Abstract
Hepatocellular carcinoma (HCC) is highly metastatic and invasive. CircRNA participates in gene regulation of multiple tumor metastases, but little is known whether it is a bystander or an actual player in HCC metastasis. We aim to explore the molecular mechanisms of novel circRNAs in HCC metastasis. RT-qPCR was used to detect the expression of 13 circRNAs derived by the ERBB3 gene. The function of circ_0098823 and DNM1L in HCC cells were estimated by CCK-8, transwell assays, flow cytometry, electron microscope, and in vivo experiments. RNA binding protein of circ_0098823 was confirmed by RNA pull-down, mass spectrometry, and RNA immunoprecipitation. The expression of DNM1L after IGF2BP3 knockdown was detected by RT-qPCR and western blot. Circ_0098823 was significantly up-regulated both in HCC tissues and HGF induced cell lines. Circ_0098823 overexpression significantly enhanced proliferation, migration, and invasion but decreased apoptosis of HCC cells, particularly promoted mitochondrial fission. Compared with the control group, the tumors in the circ_0098823 knockdown mice were significantly smaller and lighter. Circ_0098823 silencing suppressed DNM1L expression, a key molecule for fission, which enhanced proliferation, migration and invasion, and inhibited apoptosis of HCC cell. IGF2BP3 was a binding protein of circ_0098823. The expression and mRNA stability of DNM1L were down-regulated by IGF2BP3 knockdown. IGF2BP3 knockdown significantly alleviated the excessive migration, invasion and apoptosis of HCC cells caused by circ_0098823 overexpression. This study uncovered a novel circ_0098823 with tumor-promoting effect, and the mechanism by which circ_0098823 participates in HCC progression through IGF2BP3-guided DNM1L. Our study broadens molecular understanding of HCC progression.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hepatocellular carcinoma (HCC) is a common and highly malignant tumor worldwide. A study by Hyuna Sung PhD in 2020 showed that the incidence of HCC in East Asia was 26.8%, ranking first [1]. It has the third highest mortality in the world, with more than 600,000 people dying all over the world from hepatocellular carcinoma every year [2]. The high mortality of HCC in China ranks first in the world, accounting for 50% of the total number of patients worldwide, posing a serious threat to the life and health of Chinese people [3]. Up to now, the main therapeutic methods for HCC include hepatectomy, liver transplantation [4], interventional embolization, radiofrequency ablation, systemic therapy (including targeted therapy, targeted combined immunotherapy, chemotherapy) [5], and radiotherapy (stereotactic body radiotherapy, SBRT) [6]. Although these ways could improve the poor prognosis of HCC to some extent, the overall prognosis for HCC patients remains dismal primarily. The root cause is the high recurrence rate and high metastasis. Therefore, it is important to explore the molecular mechanisms and therapeutic targets of HCC metastasis.
Circular RNA (circRNA) is an endogenous non-coding RNA with a closed–loop structure, which has tissue-specific expression. Accompanied by a covalent closed loop structure and a lack of 3' and 5' segments, circRNA has a more stable and anti-decay mechanism than linear RNA [7]. CircRNA has been involved in various biological regulation of tumor microenvironments (TME) by its abundance [8,9,10], stability [11, 12], conservation [13], and specificity [14, 15]. For instance, Sun et al. [16] identified circ_0000105 was up-regulated in liver cancer tissues and cell lines compared with tumor-adjacent tissues. Circ_002136 blocked miR-19a-3p expression, elevated RAB1A expression activity and stimulated HCC development [17]. In mechanism, circRNA can not only act as a sponging factor of miRNA, but also bind to RNA binding protein (RBP) to regulate splicing patterns or mRNA stability. For example, Wang et al. [18] revealed that circRHOT1 inhibits the development and progression of HCC by initiating NR2F6 expression. Circular RNA MTCL1 inhibits the degradation of C1QBP ubiquitin, thus promoting the development of advanced laryngeal cancer [19]. circRNA CCNB1 has been reported to bind and stabilize the pathway of TJP1 mRNA, inhibiting the invasion and migration of nasopharyngeal carcinoma [20]. However, the regulatory mechanism of circRNAs’ involvement in HCC progression in combination with RBP is not comprehensive enough.
Mitochondria are semi-autonomous organelles. It not only supplied energy to cells but also participated in lots of biological processes such as apoptosis, differentiation, and information transmission. There exist two opposing processes named fusion and fission precisely regulating the mitochondrial network [21]. Mitochondrial dynamics is the basic basis for mitochondrial quality control in tissues [21]. Notably, DNM1L which can encode members of the dynamin superfamily of GTPases is the central mediator of mitochondrial fission and fusion [22]. In glioblastoma, DNM1L-mediated mitochondrial fission plays a mediating role and enhances the promotion effect of BCL2L13 on mitochondrial autophagy [23]. In enhancing chemotherapy sensitivity of breast cancer cells, DNM1L-mediated mitochondrial fission acts as a bridge of communication [24]. Furthermore, the abnormal mitochondrial structure in mice lacking DNM1L further indicated the relationship between DNM1L and mitochondria [25]. Accumulative evidences are going to reveal dysregulation of DNM1L in cancers of the lung, and HCC [26,27,28]. However, the mechanism of DNM1L-mediated mitochondrial fission and postoperative invasion and metastasis of HCC cells is still unclear.
Hepatocyte growth factor (HGF), synthesized and released by liver regeneration after hepatectomy, can change the tumor microenvironment and induce invasion and metastasis of liver cancer [29]. Our research group previously found that after HCC resection, liver parenchymal cells synthesized and released a large amount of HGF, and the expression of ERBB3 in HCC cells was significantly increased under the stimulation of HGF, and in vitro and in vivo experiments verified that the increase of ERBB3 promoted the invasion and metastasis of HCC cells [30]. Based on these, we speculated that ERBB3-derived circERBB3 transcription increased after liver cancer surgery and was involved in the regulation of invasion and metastasis of liver cancer cells. In our study, we identified the expression of 13 circRNAs derived from ERBB3 in HCCLM3 cells and determined circ_0098823 as the object of study. Subsequently, RNA pull-down and mass spectroscopy revealed the binding protein IGF2BP3 interacting with circ_0098823 and investigated the effect of circ_0098823 on tumor phenotypes by IGF2BP3 guiding DNM1L. Our study provides strong evidence to support a novel strategy for HCC therapeutic methods of targeting proteins of the mitochondrial fission mechanism.
Methods and materials
Patients and tissues
Tumor and adjacent normal tissues were provided by the Shanghai General Hospital of Shanghai Jiao Tong University School of Medicine. A total number of 10 pairs of HCC tissues and adjacent normal tissues were included. The informed consent was signed by all individuals before the experimental study. The patients information is shown in Supplementary Table 1. The study was approved by the Ethics Committee of the Shanghai General Hospital of Shanghai Jiao Tong University School of Medicine [2020SQ265].
Cell culture
The HCC cell line of HCCLM3 (iCell-h259) and hepatoblastoma cell line HepG2 (iCell-h092) was purchased by iCell Bioscience Inc (Shanghai, China). The HepG2 cells were inoculating in MEM (Minimum Essential Medium) supplemented with 10% heat-inactivated fetal bovine serum (FBS) and 1% Peptococcus Selective (PS). The HCCLM3 cells were cultured in Dulbecco’s Modified Eagle medium (DMEM) supplemented with 10% heat-inactivated FBS and 1% PS. HCCLM3 and HepG2 cells were maintained in a humidified atmosphere containing 5% CO2 at 37 °C.
Knockdown, forced expression of target genes, and cell transfection
For the knockdown of circ_0098823, siRNA-1 and siRNA-2 were introduced into HepG2 cells to knock down circ_0098823, and si-NC was introduced as the control group. Lentiviral plasmid pLVX-circERBB3-EGFP-IRES-Puro was transfected into HepG2 cells to stable knockdown circ_0098823. For the overexpression of circ_0098823, circ_0098823 was recombined into pcDNA plasmid to form circ_0098823 overexpression vector and transferred into the HCCLM3 cells. DNM1L knockdown was achieved by introducing siRNA-202, siRNA-479, and siRNA-760 in HCCLM3 and HepG2 cells. All of siRNA sequences were provided in Table S2. For the overexpression of DNM1L, DNM1L sequence was cloned into pcDNA3.1 vector. The pcDNA3.1-DNM1L vector was constructed and then introduced into HCCLM3 and HepG2 cells.
Lipofectamine™ 2000 (11,668,500) in this study provided by Invitrogen (China) were used for transfection. Cells were inoculated into six-well plates. When the fusion of cells reached 80–90%, the fresh medium was replaced. For the circ_0098823, diluted siRNA-1, siRNA-2 (100 nM) were transfected into HepG2 cells, and overexpression vector (100 nM) were transfected into HCCLM3 cells. For the DNM1L, diluted siRNA sequences (100 nM), and overexpression vector (100 nM) were transfected into HepG2 cells and HCCLM3 cells, respectively. OPTI-MEM was used to dilute plasmid. Subsequently, the diluted transfection reagent and plasmid were mixed and incubated at room temperature for 15 min. The incubated mixture was added to the cell sample, and the fresh medium was replaced 6 h later. The transfected cells could be used for follow-up experiments after 18 h culture in fresh medium.
Nucleocytoplasmic separation, RNA extraction and Real-time quantitative PCR (RT-qPCR)
HepG2 cells were divided into two parts after PBS resuspension, one part was used for total RNA extraction and the other part was used for nucleocytoplasmic separation. The isolated cytoplasm was obtained by cell fractionation buffer (cold) resuspension of HepG2 cells, and the isolated nucleus was obtained by cell disruption buffer resuspension of HepG2 cells. Total RNA derived from HCC tissues and cells was extracted using TRIzol (Invitrogen/Life Technologies) following the manufacturer’s instructions. The quality inspection of RNA was verified by microspectrophotometer (Tiangen) and agarose gel electrophoresis. The cDNA was synthesized from total RNA by using the K1622 Revert Aid First Strand cDNA Synthesis Kit (Thermo). RT-qPCR was performed using a 2 × PCR Master Mix (Roche) on the ABI Q6 Flex Real-time PCR system (Applied Biosystems). All gene expression values were normalized to β-actin and calculated using 2−ΔΔCt method. Primer sequences were provided in Table S2.
Actinomycin D treatment
To block transcription, HCC cells were handled with 2 mg/mL actinomycin D (Merck, Germany). Then, the remaining RNAs extracted from treated cells were assessed by qRT-PCR at 0 h, 1 h, 3 h, 6 h, 9 h, and 12 h.
Western blotting analysis
HCCLM3 and HepG2 cells were treated with a lysis buffer. Cell extracts were boiled in a loading buffer, and equal amounts of cell extracts were separated on 15% SDS-PAGE gel. After transferring separated protein bands into PVDF (Millipore, Billerica, MA, USA), membranes were blocked in TBST with 5% fat-free milk and incubated with primary antibodies overnight at 4 °C. Next, the membranes were incubated with secondary antibodies (ProteinTech, Chicago, Illinois, USA). Finally, the bands were visualized through a chemiluminescence ECL kit (Beyotime Biotechnology, Beijing, China) in accordance with the manufacturer’s introduction. CheniScope (CLiNX; Shanghai) was used to detect the bands and then analyzed on Image J (National Institutes of Health, Bethesda, MD, USA). The primary antibodies GAPDH (60,004-1-Lg) and DNM1L (12,957-1-AP) in this study were purchased from Proteintech. Goat Anti-Mouse IgG H&L(HRP) (ab205719, abcam) and Goat Anti-Rabbit IgG H&L(HRP) (ab6721, abcam) were set as secondary antibodies.
Flow cytometry assay
In this study, flow cytometry was used to detect the apoptosis of HCC cells after overexpression/knockdown of circ_0098823 and DNM1L, and simultaneous overexpression of circ_0098823 and knockdown of DNM1L. Apoptosis was determined using Annexin V-fluorescein isothiocyante (Annexin V-FITC) Apoptosis Detection reagents (C1062, Biyuntian). After washing and resuspending, the tumor cells were stained by propidium iodide (PI) at room temperature in the dark for 15 min. Finally, the washed precipitation was resuspended in PBS solution and subjected to flow cytometry analysis within 4 h by CytoFLEX LX Flow Cytometer (Beckman).
Transwell migration and invasion assay
The transwell chamber covered with or without Matrigel Mix (Corning, NY, USA) was performed to detect the invasion and migration ability of HCCLM3 and HepG2 cells respectively. HCCLM3 (6 × 104) and HepG2 cells (6 × 104) suspended in 500 µL medium containing 10% serum were suspended in the upper chamber. And filled 700 µL 10% FBS medium for lower chamber culture. After incubation for 24 h for the migration assay and 48 h for the invasion assay at 37 °C, non-migrating or non-invading cells were gently removed, and the invaded cells in the lower filters were fixed with 4% poly methanol for 20 min, stained with crystal violet (Sigma, MO, USA) and counted under a microscope.
Electron microscope
The tumor cells were rinsed in PBS and fixed up with 0.05% glutaraldehyde for 1–3 h at 4 °C, and then immobilized with 1–2% osmic acid for 15–40 min after being washed by PBS for three times. After fixation, ethanol with different concentration gradients was used to remove excess water from HCCLM3 and HepG2 cells.
After finishing the operation of washing and drying, the cells were placed in a mixture of 100% dehydrating agent and an equal amount of epoxy embedding agent for 30 min at room temperature. The sample was completely embedded in epoxy resin and placed in a 60 °C temperature box for 24 h to make the embedded block. Used Leica ultra-thin microtome to cut 50–70 mm slices. After staining with uranium acetate and lead citrate, washing, and drying, they were observed under transmission electron microscope (JEM-1200EX, Nippon Electronics Corporation).
RNA pull-down and mass spectrometry (MS) analysis
The purpose of RNA pull-down and MS was to identify RBP of circ_0098823. The HepG2 cells were cleaved by cell lysate. Subsequently, labeled RNA probes were bound to streptavidin magnetic beads. The same volume of 20 mM Tris (PH7.5) magnetic beads was used. After repeating the above steps two or three times, diluting the 10X Protein-RNA Binding Buffer to 1X Protein-RNA Binding Buffer and then the mixed solution of RNA–protein binding system was poured into the magnetic bead of RNA binding and incubated at 4 °C for 30–60 min. The reaction liquid was placed in the magnetic rack for 5 min, and then magnetic beads were collected at the same time and the supernatant was analyzed. Subsequently, the magnetic beads and the Elution Buffer were stirred at 37 °C and incubated for 15–30 min. After the magnetic beads were precipitated, the supernatant was collected for 10% SDS-PAGE electrophoresis. After electrophoresis, silver staining experiment was conducted. After silver staining, different protein bands between Control and circ_0098823 probe lanes were cut for MS analysis. MS analysis was completed by APTBIO company (Shanghai, China).
RNA immunoprecipitation (RIP)
This experiment was performed to estimate the association between circ_0098823 and IGF2BP3. The HCCLM3 and HepG2 cells were firstly treated with RIP lysis buffer for 5 min and then stored at -80 °C after being collected by centrifugation. Next, the suspended magnetic beads were incubated with IGF2BP3/IgG antibody at room temperature for 30 min. The liquid after the centrifugal reaction is collected and precipitated. RIP Immunoprecipitation Buffer was added into the magnetic bead-antibody mixture and incubated overnight at 4 °C for co-immunoprecipitation reaction. Absorb 10 µL supernatant as "input" and store it at − 80 °C. Finally, RNA was incubated with proteinase K buffer at 55 °C for 30 min, and RNA was extracted by Trizol method and purified overnight.
Animal experiments
Nude female BALB/c-nu mice (aged 4–6 weeks) were placed in an environment of free pathogens, with an ambient temperature of 24 ± 2 °C, a humidity of 55 ~ 60%, and a light/dark cycle of 12 h. All mice used in this study were purchased from SPF (Beijing) Biotechnology Co., Ltd. In the subcutaneous tumor model, mice were randomly divided into two groups: shNC groups (n = 5) and sh-circ_0098823 groups (n = 5). HepG2 cells (5 × 106) stably transfected with sh-circ_0098823 were injected into the right underarm of female nude mice. Tumor size was measured at 3-day intervals 19 days after cell injection. After 4 weeks, the mice were sacrificed by euthanasia (150 ~ 200 mg/kg pentobarbital sodium). Crysections of the harvested livers were stained using Haematoxylin and eosin (H&E) and IHC for histological assessment.
For the liver metastasis study, fluorescent-labeled HepG2 cells (5 × 106) with sh-circ_0098823 were injected through the lateral tail vein of mice. After 4 weeks, mice were first used in vivo imaging to observe the fluorescence intensity of tumor metastasis, and then were anesthetized with euthanasia (150–200 mg/kg pentobarbital sodium). The tumor and lung tissues were isolated for subsequent staining observations. The procedures for care and use of animals were approved by the Ethics Committee of the Shanghai General Hospital of Shanghai Jiao Tong University School of Medicine.
HE staining
The fresh tumor and lung tissue were fixed with 4% paraformaldehyde over 24 h. Dehydration was carried out with gradient alcohol and xylene in turn. Paraffin was used for tissue embedding. After cooling, the trimmed wax blocks are cut into 4 µm slices on a microtome. The sections were then dewaxed and stained with hematoxylin in the nucleus and eosin in the cytoplasm. After dewatering and sealing, a light microscope (100 nm) was used to observe the histopathological injury.
Immunohistochemistry (IHC) analysis
Detailed operations of IHC staining were shown in the previous report [31]. In brief, HCC tissues and normal tissues were treated with 4% paraformaldehyde, paraffin-embedded, and then were behind been made into slices of 4 µm which were placed on the slides. After deparaffinization, rehydration was following. And microwave antigen retrieval, the slides were put into a hydrogen peroxide solution to block endogenous catalase and then blocked at the room temperature for 30 min. Subsequently, the slides were incubated with DNM1L antibody (12,957-1-AP, Proteintech) at 1:200 dilution at 4 °C overnight. Afterward, the slides were incubated with None-Linked Caprine Anti-Mouse IgG Polyclonal Antibody (SAA544Mu10, CLOUD-CLONE Corp.wuhan) at room temperature for 30 min and stained with DAB substrate, followed by hematoxylin counterstaining. Microscope was used to observe and images were collected to analyze.
Statistical analysis
The statistical analysis was executed with GraphPad Prism 8.0 (GraphPad Software Inc, San Diego, CA, USA). The comparisons of means among groups were analyzed by Student’s test and analysis of variance. Differences among the three or more groups were assessed using univariate analysis of variance (ANOVA) and Tukey post hoc tests. Pearson Correlation Analysis was used to analyze the correlation of circ_0098823 with DNM1L. A P-value < 0.05 was indicated as statistically significant.
Results
Identification and characterization of circ_0098823 derived from ERBB3 in HCC tissues
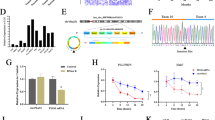
After HCC resection, HCC cells will release HGF, which promotes the invasion and migration of HCC cells [29, 32]. In the previous study, we found HGF promoted ERBB3 expression and the increase of ERBB3 promoted the invasion and metastasis of HCC cells both in vivo and in vitro [30]. Based on bioinformatics analysis, we revealed 13 circRNAs derived from ERBB3, promoting us to examine whether these circRNAs are also involved in migration and invasion. To verify this idea, we detected the expression of 13 circRNAs in HepG2 cells treated with HGF for 24 h by RT-qPCR. Compared with the control group, HGF stimulation led to 2 up-regulated circRNAs, 3 down-regulated circRNAs, while 7 circRNAs whose expression was not statistically significant, and the expression of circ0026817 could not be detected due to quite low background expression (Fig. 1A). Particularly, circ_0098823 was the most differentially expressed circRNAs (Fig. 1A). This result was supported by experiments in HCC patients. The expression of circ_0098823 in HCC tissues was significantly up-regulated compared with para-cancer tissues (Fig. 1B). Moreover, the diagnostic value of circ_0098823 expression level was demonstrated by ROC curve. The results showed that the area under the ROC curve (AUC) of circ_0098823 in HCC patients and healthy controls was 0.98 (p = 0.0003, Fig. 1C). Therefore, we preliminarily selected circ_0098823 for subsequent studies.
Identification and characterization of circRNAs derived from ERBB3 in HCC tissue. A The expression levels of 12 circRNAs derived from ERBB3 in HepG2 cells treated with HGF (40 ng/mL) for 24 h were detected by RT-qPCR (n = 3). NC, negative control. NC was treated with PBS as control. B The expression of circ_0098823 in tumor tissues and normal tissues adjacent to cancer from 10 HCC patients was detected by RT-qPCR.T: HCC tumor tissue. The internal control gene is Actin. C ROC curve illustrating performance of the circ_0098823 expression in distinguishing HCC from normal subjects. D Diagram illustrating back-splicing of circ_0098823 from the ERBB3 host gene. E The ring structure of circ_0098823 was verified by agarose electrophoresis. The arrows represent the direction of primer amplification. Divergent primers for detecting circ_0098823, and convergent primers for detecting linear ERBB3. F The specific sequences of circ_0098823 were validated by sanger sequencing. The arrow indicates the splicing site of circ_0098823. back-splicing of ERBB3 exon 14 to exon 21 confirmed by Sanger sequencing. G The localization of circ_0098823 in HepG2 cells was verified by karyoplasmic isolation. ns means P > 0.05, *means P < 0.05, ** means P < 0.01, *** means P < 0.001
Circ_0098823 is a circRNA formed by the free combination of exon14 and exon21 on Chr12 (Fig. 1D). The back splices junction site of circ_0098823 was verified using divergent primers. Divergent primers were applied to amplify circ_0098823 transcripts, while the convergent primers were used to detect the linear transcripts. The results showed that circ_0098823 could be amplified by polymerizing or diverging primers of cDNA. As for gDNA, circ_0098823 can only be amplified with its polymeric primers and cannot be amplified with its divergent primers (Fig. 1E). Sanger sequencing further confirmed the ring structure of circ_0098823 (Fig. 1F). Furthermore, the localization of circ_0098823 was detected by nucleocytoplasmic separation experiment (Fig. 1G). The result demonstrated that circ_0098823 was mainly expressed in cytoplasm in HepG2 cell. These results suggested that circ_0098823 is a bona fide circRNA, and was significantly up-regulated in HCC.
Interference with circ_0098823 significantly inhibited HCC cells migration, invasion, and mitochondrial fission
To explore the function of circ_0098823 in HCC progress, we check the invasion, migration, and apoptosis of HCC cells under knockdown and overexpression of circ_0098823. RT-qPCR showed that circ_0098823 was significantly up-regulated in HepG2 cells compared with HCCLM3 cells (Fig. 2A). Based on the result, we selected HepG2 cells to knockdown circ_0098823 and overexpress circ_0098823 in HCCLM3 cells to verify its functional effect. According to the back-splice sequence of circ_0098823, two siRNAs interference targets were designed. The expression of circ_0098823 significantly declined after si-circ_0098823 transfection compared with si-NC transfection (Fig. 2B). The result of CCK-8 showed that si-circ_0098823 inhibited the viability of HepG2 cells (Fig. 2C). Transwell showed that si-circ_0098823, HepG2 cell migration, and invasion were significantly reduced (Fig. 2D–F). Flow cytometry showed that the knockdown of circ_0098823 significantly promoted HepG2 cell apoptosis (Fig. 2G–H). Mitochondrial fission-induced mtDNA stress can promote HCC progression [33]. Besides exploring cell function, we reveal the mitochondrial fission situation after circ_0098823 knockdown by observing the number of mitochondria. Electron microscopy showed that the number of mitochondria in HepG2 cells was significantly reduced after circ_0098823 knockdown (Fig. 2I). Interference with circ_0098823 significantly impaired mitochondrial fission in HCC cells.
Interference with circ_0098823 significantly inhibited HCC cells migration, invasion, and mitochondrial fission. A Background expression of circ_0098823 in HCCLM3 cells and HepG2 cells (n = 3). The standard of the relative expression is with respect to actin. B Silencing efficacy of circ_0098823 was confirmed by RT-qPCR (n = 3). C The viability of HepG2 cells after silencing of circ_0098823 was measured by CCK-8 assays (n = 6). D The migration and invasion of HepG2 cells after silencing of circ_0098823 were measured by transwell assays. Scale bars 50 μm. E The migration statistics of HepG2 cells after circ_0098823 silencing (n = 3). F The invasion statistics of HepG2 cells after circ_0098823 silencing (n = 3). G The apoptosis of HepG2 cells after silencing of circ_0098823 was measured by flow cytometry assays. H The apoptosis statistics of HepG2 cells after circ_0098823 silencing (n = 3). I Electron microscopy observed the number of mitochondria in HepG2 cells after silencing circ_0098823 (n = 3). ns means P > 0.05, * means P < 0.05, ** means P < 0.01, *** means P < 0.001. Scale bars, 2 μm and 50 μm
To further verify the effect of circ_0098823 in HCC, we conducted the effect of circ_0098823 overexpression in HCCLM3 cells. Circ_0098823 was successfully overexpressed in HCCLM3 cells (Fig. S1A). CCK-8, transwell and flow cytometry showed that OE-circ_0098823 significantly increased HCCLM3 cell viability, cell migration and invasion, and significantly reduced cell apoptosis (Fig. S1B-G). All the results of the overexpression group were the opposite of the knockdown group. Especially, under the condition of circ_0098823 overexpression, the number of mitochondrial in HCCLM3 cells was significantly increased (Fig. S1H). This suggests that overexpression of circ_0098823 is significantly advantageous to mitochondrial fission in HCC cells. All the above results suggested that circ_0098823 promoted the viability, migration, invasion and impeded apoptosis of HCC.
Interference with circ_0098823 inhibits HCC tumor growth and metastasis in vivo
To further validate the function of circ_0098823 in HCC development, a subcutaneous tumor model was established through subcutaneously injected HepG2 cells stably transfected with sh-circ_0098823 and sh-NC into BALB/c-nu mice. The morphology and size of the tumor are shown in Fig. 3A. Knockdown of circ_0098823 resulted in smaller tumor volumes and lighter tumor weight (Fig. 3B–C). Subsequently, we conducted H&E staining to observe the tumor pathological characteristics of sh-circ_0098823 group in vivo (Fig. 3D). The effect of circ_0098823 on HCC metastasis was observed in vivo. Live imaging of the control group and the knockdown group was presented in Fig. 3E, which displayed knockdown of circ_0098823 obviously attenuated the tumor metastasis in mice. Additionally, compared with sh-NC group, the sh-circ_0098823 group had fewer and smaller metastatic nodules and more normal alveoli (Fig. 3F). These results strongly demonstrated that interference with circ_0098823 inhibit HCC tumor growth and metastasis in vivo.
Interference with circ_0098823 can inhibit HCC tumor growth and metastasis in vivo. A Pictures of mice and tumor morphology in control group and knockdown group (n = 5). B Tumor volume statistics at days 19, 22, 25, and 28 after successful transplantation (n = 5). C Difference in tumor weight between sh-circ_0098823 group and sh-NC group (n = 5). D H&E staining was used to detect tumor pathology. Scale bars, 50 μm. E The bioluminescence imaging in the sh-NC and sh-circ_0098823 groups before sacrifce. F Lung metastasis of HCC was detected by H&E staining (n = 3). Scale bars, 50 μm. *** means P < 0.001
Circ_0098823 promotes the expression of DNM1L
DNM1L is an evolutionarily conserved protein that mediated mitochondrial and peroxisomal division [34]. Previous studies have found that based on the TCGA database, the expression of DNM1L was up-regulated in HCC tissues compared with neighboring HCC tissues and DNM1L is closely related to mitochondrial fission [28]. Therefore, we investigated the regulatory effect between circ_0098823 and DNM1L by RT-qPCR and western blot (WB). DNM1L was significantly up-regulated after circ_0098823 overexpression in HCC cells (Fig. 4A–B), while knockdown of circ_0098823 significantly down-regulated DNM1L (Fig. 4C–D). Moreover, to further verify the regulation of circ_0098823 on DNM1L expression in vivo, we detected DNM1L expression using sh-circ_0098823 mouse model by IHC assays. The results showed that compared with shRNA controls, silencing of circ_0098823 significantly down-regulated the expression of DNM1L (Fig. 4E). These results suggested circ_0098823 may enhance the expression level of DNM1L.
Circ_0098823 maybe regulate mitochondrial fission through DNM1L. A Circ_0098823 was overexpressed in HCCLM3 cells, and the expression of DNM1L was detected by RT-qPCR. B The expression of DNM1L was detected by western blot (WB) when overexpression of circ_0098823 in HCCLM3 cells. C The expression of DNM1L was detected by RT-qPCR when knockdown of circ_0098823 in HepG2 cells. D The expression of DNM1L was detected by WB when knockdown of circ_0098823 in HepG2 cells. E IHC was used to detect tumor histopathology. Scale bars, 50 μm. means P < 0.05, ** means P < 0.01, n = 3
DNM1L promotes the malignant phenotype of HCC cells
To further prove that DNM1L is the intermediate coordinator of circ_0098823 in HCC, we next investigate the functional effect of DNM1L in HCC. We detected the knockdown efficiency of DNM1L in HCCLM3 and HepG2 cells by RT-qPCR (Fig. 5A–B). Si-202 was used for DNM1L knockdown in this study because of its ideal knockout efficiency in both HCCLM3 and HepG2 cells. Furthermore, CCK-8 detected the activity of HCCLM3 cells and HepG2 cells. The data were given statistical significance at 96 h after transfection and si-DNM1L significantly reduced the activity of HCCLM3 and HepG2 cells (Fig. 5C–D). We next explored the effect of DNM1L on migration, invasion, and apoptosis of HCC cells by transwell and flow cytometry, respectively. According to Fig. 5E–I, we found that DNM1L knockdown significantly impeded HCC migration and invasion, but promoted apoptosis.
Knockdown of DNM1L significantly inhibited the migration and invasion of HCC cells and promoted apoptosis. Silencing efficacy of DNM1L in HCCLM3 cells (A) and HepG2 cells (B) were confirmed by RT-qPCR. C The viability of HCCLM3 cells after silencing of DNM1L was measured by CCK-8 assays. D CCK-8 assays measured the viability of HepG2 cells after silencing of DNM1L. E The migration and invasion of HCCLM3 and HepG2 cells after silencing of DNM1L was measured by transwell assays. Scale bars, 50 μm. F The migration statistics of HCCLM3 and HepG2 cells after DNM1L silencing. G The invasion statistics of HCCLM3 and HepG2 cells after DNM1L silencing. H The apoptosis statistics of HCCLM3 and HepG2 cells after DNM1L silencing. I The apoptosis of HCCLM3 and HepG2 cells after silencing of DNM1L was measured by flow cytometry assays. * means P < 0.05, ** means P < 0.01, *** means P < 0.001, Scale bars, 50 μm, n = 3
For the functional experiment of OE-DNM1L, we captured the exact opposite of the above experiment. The overexpression efficiency of DNM1L was shown in Fig. S2A. Transwell assays and flow cytometry were conducted to evaluate the migration, invasion, and apoptosis potential of HCCLM3 and HepG2 cells. Overexpression of DNM1L significantly promoted the migration, invasion of HCCLM3 cell, and impeded the apoptosis of HCCLM3 cells (Fig. S2B-F). These results indicated that overexpression of DNM1L was beneficial to HCC development.
Circ_0098823 regulates DNM1L protein stability through binding to IGF2BP3 to participate in HCC progression
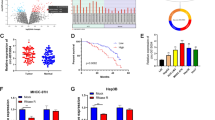
Although circRNA is well known as a ceRNA in pathogenesis studies, it is also of great research significance to explore circRNA acts by interacting with RBP. To explore the RBP of circ_0098823, we designed and synthesized a specific biotin-labeled probe for circ_0098823 to perform RNA pull-down in HepG2 cell (Fig. 6A). All bands were sent to the MS platform for protein spectrum identification (Table S3). Circ_0098823 was found to bind to several proteins, including IGF2BP1, IGF2BP2, IGF2BP3 and YTHDF1. These proteins regulate the stability of downstream mRNA [35, 36]. Therefore, we further analyzed the possibility of binding these proteins to circ_0098823. Timer database (http://timer.comp-genomics.org/timer/) was used to analyze the expression of these proteins in HCC and their correlation with the expression of circ_0098823 downstream target gene DNM1L. The results showed that IGF2BP3 had the strongest positive correlation with DNM1L (R = 0.5, p = 0) (Fig. S3), and its expression was up-regulated in HCC. It has been reported that IGF2BP3 may promote the proliferation and migration of HCC cells [37]. Therefore, we screened IGF2BP3 as the best candidate protein. The peak image of IGF2BP3 obtained by MS is shown in (Fig. 6B). Surprisingly, the results showed that IGF2BP3 was significantly enriched by the circ_0098823 probe compared to the control probe (Fig. 6C). The further verification of the binding relationship between IGF2BP3 and circ_0098823 was proved by RIP-PCR (Fig. 6D). Therefore, we regarded IGF2BP3 as the RBP of circ_0098823 in HCC. Considering that circ_0098823 maybe regulate mitochondrial fusion and fission in HCC cells through DNM1L, we explored the relationship between IGF2BP3 and DNM1L. RT-qPCR and WB showed the knockdown efficiency of IGF2BP3 (Fig. 6E–F). Then, we found the expression of DNM1L was down-regulated with IGF2BP3 knockdown (Fig. 6G–H). These data indicated that IGF2BP3 could promote the expression of DNM1L. Moreover, the experiment was conducted to investigate the effect of IGF2BP3 on the stability of DNM1L mRNA. Compared with the non-knockdown group, the expression of DNM1L mRNA began to decrease significantly after 6 h of IGF2BP3 knockdown (Fig. 6I). This result suggested that knockdown of IGF2BP3 could decrease the stability of DNM1L. The above results indicated that IGF2BP3 is the RBP of circ_0098823 and can regulated DNM1L mRNA stability.
IGF2BP3 binds to circ_0098823 and stabilizes the mRNA of DNM1L. A RNA pull-down identified binding proteins that interact with circ_0098823 in HepG2 cells. B The peak image of IGF2BP3 obtained by mass spectrometry. C The mass spectrometry results of IGF2BP3 were further verified by WB. D Circ_0098823 binding with IGF2BP3 was detected by RIP-PCR. E Silencing efficacy of IGF2BP3 on HCCLM3 cells was confirmed by RT-qPCR. F Silencing efficacy of IGF2BP3 on HCCLM3 cells was verified by western blot. G Detection of DNM1L expression in HCCLM3 cells after IGF2BP3 silencing by RT-qPCR. H Detection of DNM1L expression in HCCLM3 cells after IGF2BP3 silencing by western blot. I The expression of DNM1L mRNA after silencing IGF2BP3 with the treatment of actinomycin D at 0 h, 1 h, 3 h, 6 h, 9 h, and 12 h were measured by RT-qPCR. * means P < 0.05, *** means P < 0.001, n = 3
Circ_0098823 promotes HCC progression by regulating DNM1L through IGF2BP3
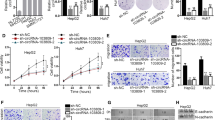
To further demonstrate the molecular mechanism of circ_0098823/IGF2BP3/DNM1L axis in HCC, we conducted rescue experiments. We found that OE-circ_0098823 significantly up-regulated the expression of DNM1L, but after IGF2BP3 knockdown, the up-regulated DNM1L was significantly reversed (Fig. 7A). The same results were displayed by WB (Fig. 7B). Simultaneous IGF2BP3 knockdown significantly reversed the increase in HCCLM3 cell viability caused by circ_0098823 overexpression (Fig. 7C). In addition, IGF2BP3 knockdown significantly inhibited circ_0098823-induced HCCLM3 cell migration and invasion (Fig. 7D–F). Flow cytometry showed that IGF2BP3 knockdown significantly restored the apoptosis of HCCLM3 cells caused by overexpression of circ_0098823 (Fig. 7G–H). Furthermore, Fig. 7I showed that IGF2BP3 knockdown significantly alleviated the stability decline of DNM1L mRNA caused by circ_0098823. These results indicated circ_0098823 regulates migration, invasion, and apoptosis of HCC cells through interacting with IGF2BP3 to enhance stability of DNM1L. The results of rescue experiments provided strong evidence for enhancing the reliability of our study.
Circ_0098823 promoted HCC progression through DNM1L. The expression of DNM1L on HCCLM3 cells in NC groups, circ_0098823 overexpression groups, and simultaneously circ_0098823 overexpression and IGF2BP3 knockdown groups were verified by RT-qPCR (A) and western blot (B), respectively. C The viability of HCCLM3 cells after NC groups, circ_0098823 overexpression groups, and simultaneously circ_0098823 overexpression and IGF2BP3 knockdown groups were measured by CCK-8 assays, respectively (n = 6). D The migration and invasion of HCCLM3 cells were measured by transwell assays, respectively. The migration (E) and invasion (F) statistics of HCCLM3 cells between NC groups, circ_0098823 overexpression groups, and simultaneously circ_0098823 overexpression and IGF2BP3 knockdown groups, respectively. G The apoptosis of HCCLM3 cells between NC groups, circ_0098823 overexpression groups, and simultaneously circ_0098823 overexpression and IGF2BP3 knockdown groups were measured by flow cytometry assays, respectively. H The apoptosis statistics of HCCLM3 cells between NC groups, circ_0098823 overexpression groups, and simultaneously circ_0098823 overexpression and IGF2BP3 knockdown groups. I The expression of DNM1L mRNA in NC group, OE-circ_0098823 group and simultaneous OE-circ_0098823 and si-IGF2BP3 treated with actinomycin D for 0, 1, 3, 6, 9 and 12 h was detected by RT-qPCR. *means P < 0.05, ** means P < 0.01, *** means P < 0.001, Scale bars, 50 μm, n = 3
Discussion
Since the stability, abundance, conservation and spatio-temporal specificity, circRNA has been a hot topic of biomedical research in recent years. The molecular mechanism of circRNA involvement in HCC has been reported. Our study further complements the research on the involvement of novel circRNA in HCC progression. In this study, we found that circ_0098823 significantly aggravates the malignant phenotype of HCC through mitochondrial fission. IGF2BP3, as the RBP of circ_0098823, can significantly save the excessive proliferation, migration, invasion, and reduced apoptosis of HCC cells caused by circ_0098823. Mechanistically, circ_0098823 promotes HCC cell invasion and metastasis through IGF2BP3-mediated DNM1L regulation of mitochondrial fission (Fig. 8). Therefore, our findings may furnish a novel therapeutic target for HCC patients.
According to the published data in the field of cancer in recent years, many occult circRNAs involved in the emergence and development of HCC are gradually being uncovered. For example, circ_0095868 promotes the occurrence of HCC through the miR-346/874-3p-ATG16L1 axis [38]. CircRNA-SORE mediates sorafenib resistance in HCC via stabilizing YBX1 [39]. The hsa_circRNA_104348 promotes HCC progression through modulating miR-187-3p/RTKN2 axis and activating the Wnt/β-catenin pathway [40]. These examples indicated that circRNA plays an important role in HCC progression. Consistent with previous studies, our study first identified a novel circRNA that is circ_0098823. High expression of circ_0098823 in HCC tumors promotes tumor progression. Different from the above examples, our study complements the research results of circRNA in HCC metastasis. This study not only broadens our understanding of the involvement of circRNA in HCC progression, but also appends a new target for targeted therapy of HCC.
ERBB3 gene is found on the long arm of chromosome 12 (12q13) [41]. ERBB3 belongs to the ERBB family of transmembrane receptor tyrosine kinases, the other members are ERBB2 and ERBB4 [42]. ERBB3 takes part in cell proliferation, differentiation, and migration in various cancers [42]. Therefore, it makes sense to explore the role of ERBB3 in HCC. Current studies only reported that ERBB3 was involved in disease progression as a target gene. Such as circ_DLGAP4 promotes the progression of diabetic nephropathy in the form of ERBB3 activation [43], circEPSTI1 promotes HER2-positive breast cancer cells via ERBB3 [44]. Notably, ERBB3 was significantly increased in HCC postoperative metastasis [30], suggesting the potential significance of ERBB3 in HCC metastasis is worth exploring. Our study is the first to report the role of ERBB3-derived circRNAs in HCC. The free combination of exons results in a variety of circRNA produced by ERBB3. Among which, we found that circ_0098823 was the most differentially expressed circRNA in HCC compared to normal, and circ_0098823 promote HCC. We are revisiting ERBB3 from circRNA's perspective for the first time.
Mitochondrial dynamic consisting of fission and fusion is complicated and important process to regulate the pro-apoptosis status of mitochondria [33]. Mitochondrial fission induced by DNM1L overexpression or MFN1 knockdown exerts an important role in apoptotic resistance in cancer and non-malignant cells [27]. DNM1L is a major cytoplasmic protein whose activation causes a change in location, translocation to the mitochondria and further initiation of mitochondrial fission [45]. Previous literature has highlighted DNM1L as enhancing the survival of HCC cells in vitro and in vivo by promoting mitochondrial fission [27]. Consistent with previous finding, we found DNM1L was highly expressed in the HCC cells, and overexpression of DNM1L significantly prohibit the migration, invasion, and impede apoptosis of HCC cells. Furthermore, we found that circ_0098823 regulate mitochondrial fission and DNM1L expression in HCC. Therefore, we conducted that circ_0098823 maybe contribute to HCC progression by DNM1L promoted mitochondrial fission.
IGF2BP3 is not only an m6A reading protein, but also a universal oncofetal protein, which has been detected to be overexpressed in a variety of cancers, especially HCC [46]. Based on RNA pull-down and MS analysis, we discovered that IGF2BP3 was RBP of circ_0098823. IGF2BP3 regulates the stability of DNM1L, and reverse the effectiveness of circ098823 overexpression on the malignant progression of HCC cells. IGF2BP protein can enhance mRNA stability and translation ability [47]. For instance, Wang et al. [48] found that RBM15 regulates the stability of TMBIM6 through IGF2BP3 dependence and facilitates the progression of laryngeal squamous cell carcinoma. Zhang et al. [49] revealed that IGF2BP3 expedites the progression of acute myeloid leukemia by enhancing the stability of RCC2. These examples suggested that IGF2BP3 could play a role by affecting mRNA stability. In general, these studies support the role of circ_0098823/IGF2BP3/DNM1L axis in HCC.
Conclusion
On the whole, our research addressed that circ_0098823 regulates DNM1L mRNA stability and mitochondrial fission through RBP IGF2BP3 to promote HCC metastasis and invasion, which provided new targets for the treatment of HCC and contributed clues to the study of the mechanisms related to HCC.
Data availability
All data are available in the main text or the supplementary materials.
References
Sung H, Ferlay J, Siegel RL (2021) Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: Cancer J Clin 71:209–249
Sayiner M, Golabi P, Younossi ZM (2019) Disease burden of hepatocellular carcinoma: a global perspective. Dig Dis Sci 64:910–917
Yang JD, Hainaut P, Gores GJ (2019) A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol 16:589–604
Sugawara Y, Hibi T (2021) Surgical treatment of hepatocellular carcinoma. Biosci Trends 15:138–141
Yang C, Zhang H, Zhang L, Zhu AX, Bernards R (2023) Evolving therapeutic landscape of advanced hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol 20:203–222
Chen B, Wu JX, Cheng SH, Wang LM (2021) Phase 2 study of adjuvant radiotherapy following narrow-margin hepatectomy in patients with HCC. Hepatology 74:2595–2604
Chen LL (2016) The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol 17:205–211
Fan X, Zhang X, Wu X et al (2015) Single-cell RNA-seq transcriptome analysis of linear and circular RNAs in mouse preimplantation embryos. Genome Biol 16:148
Shen Y, Guo X, Wang W (2017) Identification and characterization of circular RNAs in zebrafish. FEBS Lett 591:213–220
Rybak-Wolf A, Stottmeister C, Glažar P et al (2015) Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell 58:870–885
Li X, Yang L, Chen LL (2018) The biogenesis, functions, and challenges of circular RNAs. Mol Cell 71:428–442
Chen LL, Yang L (2015) Regulation of circRNA biogenesis. RNA Biol 12:381–388
Dong R, Ma XK (2017) Increased complexity of circRNA expression during species evolution. RNA Biol 14:1064–1074
Nicolet BP, Engels S, Aglialoro F, van den Akker E, von Lindern M, Wolkers MC (2018) Circular RNA expression in human hematopoietic cells is widespread and cell-type specific. Nucleic Acids Res 46:8168–8180
Westholm JO, Miura P, Olson S et al (2014) Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep 9:1966–1980
Sun Y, Sun X, Huang Q (2020) Circ_0000105 promotes liver cancer by regulating miR-498/PIK3R1. J Gene Med 22:e3256
Yuan P, Song J, Wang F, Chen B (2022) Exosome-transmitted circ_002136 promotes hepatocellular carcinoma progression by miR-19a-3p/RAB1A pathway. BMC Cancer 22:1284
Wang L, Long H, Zheng Q, Bo X, Xiao X, Li B (2019) Circular RNA circRHOT1 promotes hepatocellular carcinoma progression by initiation of NR2F6 expression. Mol Cancer 18:119
Wang Z, Sun A, Yan A et al (2022) Circular RNA MTCL1 promotes advanced laryngeal squamous cell carcinoma progression by inhibiting C1QBP ubiquitin degradation and mediating beta-catenin activation. Mol Cancer 21:1–20
Zhao M, Wang Y, Tan F et al (2022) Circular RNA circCCNB1 inhibits the migration and invasion of nasopharyngeal carcinoma through binding and stabilizing TJP1 mRNA. Sci China Life Sci 65:2233–2247
Chan DC (2020) Mitochondrial dynamics and its involvement in disease. Annu Rev Pathol 15:235–259
Nolden KA, Egner JM, Collier JJ et al (2022) Novel DNM1L variants impair mitochondrial dynamics through divergent mechanisms. Life Sci Alliance 5(12):e202101284. https://doi.org/10.26508/lsa.202101284
Wang J, Chen A (2023) BCL2L13 promotes mitophagy through DNM1L-mediated mitochondrial fission in glioblastoma. Cell Death Dis 14:585
Zhou J, Li G, Zheng Y et al (2015) A novel autophagy/mitophagy inhibitor liensinine sensitizes breast cancer cells to chemotherapy through DNM1L-mediated mitochondrial fission. Autophagy 11:1259–1279
Wakabayashi J, Zhang Z, Wakabayashi N et al (2009) The dynamin-related GTPase Drp1 is required for embryonic and brain development in mice. J Cell Biol 186:805–816
Deng X, Liu J, Liu L, Sun X, Huang J, Dong J (2020) Drp1-mediated mitochondrial fission contributes to baicalein-induced apoptosis and autophagy in lung cancer via activation of AMPK signaling pathway. Int J Biol Sci 16:1403–1416
Huang Q, Zhan L, Cao H et al (2016) Increased mitochondrial fission promotes autophagy and hepatocellular carcinoma cell survival through the ROS-modulated coordinated regulation of the NFKB and TP53 pathways. Autophagy 12:999–1014
Zhang S, Gong H, Xie H et al (2023) An integrated analysis of dynamin 1 like: a new potential prognostic indicator in hepatocellular carcinoma. Mol Carcinog 62:786–802
Ren Y, Cao B, Law S et al (2005) Hepatocyte growth factor promotes cancer cell migration and angiogenic factors expression: a prognostic marker of human esophageal squamous cell carcinomas. Clin Cancer Res Off J Am Assoc Cancer Res 11:6190–6197
Liu DL, Lu LL, Dong LL et al (2020) miR-17-5p and miR-20a-5p suppress postoperative metastasis of hepatocellular carcinoma via blocking HGF/ERBB3-NF-κB positive feedback loop. Theranostics 10:3668–3683
Zhang Y, Ma H (2023) LncRNA HOXD-AS2 regulates miR-3681-5p/DCP1A axis to promote the progression of non-small cell lung cancer. J Thorac Dis 15:1289–1301
Hao NB, Tang B, Wang GZ et al (2015) Hepatocyte growth factor (HGF) upregulates heparanase expression via the PI3K/Akt/NF-κB signaling pathway for gastric cancer metastasis. Cancer Lett 361:57–66
Bao D, Zhao J, Zhou X et al (2019) Mitochondrial fission-induced mtDNA stress promotes tumor-associated macrophage infiltration and HCC progression. Oncogene 38:5007–5020
Hogarth KA, Costford SR, Yoon G, Sondheimer N, Maynes JT (2018) DNM1L variant alters baseline mitochondrial function and response to stress in a patient with severe neurological dysfunction. Biochem Genet 56:56–77
Liu L, He J, Sun G et al (2022) The N6-methyladenosine modification enhances ferroptosis resistance through inhibiting SLC7A11 mRNA deadenylation in hepatoblastoma. Clin Transl Med 12:e778
Zhang S, Guan X, Liu W et al (2022) YTHDF1 alleviates sepsis by upregulating WWP1 to induce NLRP3 ubiquitination and inhibit caspase-1-dependent pyroptosis. Cell Death Discov 8:244
Degrauwe N, Suvà ML, Janiszewska M, Riggi N, Stamenkovic I (2016) IMPs: an RNA-binding protein family that provides a link between stem cell maintenance in normal development and cancer. Genes Dev 30:2459–2474
Du A, Li S, Zhou Y et al (2022) M6A-mediated upregulation of circMDK promotes tumorigenesis and acts as a nanotherapeutic target in hepatocellular carcinoma. Mol Cancer 21:109
Xu J, Ji L, Liang Y, Wan Z, Zheng W (2020) CircRNA-SORE mediates sorafenib resistance in hepatocellular carcinoma by sta bilizing YBX1. Signal Transduct Target Ther 5:298
Huang G, Liang M, Liu H et al (2020) CircRNA hsa_circRNA_104348 promotes hepatocellular carcinoma progression through modulating miR-187-3p/RTKN2 axis and activating Wnt/β-catenin pathway. Cell Death Dis 11:1065
Hafeez U, Parslow AC, Gan HK, Scott AM (2020) New insights into ErbB3 function and therapeutic targeting in cancer. Expert Rev Anticancer Ther 20:1057–1074
Kiavue N, Cabel L, Melaabi S et al (2020) ERBB3 mutations in cancer: biological aspects, prevalence and therapeutics. Oncogene 39:487–502
Bai S, Xiong X, Tang B et al (2020) Exosomal circ_DLGAP4 promotes diabetic kidney disease progression by sponging miR-143 and targeting ERBB3/NF-κB/MMP-2 axis. Cell Death Dis 11:1008
Zhang Y, Tan D, Xie Y et al (2022) CircEPSTI1 Promotes the Proliferation of HER2-Positive Breast Cancer Cells via circEPSTI1/miR-145/ERBB3 Axis. J Oncol 2022:1028851
Wang Q, Wu S, Zhu H et al (2017) Deletion of PRKAA triggers mitochondrial fission by inhibiting the autophagy-dependent degradation of DNM1L. Autophagy 13:404–422
Ding WB, Wang MC, Yu J et al (2021) HBV/pregenomic RNA increases the stemness and promotes the development of HBV-related HCC through reciprocal regulation with insulin-like growth factor 2 mRNA-binding protein 3. Hepatology 74:1480–1495
Huang H, Weng H, Sun W et al (2018) Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol 20:285–295
Wang X, Tian L, Li Y et al (2021) RBM15 facilitates laryngeal squamous cell carcinoma progression by regulating TMBIM6 stability through IGF2BP3 dependent. J Exp Clin Cancer Res CR 40:80
Zhang N, Shen Y, Li H et al (2022) The m6A reader IGF2BP3 promotes acute myeloid leukemia progression by enhancing RCC2 stability. Exp Mol Med 54:194–205
Acknowledgements
Not applicable.
Funding
This study was supported by the National Natural Science Foundation of China [82003117].
Author information
Authors and Affiliations
Contributions
Conceptualization: JY, XW, ZF, LG, and DL; Methodology: JY, XW, ZF, and YX; Formal analysis and investigation: JY, XW, ZF, YX, YZ, and YL; Writing—original draft preparation: JY, XW, ZF, and YX; Writing—review and editing: YZ, YL, LG, and DL; Funding acquisition: DL; Resources: YX; Supervision: YZ.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
10495_2023_1903_MOESM1_ESM.tif
Fig. S1. Overexpression of circ_0098823 significantly promoted HCC cells migration, invasion, and mitochondrial fission(A) Overexpressing efficacy of circ_0098823 in HCCLM3 cells was confirmed by RT-qPCR (n=3).(B) The viability of HCCLM3 cells after overexpression of circ_0098823 was measured by CCK-8 assays (n=6). (C) The migration and invasion of HCCLM3 cells after overexpressing of circ_0098823 was measured by transwell assays. Scale bars, 50 μm.(D) The migration statistics of HCCLM3 cells after circ_0098823 overexpressing (n=3).(E) The invasion statistics of HCCLM3 cells after circ_0098823 overexpressing (n=3).(F) The apoptosis of HCCLM3 cells after overexpressing of circ_0098823 was measured by flow cytometry assays.(G) The apoptosis statistics of HCCLM3 cells after circ_0098823 overexpressing (n=3).(H) Electron microscopy observed the number of mitochondria in HCCLM3 cells after overexpressing of circ_0098823 (n=3). Scale bars, 2 μm and 50 μm.ns means P > 0.05, * means P < 0.05, ** means P < 0.01, *** means P < 0.001. (TIF 9030 kb)
10495_2023_1903_MOESM2_ESM.tif
Fig. S2. Overexpression of DNM1L significantly promotes the migration and invasion of HCC cells and inhibited apoptosis.(A) Overexpressing efficacy of DNM1L in HCCLM3 and HepG2 cells were confirmed by RT-qPCR.(B) The viability of HCCLM3 cells after overexpressing of DNM1L was measured by CCK-8 assays (n=6).(C) The migration and invasion of HCCLM3 and HepG2 cells after overexpressing of DNM1L was measured by transwell assays. Scale bars, 50 μm.(D) The migration statistics of HCCLM3 and HepG2 cells after DNM1L overexpressing.(E) The invasion statistics of HCCLM3 and HepG2 cells after DNM1L overexpressing.(F) The apoptosis statistics of HCCLM3 and HepG2 cells after DNM1L overexpressing.(G) The apoptosis of HCCLM3 and HepG2 cells after overexpressing of DNM1L were measured by flow cytometry assays.** means P < 0.01, *** means P < 0.001, n=3. (TIF 6464 kb)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yan, J., Wang, X., Fan, Z. et al. Circ_0098823 binding with IGF2BP3 regulates DNM1L stability to promote metastasis of hepatocellular carcinoma via mitochondrial fission. Apoptosis 29, 709–725 (2024). https://doi.org/10.1007/s10495-023-01903-8
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10495-023-01903-8