733), anti-GAPDH (Proteintech, #60004-1-Ig), anti-Cdc42 (Santa Cruz, sc-8401), anti-RhoA (CST, #8820), anti-RhoB (CST, #8820), anti-RhoC (CST, #8820), anti-Twist (Santa Cruz, sc-81417), anti-MMP2 (CST, #13132), anti-MMP9 (CST, #13667), anti- E-cadherin (CST, #14473), anti-N-cadherin (CST, #14215), anti-vimentin (CST, #12826), anti-Snail (CST, #3879), anti-histone 3 (CST, #4499) and anti-HA (Santa Cruz, #7392). The secondary antibodies used were anti-rabbit and anti-mouse secondary antibodies (Santa Cruz, sc-2005, sc-2004).
Wound-healing assay
Cells were plated in 12-well plates overnight and then scraped with a 200 μl plastic pipette tip. Floating cells were removed by rinsing with 1X PBS. Cells were cultured at 37°C for 3 days, and the edge changes were monitored with an inverted light microscope (Olympus IX70) every day.
In vivo pulmonary metastasis assay
Five-week-old male BALB/c nude mice were obtained from the SLAC Laboratory Animal Company (Shanghai) and maintained in an animal facility with a constant temperature and a 12-h light/12-h dark cycle. Five mice in each group were injected with 1 × 106 cells through the caudal vein and sacrificed after 90 days. The mouse lungs were isolated and observed for the metastasis of tumor cells after H&E staining. The protocol for animal experiments was approved by the Ethics Committee of the Eastern Hepatobiliary Surgery Hospital.
Cell migration and invasion assay
Cells were placed onto transwell filters (Costar, Corning, NY) or BioCoat Matrigel invasion chambers (BD Biosciences) according to the manufacturer’s instructions. After a 24 h incubation, filters were fixed with 1 % crystal violet. Cells on the migrated sides in six random microscopic fields were counted. The average number of cells was expressed as the mean ± SD. These experiments were independently repeated at least three times.
Phalloidin staining
Cells were seeded in glass plates and cultured in DMEM overnight. Then, cell motility assays were performed with cells treated with or without 10 % FBS/DMEM for 6 h. At the end of the test, the cells were washed 3 times with PBS and fixed with 4 % paraformaldehyde/PBS followed by 1 % Triton X-100 for 4 min for permeabilization. Goat serum/PBS (3 %) was added to block non-specific binding. Then, phalloidin (1:40) and Hoechst were diluted in PBS and added to the well for 20 min. Images were taken with a Leica confocal microscope.
Cdc42 activity assay
Active Cdc42 was detected using an Active Cdc42 Detection Kit (CST) according to the manufacturer’s protocol. The cells were starved overnight in serum-free DMEM, washed with chilled PBS, and then lysed with cell lysis buffer containing PMSF. The supernatant was collected, GDP and GTPγS were added, and the solution was mixed at room temperature. The reaction was terminated with MgCl2, and GST-PAK1-PBD and glutathione resin were added to the samples and incubated at 4 °C for 40 min to allow GTP-bound GTPase to bind GST-PAK1-PBD. After removing the unbound proteins by centrifugation, GTP-bound GTPases were eluted, mixed with 30 μl of SDS buffer, resolved with SDS-PAGE and visualized using Western blot analysis.
Fractionation assay
Cells were washed once with cold PBS and scraped and collected in lysis buffer (20 mM Tris, 150 mM NaCl, 2.5 mM EDTA, 1 mM EGTA, 1 mM benzamidine, 4 mM leupeptin, 0.5 mM PMSF, 1 mM microcystin and 1 mM DTT at pH 7.5). After incubation for 20 min at 4°C, homogenization was performed. The nuclear pellets were collected by centrifugation for 2 min at 1000 g at 4°C. The supernatants were used for cytoplasmic and membrane fragmentation through ultracentrifugation at 100,000 g for 60 min at 4°C. Equivalent concentrations of nuclear, cytoplasmic and membrane fractions from each group were analyzed with Western blots.
Chromatin immunoprecipitation assay
Cells were processed for chromatin immunoprecipitation (ChIP) assays using a chromatin immunoprecipitation assay kit (Millipore) according to the manufacturer’s protocol. Briefly, the cells were cross-linked with 1 % formaldehyde for 10 min at 37 °C and lysed with sodium dodecyl sulfate lysis buffer. The pellets in the lysate were sonicated with a sonic dismembrator (Fisher Scientific). Protein-DNA complexes were immunoprecipitated with anti-Bif-1. The immunoprecipitates were dissolved in 20 μl distilled water for PCR analysis. Standard PCR amplification was performed using Taq PCR Master Mix (Takara) with 15 specifically designed pairs of primers targeting a region more than 2400 bp upstream of the transcription initiation site at 100–200 bp intervals. The primers are listed in Supplementary Table 1.
Statistical analysis
Statistical analyses were performed using SPSS 18.0 software. Differences among variables were analyzed for statistical significance using two-tailed Student’s t-tests, unpaired t tests, or X2 analysis. Kaplan–Meier analysis was used to assess survival. The data are presented as the mean ± SD unless otherwise indicated. P < 0.05 was considered to be statistically significant.
Results
Expression of Bif-1 in human HCC tissues is associated with poor prognosis
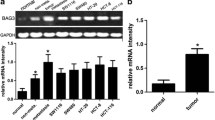
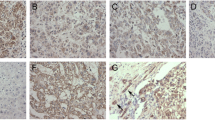
To elucidate the role of Bif-1 in HCC progression, we first determined the mRNA levels of Bif-1 in 50 paired human HCC samples with quantitative real-time polymerase chain reaction. In 84.0 % (42/50) of the samples, Bif-1 expression was higher in the HCC tissues than the adjacent non-tumor tissues (Fig. 1a). Similar results were found in Western blot assays (Fig. 1b). To determine the significance of Bif-1 in HCC, we further analyzed the relationship between Bif-1 and clinicopathological characteristics in a tissue array with 69 paired HCC samples and obtained similar results (Fig. 1c, d). Based on the median Bif-1 scores in these samples, patients were divided into two groups: the high expression group (n = 43) and the low expression group (n = 26). After statistical analysis, high expression of Bif-1 was associated with adverse clinical characteristics (Table 1), particularly with a large tumor size (≥5 cm, P = 0.005), microvascular invasion (P = 0.023), and recurrence (P = 0.040). Patients with high levels of Bif-1 had poorer DFS and OS (Fig. 1e, f). These results suggest that Bif-1 might play an important role in tumor invasion and metastasis.
Bif-1 expression is increased in human HCC tissues. a The expression of Bif-1 mRNA in 50 pairs of HCC and the adjacent non-tumor tissues was evaluated with qPCR. b Western blots showed the expression of Bif-1 in HCC tissues (T) and adjacent non-tumor tissues (N). c Selective IHC staining of Bif-1 expression in HCC and adjacent non-tumor tissues (n = 69). Magnification: ×200, ×400. d The Bif-1 expression score in 69 pairs of samples stained for IHC. ***P < 0.001 based on Student t-test. e, f The DFS and OS of 69 HCC patients with low or high Bif-1 expression
Down-regulation of Bif-1 reduces migration in HCC cells
To determine how Bif-1 affects metastasis, we designed three types of RNAi to silence Bif-1. All of them inhibited Bif-1 at both the mRNA level and the protein level (Fig. 2a, b). We used shBif-1A to prepare the lentivirus and established stable human HCC cell lines (HCC-LM3) expressing Blank and Bif-1-shRNA by pooling strongly knocked down cells from three wells. We named the cell lines LM3-shCON and LM3-shBif-1. A proliferation assay showed that knockdown of Bif-1 attenuated the proliferation of HCC-LM3 cells compared with control cells (Fig. 2c). However, there was no statistically significant difference between these two cell lines. Subsequently, wound-healing assays were performed with these cells. Silencing Bif-1 expression decreased cell proliferation (Fig. 2d), migration (Fig. 2e) and invasion (Fig. 2f) compared with LM3-shCON cells. LM3-shCON and LM3-shBif-1 cells were injected into the lateral tail vein of nude mice for a pulmonary metastatic in vivo assay, and we found that fewer and smaller nodules were formed in the lungs of the mice injected with LM3-shBif-1 than the mice injected with LM3-shCON cells (Fig. 2g). Additionally, the mice injected with LM3-shBif-1 cells showed higher survival than the mice injected with control cells (Fig. 2h, P < 0.05). The results demonstrate that Bif-1 promotes tumor metastasis.
Silencing the expression of Bif-1 inhibits the metastasis of HCC cells both in vitro and in vivo. a, b Screening of shRNA targeting Bif-1. ShBif-1A, B and C were transfected into cells. QPCR and Western blots were carried out to determine Bif-1 gene and protein expression. c The cell growth status of two stable cell lines (shCon and shBif-1) was detected with a cell counting kit 8 at the indicated time. d The migratory properties of LM3-shBif-1 cells and control cells were determined with the scratch wound-healing assay. Magnification: ×100. e The migratory properties of the cells were analyzed with transwell migration assays. The average number of migrated cells was determined from five random microscopic fields. f The invasive properties of the cell lines were analyzed with an invasion assay using BioCoat Matrigel invasion chambers. The average number of invasive cells was counted in 5 random microscopic fields. g Representative H&E images of mouse lung tissue sections from the LM3-shCON and LM3-shBif-1 groups (magnification: ×100). The number of the lung metastatic lesions in the LM3-shCON or LM3-shBif-1 group is presented as the mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001 (based on Student’s t test). h The cumulative survival of mice injected with LM3-shCON and LM3-shBif-1 cells. P values were analyzed using a 2-sided log-rank test
Bif-1 promotes cytoskeletal reorganization and cell polarity
EMT is associated with tumor metastasis. To determine how Bif-1 promotes the metastasis of hepatocellular carcinoma, we assessed the expression of EMT-related genes, such MMP2, MMP9, vimentin, Twist2, E-cadherin, N-cadherin, and Snail, in these cell lines. The results showed that there were no differences in the expression of these genes in these two types of cells (Fig. 3a, b). When observing cytoskeletal reorganization in these cells, we found that shBif-1-expressing LM3 cells showed lower actin responses regardless of whether they were cultured in medium with or without serum (Fig. 3c). Meanwhile, control cells polarized in response to serum stimulation, but knockdown of Bif-1 reduced polarization (Fig. 3d). These results suggest that Bif-1 may be involved in the regulation of cell polarization, which is important for directional migration.
Bif-1 promotes cytoskeleton reorganization and cell polarity. a LM3-shCON and LM3-shBif-1 cells were analyzed with antibodies as indicated in Western blots. b LM3-shCON and LM3-shBif-1 cells were analyzed with antibodies as indicated in the Western blots. c, d LM3-shCON and LM3-shBif-1 cells were cultured with or without serum. Cell morphology (c) and polarization (d) were observed with confocal laser scanning microscopy followed by phalloidin staining
Bif-1-mediated polarization is associated with Cdc42 expression and activity
The Rho family has been reported to modulate polarization and cytoskeletal organization. Therefore, we assessed the expression of the Rho family (RhoA, RhoB, RhoC, and Cdc42). As shown in Fig. 4a, the expression of RhoA, RhoB and RhoC did not differ in LM3-shCON and LM3-shBif-1 cells. However, the mRNA and protein levels of Cdc42 were lower in LM3-shBif-1 cells (Fig. 4a, b). Active Cdc42 was also identified with a CBD pulldown assay (Fig. 4c). The results suggest that the reduction in cell migration may be attributed to the expression and activity of Cdc42. To assess this hypothesis, we transfected either wild-type Cdc42 (pEF1-Cdc42) or a control vector into LM3-shBif-1 cells and found that the ectopic expression of Cdc42 in LM3-shBif-1 cells apparently restored migration in response to serum stimulation (Fig. 4d). To identify the effect of Bif-1 on Cdc42 expression and activity, we expressed wild-type Bif-1 in LM3 cells for a migration assay. The results showed that Bif-1 expression greatly increased the migration of LM3 cells (Fig. 4e, f). Taken together, these results suggest that Bif-1 may control cell migration by regulating Cdc42 expression and activity.
Bif-1 modulates the expression and activity of Cdc42. a The levels of Rho family proteins (RhoA, RhoB, RhoC, and Cdc42) were detected with Western blots. b The mRNA levels of Bif-1 and Cdc42 in LM3-shBif-1 cells and LM3-shCON cells were evaluated with qPCR. c Active Cdc42 proteins were analyzed with CBD pulldown assays. d LM3-shBif-1 cells and LM3-shCON cells were transfected with pEF1-Cdc42 or empty vector (pEF1). After incubation for 24 h, cells were subjected to the transwell migration assay. Migrated cells were counted in 5 random microscopic fields, and the results were expressed as the mean ± SD (right side). *P < 0.05. e Overexpression of Bif-1 in LM3 cells. Cdc42 expression was increased, as determined with Western blots. f The forced expression of Bif-1 promoted cell migration in the transwell migration assay. The average number of migrated cells was counted in 5 random microscopic fields. *P < 0.05
To elucidate the relationship between Bif-1 and Cdc42, we studied their expression in HCC specimens. Based on the levels of Bif-1 and Cdc42 in 23 HCC samples (Fig. 5a), we analyzed the association between Bif-1 and Cdc42 with a regressive curve. The results showed a significant association between Bif-1 and Cdc42 expression (r = 0.886, P < 0.01) (Fig. 5b). To determine how Bif-1 regulates cell migration via Cdc42, we performed immunofluorescence assays for both Bif-1 and Cdc42 in LM3 and CSQT2 cells. CSQT2 is a cell line derived from a portal vein tumor thrombosis. Bif-1 was shown to colocalize with Cdc42 at the leading edge of cells (Fig. 5c). These results suggest that Bif-1 may interact with Cdc42 and coordinate cell motility.
Bif-1 expression is positively related to Cdc42 expression, and Bif-1 and Cdc42 colocalize in cells. a The expression of Bif-1 and Cdc42 mRNA in 23 HCC tissues was evaluated using qPCR. b The relationship between Bif-1 and Cdc42 mRNA levels in 23 HCC tissues was analyzed by SPSS 18.0 software with a 2-sided correlation analysis. r = 0.886, P < 0.01. c The location of Bif-1 and Cdc42 in cells was detected by immunofluorescence assays, and Bif-1 and Cdc42 were shown to colocalize in LM3 and CSQT2 cells
Bif-1 is expressed in the nucleus and regulates transcription of Cdc42
Using an immunostaining assay, Bif-1 was found in the nucleus of both HepG2 and CSQT2 cells after culturing the cells in low-glucose medium. To confirm these results, we performed subcellular fractionation on three types of cells (HepG2, CSQT2 and LM3). As shown in Fig. 6a, Bif-1 was localized in the nucleus. To validate the distribution of Bif-1, we carried out subcellular fractionation with three cell lines (HepG2, CSQT2 and LM3). GAPDH and histone 3 served as endogenous markers of the cytoplasm and nucleus, respectively. The results showed that Bif-1 was present in both the cytosol and the nucleus (Fig. 6b). Because Bif-1 increased the expression of Cdc42, we examined whether Bif-1 regulated the promoter of Cdc42. The nuclear lysates of LM3 cells were precipitated with a Bif-1 antibody in a ChIP assay. The pellets were washed thoroughly and used for amplification of the Cdc42 promoter with 15 pairs of primers targeting the promoter (Supplemental Table 1). The interaction of Bif-1 with the Cdc42 promoter was confirmed by the positive band amplified from the precipitated pellets (Fig. 6c). These results suggest that Bif-1 may regulate the expression of Cdc42 via activation of the Cdc42 promoter.
Bif-1 is expressed in the nucleus and interacts with the promoter region of Cdc42. a Fluorescence staining of Bif-1 in HepG2 and CSQT2 cells showed the nuclear translocation of Bif-1. b Subcellular fragmentation assays confirmed the distribution and translocation of Bif-1 in the nucleus and cytoplasm. C cytosol, N nucleus. c Chromatin immunoprecipitation (ChIP) assays showed that Bif-1 interacts with the promoter region of Cdc42
Discussion
Studies have shown that Bif-1 plays an important role in autophagy and apoptosis in many types of cells. However, the roles of Bif-1 in cancer are less studied. In this study, Bif-1 was explored in HCC. We found that Bif-1 expression was frequently up-regulated in clinical HCC samples and invasive HCC cell lines. High expression was associated with tumor size, microvascular invasion and recurrence. Bif-1-mediated migration and invasion were found to occur via the expression and activity of Cdc42. Therefore, our data demonstrate a new mechanism of Bif-1 in promoting the metastasis of HCC.
The expression of Bif-1 in tumors is dependent on the tissue types. Low Bif-1 expression has been found in gastric cancer [13], colorectal adenocarcinoma [18], pancreatic ductal adenocarcinoma [19] and urinary bladder and gallbladder cancer [9]. In contrast, high Bif-1 expression has been reported in prostate cancer [12], HCC [14] and Merkel cell carcinoma [15]. Our results demonstrate that Bif-1 expression increased by 60 % in 142 HCC cases. Therefore, the tissue-specific expression of Bif-1 may reflect its capacity in particular roles.
Etxebarria et al. [20] showed that Bif-1 activated BAX proteins by promoting independent morphological rearrangement. The IFNγ-induced expression of immunity-related GTPase family member 1 (Irgm1) promoted melanoma tumorigenesis via the dual regulation of apoptosis and Bif-1-dependent autophagy [21]. In our study, we showed that Bif-1 was located in both the cytosol and the nucleus. Bif-1 interacts with the promoter of Cdc42, which may be the mechanism by which Bif-1 regulates the expression of Cdc42, which is responsible for actin polymerization and cell migration. Because Bif-1 interacts with Beclin 1, Vps34, and Bcl-2 and regulates autophagy [22–28], targeting Bif-1 may be a new target for inhibiting metastasis by interfering with autophagy, apoptosis [29] and migration.
Rho family proteins have important roles in cell polarity changes and movement. In 1999, Genda et al. [30] found that Rho (Rho/p160ROCK) and its related proteins had a critical effect on human hepatocellular carcinoma and its early intrahepatic metastasis. In subsequent studies, Rho family proteins were also shown to play an important role in breast cancer [31], bladder cancer [32], pancreatic cancer [33, 34], and distant metastasis [35]. Many studies have confirmed that Rho-mediated signaling is involved in various tumor metastases. Li et al. found that chemokines regulate the molecular mechanism of cell polarity and directional movement through the Gβγ-PAK1-Cdc42 signaling pathway [36], and Rho family proteins regulate cell movement via PTEN [37, 38]. In prostate cancer, RhoC promotes tumor metastasis through the sequential activation of Pyk2, FAK, MAPK, and Akt, followed by the up-regulation of MMP2 and MMP9 [39]. Our findings reveal that knockdown of Bif-1 reduced actin stress and cell motility via Cdc42, supporting the conclusion that Bif-1 is an important factor in the metastasis of HCC.
In summary, high Bif-1 expression in HCC is associated with metastasis and poor prognosis. Knockdown or expression of Bif-1 in HCC cells demonstrated its roles in tumor metastasis in a cell migration assay and a pulmonary metastatic animal model. Bif-1-mediated migration is likely dependent on the expression and activity of Cdc42. Therefore, targeting Bif-1 may be a new therapeutic approach for inhibiting HCC metastasis.
Abbreviations
- HCC:
-
Hepatocellular carcinoma
- Bif-1:
-
BAX-interacting factor 1
- EMT:
-
Epithelial-mesenchymal transition
- BAR:
-
Bin/Amphiphysin/Rvs
- SH3:
-
Src homology 3
- Irgm1:
-
Immunity-related GTPase family member 1
- OS:
-
Overall survival
- DFS:
-
Disease-free survival
References
Hendrix MJ, Seftor EA, Seftor RE, Kasemeier-Kulesa J, Kulesa PM, Postovit LM (2007) Reprogramming metastatic tumour cells with embryonic microenvironments. Nat Rev Cancer 7(4):246–255
Thiery JP, Acloque H, Huang RY, Nieto MA (2009) Epithelial-mesenchymal transitions in development and disease. Cell 139(5):871–890
Cuddeback SM, Yamaguchi H, Komatsu K, Miyashita T, Yamada M, Wu C, Singh S, Wang HG (2001) Molecular cloning and characterization of Bif-1. A novel Src homology 3 domain-containing protein that associates with Bax. J Biol Chem 276(23):20559–20565
Takahashi Y, Coppola D, Matsushita N, Cualing HD, Sun M, Sato Y, Liang C, Jung JU, Cheng JQ, Mule JJ, Pledger WJ, Wang HG (2007) Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol 9(10):1142–1151
Takahashi Y, Meyerkord CL, Hori T, Runkle K, Fox TE, Kester M, Loughran TP, Wang HG (2011) Bif-1 regulates Atg9 trafficking by mediating the fission of Golgi membranes during autophagy. Autophagy 7(1):61–73
Runkle KB, Meyerkord CL, Desai NV, Takahashi Y, Wang HG (2012) Bif-1 suppresses breast cancer cell migration by promoting EGFR endocytic degradation. Cancer Biol Ther 13(10):956–966
Aramaki Y, Ogawa K, Toh Y, Ito T, Akimitsu N, Hamamoto H, Sekimizu K, Matsusue K, Kono A, Iguchi H, Takiguchi S (2005) Direct interaction between metastasis-associated protein 1 and endophilin 3. FEBS Lett 579(17):3731–3736
de Las Heras JI, Batrakou DG, Schirmer EC (2013) Cancer biology and the nuclear envelope: a convoluted relationship. Semin Cancer Biol 23(2):125–137
Kim SY, Oh YL, Kim KM, Jeong EG, Kim MS, Yoo NJ, Lee SH (2008) Decreased expression of Bax-interacting factor-1 (Bif-1) in invasive urinary bladder and gallbladder cancers. Pathology 40(6):553–557
Ko YH, Cho YS, Won HS, Jeon EK, An HJ, Hong SU, Park JH, Lee MA (2013) Prognostic significance of autophagy-related protein expression in resected pancreatic ductal adenocarcinoma. Pancreas 42(5):829–835
Ho J, Kong JW, Choong LY, Loh MC, Toy W, Chong PK, Wong CH, Wong CY, Shah N, Lim YP (2009) Novel breast cancer metastasis-associated proteins. J Proteome Res 8(2):583–594
Coppola D, Oliveri C, Sayegh Z, Boulware D, Takahashi Y, Pow-Sang J, Djeu JY, Wang HG (2008) Bax-interacting factor-1 expression in prostate cancer. Clin Genitourin Cancer 6(2):117–121
Lee JW, Jeong EG, Soung YH, Nam SW, Lee JY, Yoo NJ, Lee SH (2006) Decreased expression of tumour suppressor Bax-interacting factor-1 (Bif-1), a Bax activator, in gastric carcinomas. Pathology 38(4):312–315
Fan R, Miao Y, Shan X, Qian H, Song C, Wu G, Chen Y, Zha W (2012) Bif-1 is overexpressed in hepatocellular carcinoma and correlates with shortened patient survival. Oncol Lett 3(4):851–854
Schlauder SM, Calder KB, Khalil FK, Passmore L, Mathew RA, Morgan MB (2009) Bif-1 and Bax expression in cutaneous Merkel cell carcinoma. J Cutan Pathol 36(1):21–25
Wang T, Hu HS, Feng YX, Shi J, Li N, Guo WX, Xue J, Xie D, Liu SR, Wu MC, Cheng SQ (2010) Characterisation of a novel cell line (CSQT-2) with high metastatic activity derived from portal vein tumour thrombus of hepatocellular carcinoma. Br J Cancer 102(11):1618–1626
Han CP, Kok LF, Lee MY, Wu TS, Ruan A, Cheng YW, Wang PH, Koo CL, Tyan YS (2010) Five commonly used markers (p53, TTF1, CK7, CK20, and CK34betaE12) are of no use in distinguishing between primary endocervical and endometrial adenocarcinomas in a tissue microarray extension study. Arch Gynecol Obstet 281(2):317–323
Coppola D, Khalil F, Eschrich SA, Boulware D, Yeatman T, Wang HG (2008) Down-regulation of Bax-interacting factor-1 in colorectal adenocarcinoma. Cancer 113(10):2665–2670
Coppola D, Helm J, Ghayouri M, Malafa MP, Wang HG (2011) Down-regulation of Bax-interacting factor 1 in human pancreatic ductal adenocarcinoma. Pancreas 40(3):433–437
Etxebarria A, Terrones O, Yamaguchi H, Landajuela A, Landeta O, Antonsson B, Wang HG, Basanez G (2009) Endophilin B1/Bif-1 stimulates BAX activation independently from its capacity to produce large scale membrane morphological rearrangements. J Biol Chem 284(7):4200–4212
Dong H, Tian L, Li R, Pei C, Fu Y, Dong X, Xia F, Wang C, Li W, Guo X, Gu C, Li B, Liu A, Ren H, Xu H (2015) IFNg-induced Irgm1 promotes tumorigenesis of melanoma via dual regulation of apoptosis and Bif-1-dependent autophagy. Oncogene 34(42):5363–5371
Juhasz G, Hill JH, Yan Y, Sass M, Baehrecke EH, Backer JM, Neufeld TP (2008) The class III PI(3)K Vps34 promotes autophagy and endocytosis but not TOR signaling in Drosophila. J Cell Biol 181(4):655–666
Ku B, Woo JS, Liang C, Lee KH, Jung JU, Oh BH (2008) An insight into the mechanistic role of Beclin 1 and its inhibition by prosurvival Bcl-2 family proteins. Autophagy 4(4):519–520
Levine B, Sinha S, Kroemer G (2008) Bcl-2 family members: dual regulators of apoptosis and autophagy. Autophagy 4(5):600–606
Takahashi Y, Meyerkord CL, Wang HG (2008) BARgaining membranes for autophagosome formation: regulation of autophagy and tumorigenesis by Bif-1/Endophilin B1. Autophagy 4(1):121–124
Yamaguchi H, Woods NT, Dorsey JF, Takahashi Y, Gjertsen NR, Yeatman T, Wu J, Wang HG (2008) SRC directly phosphorylates Bif-1 and prevents its interaction with Bax and the initiation of anoikis. J Biol Chem 283(27):19112–19118
He S, Ni D, Ma B, Lee JH, Zhang T, Ghozalli I, Pirooz SD, Zhao Z, Bharatham N, Li B, Oh S, Lee WH, Takahashi Y, Wang HG, Minassian A, Feng P, Deretic V, Pepperkok R, Tagaya M, Yoon HS, Liang C (2013) PtdIns(3)P-bound UVRAG coordinates Golgi-ER retrograde and Atg9 transport by differential interactions with the ER tether and the beclin 1 complex. Nat Cell Biol 15(10):1206–1219
Khan MM, Strack S, Wild F, Hanashima A, Gasch A, Brohm K, Reischl M, Carnio S, Labeit D, Sandri M, Labeit S, Rudolf R (2014) Role of autophagy, SQSTM1, SH3GLB1, and TRIM63 in the turnover of nicotinic acetylcholine receptors. Autophagy 10(1):123–136
Xie T, Li SJ, Guo MR, Wu Y, Wang HY, Zhang K, Zhang X, Ouyang L, Liu J (2015) Untangling knots between autophagic targets and candidate drugs, in cancer therapy. Cell Prolif 48(2):119–139
Genda T, Sakamoto M, Ichida T, Asakura H, Kojiro M, Narumiya S, Hirohashi S (1999) Cell motility mediated by rho and Rho-associated protein kinase plays a critical role in intrahepatic metastasis of human hepatocellular carcinoma. Hepatology 30(4):1027–1036
Liu S, Goldstein RH, Scepansky EM, Rosenblatt M (2009) Inhibition of rho-associated kinase signaling prevents breast cancer metastasis to human bone. Cancer Res 69(22):8742–8751
Kamai T, Tsujii T, Arai K, Takagi K, Asami H, Ito Y, Oshima H (2003) Significant association of Rho/ROCK pathway with invasion and metastasis of bladder cancer. Clin Cancer Res 9(7):2632–2641
Kusama T, Mukai M, Endo H, Ishikawa O, Tatsuta M, Nakamura H, Inoue M (2006) Inactivation of Rho GTPases by p190 RhoGAP reduces human pancreatic cancer cell invasion and metastasis. Cancer Sci 97(9):848–853
Li Z, Chang Z, Chiao LJ, Kang Y, Xia Q, Zhu C, Fleming JB, Evans DB, Chiao PJ (2009) TrkBT1 induces liver metastasis of pancreatic cancer cells by sequestering Rho GDP dissociation inhibitor and promoting RhoA activation. Cancer Res 69(19):7851–7859
Titus B, Schwartz MA, Theodorescu D (2005) Rho proteins in cell migration and metastasis. Crit Rev Eukaryot Gene Expr 15(2):103–114
Li Z, Hannigan M, Mo Z, Liu B, Lu W, Wu Y, Smrcka AV, Wu G, Li L, Liu M, Huang CK, Wu D (2003) Directional sensing requires G beta gamma-mediated PAK1 and PIX alpha-dependent activation of Cdc42. Cell 114(2):215–227
Meili R, Sasaki AT, Firtel RA (2005) Rho Rocks PTEN. Nat Cell Biol 7(4):334–335
Li Z, Dong X, Wang Z, Liu W, Deng N, Ding Y, Tang L, Hla T, Zeng R, Li L, Wu D (2005) Regulation of PTEN by Rho small GTPases. Nat Cell Biol 7(4):399–404
Iiizumi M, Bandyopadhyay S, Pai SK, Watabe M, Hirota S, Hosobe S, Tsukada T, Miura K, Saito K, Furuta E, Liu W, Xing F, Okuda H, Kobayashi A, Watabe K (2008) RhoC promotes metastasis via activation of the Pyk2 pathway in prostate cancer. Cancer Res 68(18):7613–7620
Acknowledgments
We thank Huanlin Sun, Dongping Hu, Liang Tang, LinnaGuo, and Shanna Huang for their technical assistance. This work was supported by Grants from the National Natural Science Foundation of China (81221061, 81272427 and 31471339); The Ministry of Science and Technology Key Program (2012ZX10002-009, -011); The State Key Project on Diseases of China (2012zx10002016016003); The China National Funds for Distinguished Young Scientists (81125018); the Chang Jiang Scholars Program (2013) of the China Ministry of Education; and The National Key Basic Research Program “973 project” (2015CB554000).
Authors’ contributions
ZL, HYW, SQC and CZZ conceived the study. ZL, CZZ, and FHL designed and coordinated the study. CZZ and FHL performed most experiments and the statistical analysis. HYC and NL performed several of the cellular experiments. ZLL, DDH, WXG and SHT provided the clinical parameters and survival information. HGW provided a critical review. CZZ and ZL drafted the manuscript. ZL, SQC and HYW provided funds and revised the manuscript. All authors read and approved the final manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Cunzhen Zhang and Fenghua Liu have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, C., Liu, F., Chen, H. et al. Bif-1 promotes tumor cell migration and metastasis via Cdc42 expression and activity. Clin Exp Metastasis 34, 11–23 (2017). https://doi.org/10.1007/s10585-016-9825-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10585-016-9825-7