Abstract
It is well established that chromosomes occupy distinct positions within the interphase nuclei, conferring a potential functional implication to the genome. In addition, alterations in the nuclear organisation patterns have been associated with disease phenotypes (e.g. cancer or laminopathies). The human sperm is the smallest cell in the body with specific DNA packaging and the mission of delivering the paternal genome to the oocyte during fertilisation. Studies of nuclear organisation in the sperm have postulated nonrandom chromosome position and have proposed a chromocentre model with the centromeres facing toward the interior and the telomeres toward the periphery of the nucleus. Most studies have assessed the nuclear address in the sperm longitudinally predominantly using centromeric or telomeric probes and to a lesser extent with whole chromosome paints. To date, studies investigating the radial organisation of human sperm have been limited. The purpose of this study was to utilise whole chromosome paints for six clinically important chromosomes (18, 19, 21, 22, X, and Y) to investigate nuclear address by assessing their radial and longitudinal nuclear organisation. A total of 10,800 sperm were analysed in nine normozoospermic individuals. The results have shown nonrandom chromosome position for all chromosomes using both methods of analysis. We present novel radial and polar analysis of chromosome territory localization within the human sperm nucleus. Specifically, a hierarchical organisation was observed radially with chromosomes organised from the interior to the periphery (chromosomes 22, 21, Y, X, 19, and 18 respectively) and polar organisation from the sperm head to tail (chromosomes X, 19, Y, 22, 21, and 18, respectively). We provide evidence of defined nuclear organisation in the human sperm and discuss the function of organisation and potential possible clinical ramifications of these results in regards to male infertility and early human development.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The spermatozoon is a highly elaborate, specialised cell. Spermatozoa are formed through the process of spermatogenesis, which can be summarised in three phases: (1) mitotic proliferation of spermatogonia to produce spermatocytes; (2) meiotic divisions to produce genetically diverse haploid spermatids; and (3) cytodifferentiation/spermiogenesis to package the paternal chromosomes for effective delivery to the oocyte as immature spermatids become mature sperm cells. Unlike other mammalian cells, mature spermatozoa are unique because they are the smallest cell in the human body, they possess a flagellum (essential for motility), they are functionally silent with no gene transcription (Ward and Coffey 1991) and their chromatin is remodelled resulting in highly condensed packaging of haploid DNA. During the latter stages of spermiogenesis, sperm chromatin is reorganised with histones being replaced first by transition proteins (Meistrich et al. 2003) followed by protamines (Ward and Coffey 1991). Quantitatively, 15 % of chromatin remains bound to histones (Wykes and Krawetz 2003) with the remainder of the DNA bound by protamines, which allow a higher degree of chromatin compaction (10−6-fold compared to 10−5 offered by histones) (Bjorndahl and Kvist 2009). The functional significance of histone-intact chromatin is believed to play a vital role in early embryo development (Carrell and Hammoud 2009). Despite the unique properties of the human sperm cell and its importance in human fertilisation and embryo development, relatively few studies have been published examining the organisation of chromosomes in human sperm (Haaf and Ward 1995; Zalensky et al. 1995; Hazzouri et al. 2000; Tilgen et al. 2001; Mudrak et al. 2005; Finch et al. 2008b; Ioannou et al. 2011).
Irrefutable evidence has been provided to demonstrate that chromosomes occupy distinct positions within the nucleus termed chromosome territories (CTs; Manuelidis 1990; Cremer and Cremer 2001; Parada and Misteli 2002; Meaburn and Misteli 2007). Two models have emerged to explain the radial position of chromosomes in the interphase nucleus. These are largely based on (1) gene density and (2) chromosome size. The gene density model suggests gene poor chromosomes orient themselves toward the nuclear periphery (associated with gene repression), with gene-rich chromosomes (associated with gene activation) being located toward the nuclear interior (Croft et al. 1999; Boyle et al. 2001; Lukasova et al. 2002; Federico et al. 2008; Foster and Bridger 2005; Meaburn and Misteli 2007; Takizawa et al. 2008; Szczerbal et al. 2009). The chromosome size model suggests small CTs are located toward the centre, with larger chromosomes being located toward the periphery (Bolzer et al. 2005; Bridger et al. 2000). It is clear each model may be cell type specific, with some systems that fit both models (chicken and New World monkeys) (Mora et al. 2006; Habermann et al. 2001; Skinner et al. 2009) and others such as murine which do not follow either model (Meaburn et al. 2008). It has been proposed that these two models are not mutually exclusive and chromosome position may depend on the status of the cell or the specific chromosome and the chromosomal neighbourhood (Foster and Bridger 2005; Cavalli 2007).
Cell differentiation has been an attractive process to study nuclear organisation in relation to functional implications. Several studies have shown proximity associations in differentiated cells (Kuroda et al. 2004) tissue-specific organisation (Szczerbal et al. 2009), even an advantage toward a specific nuclear architecture (Solovei et al. 2009) to accommodate certain functionality. Organisation of the human sperm nucleus appears to be very different than other somatic cells. Chromosomes appear to be clustered via their centromeres to form a chromocentre in the interior of the nucleus, while the telomeres are preferentially located toward the nuclear periphery where they form dimers (Zalensky et al. 1993, 1995; Zalenskaya and Zalensky 2004; Solov’eva et al. 2004; Luetjens et al. 1999). The chromocentre appears to be formed by pericentric heterochromatin from different chromosomes that have a tendency to aggregate (Martin et al. 2006; Zalensky et al. 1993; Ioannou et al. 2011). A similar spatial organisation in sperm is retained in other mammals, as data from mouse (Haaf and Ward 1995; Meyer-Ficca et al. 1998), bovine, pig, horse, and rat (Zalenskaya and Zalensky 2004) indicates. It has been proposed that this configuration could be in favour of an ordered withdrawal of chromosomes through their association with the sperm microtubule machinery (Zalensky and Zalenskaya 2007).
Thus far, only a handful of studies have examined the position of chromosomes in human sperm with the vast majority of studies utilising centromeric, telomeric and/or locus-specific fluorescence in situ hybridization (FISH) probes (Haaf and Ward 1995; Zalensky et al. 1995; Hazzouri et al. 2000; Tilgen et al. 2001; Mudrak et al. 2005; Finch et al. 2008b; Ioannou et al. 2011). Only a few studies have utilised whole chromosome paints (WCPs) (chromosomes 1, 6, 13, and 18) to evaluate the longitudinal organisation in sperm (Hazzouri et al. 2000; Zalenskaya and Zalensky 2004; Luetjens et al. 1999). The functional implication of longitudinal organisation is postulated to be related to the specific order that chromosomes are affected by the maternal cytoplasmic environment after fertilisation (Zalensky and Zalenskaya 2007). Typically, nuclear organisation is addressed using radial assessment; despite this, few studies in sperm have performed this type of assessment (Ioannou et al. 2011; Finch et al. 2008b).
The possible functional implication of CTs is strengthened by observations that nuclear architecture is altered in disease states. More specifically, different patterns of organisation have been observed in cancer (Marella et al. 2009) and laminopathies (Foster and Bridger 2005; Misteli 2005; Elcock and Bridger 2010). In addition, findings have suggested nuclear changes in the development of diseases related to polysomies (Petrova et al. 2007), promyelocytic leukaemia, X-linked mental retardation and Huntington disease (Misteli 2005). Thus, it seems reasonable to hypothesise that a “healthy state” of nuclear organisation may become compromised in disease states or potentially that perturbation of the “healthy state” may lead to disease states. Given that nuclear organisation of chromosomes appears to be linked with cellular function, the unusual properties of the sperm cell and the vital role it plays in fertilisation and early human embryo development it would be of benefit to further our understanding of the nuclear architecture of the sperm cell. It would seem reasonable to suggest that alteration in CTs in sperm could also be associated with male infertility, which could have important clinical ramifications and possible implications for diagnosis and therapy (Ioannou and Griffin 2011).
In this study, WCPs for 18, 19, 21, 22, X, and Y have been used to analyse their nuclear address in nine males with normal semen parameters (as per World Health Organisation definitions) using both radial and longitudinal analysis. The aforementioned chromosomes were specifically selected as they are clinically significant being among the most commonly observed aneuploides in human development (Hassold and Hunt 2001). Chromosomes tested also represent gene poor chromosomes (18, 21, and Y) and relatively gene-rich chromosomes (19, 22, and X). Our findings will be discussed in the context of whether nonrandom organisation exists for the chromosomes chosen in our cohort and the potential implications for fertility and early human embryonic development.
Materials and methods
Patient recruitment and semen sample analysis
Informed written consent was provided by nine individuals undergoing routine semen analysis for assessment of fertility at IVF Florida Reproductive Associates in Margate, FL, USA. The fertility status of the female partner is unknown. However, we can conclude that presentation at a fertility clinic would indicate that the couples are having difficulties in conceiving. The average age of the individuals recruited to this study was 35.8 years (range, 29–48). Semen samples were collected via masturbation; subsequently, the semen parameters were evaluated using the World Health Organization criteria (World Health Organization 2010). The semen parameter assessment for each of the nine individuals is provided in Table 1. All samples were classified as normozoospermic, and hence are presumed to have normal reproductive function. Following the semen parameter assessment, the remainder of the semen sample surplus to requirements was snap frozen and stored in liquid nitrogen until use. This research study was approved by the Florida International University Institutional Review Board.
Semen sample preparation
Semen samples were thawed at room temperature, followed by repeated washings in 10 mM NaCl/10 mM Tris pH 7.0 (sperm wash buffer). This was followed by centrifugation for 7 min at 1,900 rotations per minute (rpm). The supernatant was removed and further resuspended in wash buffer three to five times, dependent on pellet size and supernatant colour to remove the seminal fluid. The sperm sample was then fixed using 3:1 methanol/acetic acid to a final volume of 5 ml followed by centrifugation for 7 min at 1,900 rpm and the supernatant was removed. The pellet was resuspended in fixative and the process above was repeated depending on the pellet size up to five times. Between 3 and 5 μl of the sample was placed on a glass slide (Fischer Brand®) and allowed to air dry. Using differential interference contrast microscopy (Olympus BX53), the area with the optimal density of cells was identified and marked with a diamond pen. Slides were stored overnight at room temperature to allow improved adherence of the sperm cells to the glass slide. All samples were stored at −20 °C in fixative when not used for experiments.
Sperm fluorescent in situ hybridisation
Following overnight storage of slides, sperm cells were dehydrated in ethanol series (70, 80, and 100 %) for 3 min each and allowed to air dry. This was followed by a 20 min incubation in 0.1 % DTT (Sigma Aldrich, St Louis, MO, USA), 0.1 % Tris solution (pH 8.0) in the dark, to swell the cells to allow the FISH probes to penetrate. Slides were then rinsed in 2× saline sodium citrate (SSC; Fisher Scientific, Pittsburgh, PA, USA) followed by another dehydration series in ethanol for 1 min and subsequent air drying. Aliquots of whole chromosome paints (Rainbow Scientific Inc., CO, USA) for the chromosomes of interest were prepared. The probes utilised included chromosomes 18 (LPP18G), 19 (LPP19R), 21 (LPP21R), 22 (LPP22G), X (LPPOXR), and Y (LPPOYG). When a dual colour combination was used, 0.2 μl from each probe was mixed per area of interest (alternatively, the amount of a single probe was 0.4 μl). The probe mix was added to the area of interest on the slide with a glass coverslip (5 mm) which was sealed with a Parafilm® coverslip. Co-denaturation of probe and sperm cells was performed at 75 °C for 10 min followed by hybridisation at 37 °C for approximately 24 h, using a Thermobrite® Statspin (Abbott Molecular, Illinois, IL, USA). Following completion of the hybridisation, the slides were removed from the Thermobrite® and placed in a 2× SSC/0.1 % Tween-20 (Sigma-Aldrich) solution at room temperature until the coverslips had floated off the slides. The slides were then washed in 0.4× SSC/0.3 % Tween-20 for 2 min at 72 °C (±1 °C) without agitation. Slides were immediately removed after the 2 min elapsed and placed in 2× SSC/0.1 % Tween-20 for 1 min at room temperature. A dehydration step followed for 1 min each. Slides were then mounted using 4′,6′-diamidino-2-phenylindole (DAPI; Vector Labs, Burlingame, CA, USA) under a 24 × 55 mm coverslip. Image acquisition was performed using an Olympus BX61 epifluorescence microscope equipped with a cooled charged coupled device camera (Hamamatsu ORCA—R 2 C10600) and single-band pass filters for the fluorochromes (fluorescein isothiocyanate, tetramethyl rhodamine isothiocyanate (TRITC), DAPI) of interest (Chroma Technology, Bellows Falls, VT, USA). All images were acquired using SmartCapture three software (Digital Scientific, UK) and exported as .tiff files for subsequent analysis. For each assay (radial and polar), a minimum of 100 cells per chromosome per patient were analysed.
Radial analysis of nuclear organisation
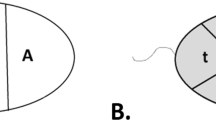
The principles of previously published methodologies (Boyle et al. 2001; Croft et al. 1999) were used to ascertain the radial chromosome position of the WCPs of interest. This was performed using a custom macro plug-in written for ImageJ software. The macro splits each captured image into separate red, green, and blue channels, which correspond to the two probe colours and the DAPI counterstain. The image representing the DAPI was then converted to a binary mask, from which concentric regions of interest (rings) of equal area were created (ring 1 representing the most interior part of the nucleus and ring 5 representing the most peripheral part of the nucleus; Fig. 1). The proportion of each of the WCP signal within each ring for each channel (i.e. red or green) was measured relative to the total signal for that channel contained within the area covered by the binary mask and output in a separate log file. These values were transferred to an excel spread sheet for statistical analysis. To compensate for the fact that we were analysing information of a 3D object under 2D, the proportion of signal observed in each ring was normalised against the measurements of the blue channel (DAPI density), that represent the amount of DNA present in that ring. Thus, signals in ring 5 (peripheral ring) received proportionally higher “scores” than those in the interior (ring 1) to adjust for the fact that centrally located observations were statistically more likely in a random distribution (Ioannou et al. 2011).
Polar analysis of nuclear organisation
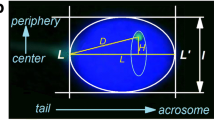
Sperm tails were visualised as background fluorescence on the TRITC filter in order to determine sperm polarity. The point of attachment of the tail to the nucleus was marked for each cell captured. ImageJ was utilised to measure (in pixels) the exact location of the CT within the sperm nuclei in reference to its location in relation to the sperm tail. A total of three measurements were performed for each chromosome per cell. The sperm cell has an ellipsoid shape; therefore, a transverse axis can be drawn along the nucleus from the point of attachment of the sperm tail to the opposite end (the “head” of the sperm cell). Thus, the first measurement provided the length of the sperm cell (from the sperm tail to the sperm head) in pixels. The second measurement was the distance in pixels from the sperm tail to the start of the CT. The third measurement was the distance in pixels between the sperm tail to the end of the CT (Fig. 2). These three measurements enable the localisation of the CT within the sperm nucleus. Specifically, the midpoint of the CT was calculated and used to describe the location of a given CT in relation to its location within the sperm nucleus. The midpoint of the CT was determined by addition of the measurement obtained for the distance from the sperm tail to the end of the CT to the measurement obtained from the sperm tail to the start of the CT. Subsequently, this value was divided by two giving the exact measurement of the midpoint of the CT from the sperm tail in pixels. In order to determine the location of the centre of the CT within the sperm nucleus the midpoint value was divided by the total length of the sperm. In doing so, this provided a value between 0 and 1. A value close to zero would indicate the midpoint of the CT resided close to the sperm tail; a value close to 1 would denote the CT resided in the apical region (“head”) of the sperm nucleus. In addition, to identifying the exact location of the CT within the sperm, it is also possible to subdivide the sperm nucleus into multiple regions to determine whether the CT preferentially occupied a specific region of the nucleus. Using the 0–1 values obtained, the sperm was divided into three regions to produce the tail, mid, and head region. Thus, if the midpoint of the CT was between the following values 0–0.33, 0.33–0.66, and 0.66–1 it would be designated to reside within the tail, mid, and head region, respectively.
The principle of the polar analysis. Whole chromosome paints for 19 (red) and 18 (green) hybridised to a sperm nucleus and counterstained with 4′,6-diamidino-2-phenylindole. The tail attachment was identified using Tritc filter and for presentational purposes is drawn in blue. The sperm cell periphery is depicted in white. A transverse axis was drawn on ImageJ (A) from the site of the tail attachment to the opposite side of the nucleus. Measurements for the tail attachment to the start of the chromosome territory (B) and the end of the chromosome territory (C) were also obtained. Subsequently, the value obtained for B and C was added and divided by two giving the exact measurement of the middle of the chromosome territory from the sperm tail in pixels. In order to determine the location of the midpoint of the chromosome territory within the sperm nucleus, this value was divided by the total length of the sperm. This provided a value between 0 and 1. A value close to 0 would suggest the probe resided close to the tail whereas a value close to 1 would mean the centre of the chromosome territory resided in the apical “head” region
Statistical analysis
Both the radial and polar analysis utilised the chi-squared (χ 2) goodness-of-fit statistical test to test for nonrandom distribution. The chi-squared goodness-of-fit test allows for a comparison to be made between the experimental data against an expected theoretical distribution (Ioannou et al. 2011). A p value of <0.05 was deemed to be significant, and in this case provided evidence to suggest that CTs exhibited non-random distribution and thus preferentially occupied a specific region of the sperm nucleus either radial, polar, or both. Further statistical analysis was undertaken for both assays to investigate interchromosomal differences using a cutoff p value of <0.05 to establish statistically significant differences.
Results
A total of 10,800 cells in nine individuals were analysed in this study, a minimum of 100 cells per patient, per chromosome were analysed for both radial and polar analysis. For each chromosome investigated 1,800 cells were analysed (900 for radial analysis and 900 for polar analysis) and for each patient 1,200 cells were analysed (600 for radial analysis and 600 for polar analysis).
Radial analysis of nuclear organisation
All investigated chromosomes (18, 19, 21, 22, X, and Y), showed evidence of nonrandom organisation as determined by the χ 2 goodness-of-fit (p < 0.05) in virtually all cases (Table 2). In only one instance was random organisation of the CT observed for (chromosome 18 in P1; Table 2). Contrary to what was observed for all other chromosomes and subjects, this suggests that in this case roughly equal amounts of fluorescence were observed in all five rings. The data output from the radial analysis is presented in two ways. Firstly, it provides a median location of the CT within the ring providing a reference for where the highest proportion of the fluorescence is observed. Secondly, it provides the actual percentage of fluorescence observed within each ring. Table 2 presents the median ring location of the CT for each chromosome for all nine individuals (ring 1 being the innermost and ring 5 being the outermost). For the most part, the data for all chromosomes demonstrated a median radial position at the border of rings 1–2 and 2, with an average median location of the six investigated CTs of between 2.05 and 2.49 (Table 2). The mean of the values presented in Table 2 provides the approximate radial order of the six chromosomes. Based on this data, the radial positioning from the innermost to the most peripheral, chromosomes were arranged as follows: 22, 21, Y, X, 19 and 18, respectively. Figure 3 presents the average nuclear address (pooled data from all nine individuals) for each of the chromosomes studied and illustrates the average amount of fluorescence for all nine subjects recorded in each ring for the six chromosomes investigated. These data clearly demonstrate preferential localisation of the CTs toward the nuclear interior with increased proportions of fluorescence in rings 1 and 2 with decreasing levels of fluorescence in rings 3–5. This data was compared to determine whether significant differences in the location of the six investigated chromosomes were observed. For the most part, no significant differences were observed between the chromosomes with the exception of chromosomes 18 and 22 and chromosomes 18 and 21. Chromosome 22 followed by chromosome 21 were the most internal with chromosome 18 being the most peripheral.
Presents an overview of the radial analysis. Each histogram shows the proportional distribution of fluorescence intensity (expressed as a percentage) in each of the rings 1–5 (with ring 1 being the innermost and ring 5 the outermost). The data represents pooled data for the nine subjects for each of the six investigated chromosomes. The error bars on each of the histograms depict the standard error of the mean
Polar analysis of nuclear organisation
In terms of polar analysis, evidence of nonrandom organisation was observed for all the investigated chromosomes in all subjects with the exception of chromosome 18 in two subjects (P4 and P5) and chromosome 21 in one subject (P5). The polar analysis measurements of the sperm cell length, the start and end of the CT relative to the sperm tail allowed the location of the CT to be established (Fig. 2). Specifically, the midpoint of the CT was utilised to determine the location of the CT within the sperm nucleus in relation to the sperm tail by dividing this value by the sperm cell length. For each cell, this produced a value between 0 and 1 (0 being the sperm tail and 1 being the sperm head). Table 3 provides the pooled polar data from nine patients for the six chromosomes. This table summarises the data from the 5,400 cells analysed. The average sperm cell length was 157.8 pixels (range, 137.35–170.17). Table 3 includes the average location of the midpoint of the CT, this value was utilised for subsequent analysis and the average location of the start and end of the CT is provided to demonstrate the size of the CT and the physical location of the entire CT in relation to the sperm tail. The polar organisation of the CTs can be ordered from the sperm head to the tail (chromosome X, 19, Y, 22, 21, and 18, respectively). Further analysis of the polar data allowed the sperm nucleus to be subdivided into sections and the location of the midpoint of the CT was assigned to the appropriate subdivision. In this study, we divided the sperm nucleus into three, four, and five subdivisions. Subdivision of the nuclei allowed the percentage of CTs which fall within these regions to be calculated rather than just providing an average location, the data for three subdivisions (head, mid, and tail region) is presented in Fig. 4. These data clearly demonstrate that chromosome X, 19, and Y were found predominantly within the head region with very low occurrences within the tail region. For chromosomes 22, 21, and 18, a lower proportion of the midpoint of the CT occurred within the head region favouring an increasing proportion within the mid and tail regions, respectively. The same pattern was observed when further dividing the sperm nucleus into four and five subdivisions (data not shown), but owing to the large size of the CTs further subdivision greater than three did not provide any further distinction between the chromosomes. Statistical analysis was performed to identify whether differences between the polar localisation of the six CTs was identified. A significant difference was observed between all the tested chromosomes with the exception of chromosomes 19 and Y and chromosomes 22 and 21. Thus, suggesting these particular chromosomes are within close proximity of each other, which is the case when they are ordered longitudinally from the sperm head to the sperm tail as per the suggested hierarchy (chromosomes X, 19, Y, 22, 21, and 18, respectively).
A summary of the data from polar analysis for all six investigated chromosomes in nine subjects. Each of the different coloured segments of the bar represents the mean location of the midpoint of the chromosome territory. Allocation of chromosome territories to head, mid, and tail is based on dividing the sperm nucleus into three regions using as a reference the point of the tail attachment to the nucleus. Chromosomes are ordered based on their proximity from the apical region to the tail
Discussion
In this study, we tested the hypothesis of nonrandom organisation of chromosomes 18, 19, 21, 22, X, and Y within the sperm nucleus in nine normozoospermic males using WCPs. We present strong evidence to support preferential organisation of the CTs for these chromosomes. In terms of radial analysis all chromosomes were found to be located more toward the nuclear interior than the nuclear periphery. We identified a hierarchical order with chromosomes arranged radially from the interior toward the periphery (chromosomes 22, 21, Y, X, 19, and 18, respectively) and longitudinally from the sperm head to the tail (chromosomes X, 19, Y, 22, 21, and 18, respectively). The interior location as determined by the radial analysis seems to affirm the presence of a chromocentre and the internal nuclear address of the sex chromosome territories (Zalensky et al. 1995; Zalenskaya and Zalensky 2004; Ioannou et al. 2011).
To the best of our knowledge, this is the first study to examine the radial organisation of whole chromosomes in human spermatozoa. To date, longitudinal analysis has arranged 11 chromosomes using WCPs and locus-specific probes in the order of X, 7, 6, 15, 16, 17, 1, Y, 18, 2, 5 (head toward the tail), with chromosome 13 occupying a random organisation (Zalenskaya and Zalensky 2004; Mudrak et al. 2005; Luetjens et al. 1999; Hazzouri et al. 2000). Longitudinal organisation of chromosomes in sperm has also been demonstrated to be evolutionarily conserved in marsupials and monotremes (Greaves et al. 2003). The polar analysis results from this study agree with current literature with regards to the location of chromosomes X and 18. However, our results for chromosome Y suggest a more apical localisation than previously reported (Zalenskaya and Zalensky 2004). This study presents novel data for polar nuclear organisation of CTs (chromosomes 19, 21, and 22) in human sperm. Chromosome 19 demonstrates an apical localisation with chromosomes 21 and 22 displaying preferential localisation in the mid region of the sperm nucleus.
Chromosome territories occupy a large region of the sperm nucleus (Figs. 1 and 2); in some instances, the probe size was equivalent to approximately one third of the length of the sperm nucleus. Due to the large size and diffuse nature of the hybridisation signals produced by WCPs, it is difficult to accurately evaluate the chromosome position in quantitative terms (Zalenskaya and Zalensky 2004). Despite the large size of the CTs, we were able to observe significant differences in the localisation of CTs by radial and polar analysis. The ability to divide the nucleus into five equal regions and measure the fluorescence intensity within each ring allows accurate assignment of the radial localisation within the nucleus. Radial analysis detects presence or absence of a CT and allows quantification of the amount of DNA within each ring. This mitigates the large probe size and allows accurate determination for the first time of the radial organisation of entire chromosomes in human sperm nuclei. The issues with CT size are more problematic for the polar analysis; at present, we are unable to quantify the amount of fluorescence within the nucleus in the same manner. Despite the CT size, we were able to observe significant differences in the location of different CTs, based on the midpoint of the CT. The centre of the CT was chosen as the reference point to provide a more accurate representation of the CT location within the nucleus. Novel data for three chromosomes is presented as well as confirmation of the location of two chromosomes from previously published studies. Based on our results, we believe that this method allows for an accurate identification of CT position and is capable of detecting differences in chromosome location. Therefore, this is a useful method to address CT polar organisation in sperm.
It should be clarified that the two types of analysis performed are mutually exclusive both in mode of assessment and the fact that they address different questions. However, this does not preclude the two methods from complementing each other. If one looks at the hierarchical organisation as determined by the radial analysis (chromosomes 22, 21, Y, X, 19, 18—innermost to outermost, respectively) and polar analysis (chromosomes X, 19, Y, 22, 21, 18—head to tail, respectively) these results seem conflicting. However, if Fig. 1 is considered, which demonstrates the radial analysis, we can see that chromosome 18 occupies rings 1–3 and chromosome 19 occupies rings 1–2. If this sperm is divided into three areas along the transverse axis from the sperm tail, each of the sections will contain rings 1 through 5. Therefore, chromosome 18, which occupies rings 1–3 will be found in the tail region but equally a chromosome occupying the same radial rings could be located in the mid or head region.
It has been established that the sperm chromatin undergoes reorganisation during fertilisation by removal of protamines, which are replaced by oocyte derived histones (Gao et al. 2004). In sperm, it has been hypothesised that paternal chromosomes are withdrawn from the nucleus in a sequential order, thus allowing a gradual exposure of chromosomes and chromosomal regions to the ooplasm (Zalensky and Zalenskaya 2007). This theoretically allows the gradual decondensation and remodelling of specific regions at a time, which could contribute to the differential gene activation patterns within the early embryo (Zalensky and Zalenskaya 2007; Foster and Bridger 2005). Organisation of CTs has important ramifications given the proposed sequential withdrawal of chromosomes from the sperm nucleus in the ooplasm, suggesting those located at the periphery and at the apical region are more likely to be exposed and activated first. Consequently, it is clear that more internally located chromosomes will be remodelled and activated at a later stage, thus providing a potential functional significance for the polar and radial positioning (Zalensky and Zalenskaya 2007). Conservation of chromosome arrangement and apical position of the X chromosome in the sperm of marsupials, monotremes and eutherians (with the exception of birds) suggests nuclear architecture is important and has a functional role. It is hypothesised that this may be crucial for spatial chromatin differentiation, normal development of fertilised eggs and may have evolved with other mammalian regulatory systems including genomic imprinting and X inactivation (Greaves et al. 2003). Thus, organisation within the sperm nucleus may not conform to either the gene density or chromosome size model as observed in other cell types and species. But will likely reflect important differences between the gene content of specific chromosomes required to be activated first and the requirement to occupy specific regions during spermatogenesis, fertilisation, and early embryo development. Therefore, the radial and polar localisation in terms of interior to periphery and head to tail will be crucial. Our polar analysis data suggest that gene rich chromosomes tend to occupy the apical and mid region (chromosomes 19, X, and 22) with gene poor chromosomes being preferentially located toward the mid and tail region. The one exception includes the Y chromosome, which is very gene poor but contains essential genes for sex determination and thus may have a selective advantage to be located toward the apical region of the sperm. This study provides little evidence to support the gene density model for radial organisation in human sperm.
Polar and radial organisation as shown in this study and previous studies may have important functional and clinical ramifications in humans. It is currently estimated that one in six couples are infertile, with up to 40 % attributed to male factors (Harton and Tempest 2012; Hann et al. 2011). The underlying cause of infertility in many remains idiopathic, with more genetic factors being identified. Male infertility has been associated with karyotype aberrations including structural and numerical (Hann et al. 2011; Harton and Tempest 2012; Shah et al. 2003), increased levels of sperm aneuploidy (Tempest 2011; Tempest and Griffin 2004), DNA damage (Barratt et al. 2010), mitochondrial dysfunction (Sharbatoghli et al. 2012), and aberrant DNA packaging (Hammoud et al. 2009) amongst others. Given the current hypothesis of gradual incorporation and remodelling of paternal DNA in the oocyte, any alteration in the organisation of the genome may have catastrophic consequences if the timing of gene activation is required in a specific sequence for successful fertilisation and early embryo development. Preliminary evidence of nuclear organisation has been provided for all chromosomes using locus-specific probes in human preimplantation embryos (Ioannou et al. 2012; Finch et al. 2008a). Therefore, altered sperm nuclear organisation may lead to aberrant organisation during embryo development which may manifest as infertility, recurrent fertilisation failure, poor embryo development, and recurrent miscarriage. In addition, concerns have also been raised regarding the increased levels of sex chromosome aneuploidies observed after intracytoplasmic sperm injection (ICSI; Harton and Tempest 2012; Hann et al. 2011). ICSI bypasses the acrosome reaction between the sperm and the oocyte, which has been shown to result in delayed decondensation of the apical region in sperm (Luetjens et al. 1999; Terada et al. 2000). This, in turn, could have negative effects on the first mitotic division of the zygote. Given the preferential localisation of the sex chromosomes and chromosome 19, this could result in mitotic errors that would be translated as viable chromosome aneuploidies and miscarriages in ICSI conceptions.
A handful of studies have investigated whether centromere organisation is altered in men with compromised semen parameters (Finch et al. 2008b; Ioannou et al. 2011; Olszewska et al. 2008; Sbracia et al. 2002). For the most, part no (Olszewska et al. 2008; Sbracia et al. 2002) or limited (Finch et al. 2008b; Ioannou et al. 2011) alterations in centromere position were observed between the control and patient group. Significant alteration in nuclear organisation in infertile males will likely depend on patient selection. Targeting those patients with recurrent fertilisation failure or poor embryo development may be the most appropriate patient cohort to investigate, rather than just reduced semen parameters. To date, only centromere organisation has been tested in infertile men. Perhaps testing centromere organisation alone will be less informative than assessing alteration of entire chromosomes given that the centromeres aggregate at the chromocentre. In order, for any alteration to be observed, this could require a major aberration to cause the centromeres to disaggregate from the chromocentre. Hence, alteration in the nuclear organisation of the CT may occur in terms of radial or polar location with potentially little measurable change in centromere location. One published study has reported a disruption in telomere–telomere interaction with loss of the looped chromosome configuration, associated with high levels of DNA damage in human spermatozoa (Moskovtsev et al. 2010). Therefore, assessing both CTs and telomeres may be a more informative approach to assess nuclear organisation in sperm.
In summary, we have provided evidence of nonrandom organisation of CTs in sperm of normozoospermic individuals by both means of radial and polar assessment. We plan to develop novel strategies for the polar analysis that may be able to quantify the distribution of the fluorescence and allow for automation of this type of analysis. Future studies involving more chromosomes and different chromosome regions could provide an understanding of the mechanistic basis of some causes of infertility, poor embryo development and early miscarriage. Ultimately, this could provide individuals and clinicians with valuable information to make more informed choices regarding assisted reproductive technologies.
Abbreviations
- CTs:
-
Chromosome territories
- DAPI:
-
4′,6-Diamidino-2-phenylindole
- DNA:
-
Deoxyribonucleic acid
- DTT:
-
Dithiothreitol
- FISH:
-
Fluorescence in situ hybridization
- HCl:
-
Hydrochloric acid
- ICSI:
-
Intracytoplasmic sperm injection
- NaCl:
-
Sodium chloride
- PBS:
-
Phosphate-buffered Saline
- Rpm:
-
Rotations per minute
- SSC:
-
Saline sodium citrate
- TRITC:
-
Tetramethyl rhodamine isothiocyanate
- Tris:
-
2-Amino-2-hydroxymethyl-propane-1,3-diol
- WCP:
-
Whole chromosome paint
- 2D:
-
Two dimensional
- 3D:
-
Three dimensional
References
Barratt CL, Aitken RJ, Bjorndahl L et al (2010) Sperm DNA: organization, protection and vulnerability: from basic science to clinical applications—a position report. Hum Reprod 25(4):824–838
Bjorndahl L, Kvist U (2009) Human sperm chromatin stabilization—a proposed model including zinc bridges. Mol Hum Reprod 16(1):23–29
Bolzer A, Kreth G, Solovei I et al (2005) Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes. PLoS Biol 3:e157
Boyle S, Gilchrist S, Bridger JM et al (2001) The spatial organization of human chromosomes within the nuclei of normal and emerin-mutant cells. Hum Mol Genet 10:211–219
Bridger JM, Boyle S, Kill IR, Bickmore WA (2000) Re-modelling of nuclear architecture in quiescent and senescent human fibroblasts. Curr Biol 10:149–152
Carrell DT, Hammoud SS (2009) The human sperm epigenome and its potential role in embryonic development. Mol Hum Reprod 16(1):37–47
Cavalli G (2007) Chromosome kissing. Curr Opin Genet Dev 17:443–450
Cremer T, Cremer C (2001) Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet 2:292–301
Croft JA, Bridger JM, Boyle S et al (1999) Differences in the localization and morphology of chromosomes in the human nucleus. J Cell Biol 145:1119–1131
Elcock LS, Bridger JM (2010) Exploring the relationship between interphase gene positioning, transcriptional regulation and the nuclear matrix. Biochem Soc Trans 38:263–267
Federico C, Cantarella CD, Di Mare P, Tosi S, Saccone S (2008) The radial arrangement of the human chromosome 7 in the lymphocyte cell nucleus is associated with chromosomal band gene density. Chromosoma 117:399–410
Finch KA, Fonseka G, Ioannou D et al (2008a) Nuclear organisation in totipotent human nuclei and its relationship to chromosomal abnormality. J Cell Sci 121:655–663
Finch KA, Fonseka KG, Abogrein A et al (2008b) Nuclear organization in human sperm: preliminary evidence for altered sex chromosome centromere position in infertile males. Hum Reprod 23:1263–1270
Foster HA, Bridger JM (2005) The genome and the nucleus: a marriage made by evolution. Genome organisation and nuclear architecture. Chromosoma 114:212–229
Gao S, Chung YG, Parseghian MH et al (2004) Rapid H1 linker histone transitions following fertilization or somatic cell nuclear transfer: evidence for a uniform developmental program in mice. Dev Biol 266:62–75
Greaves IK, Rens W, Ferguson-Smith MA, Griffin D, Marshall Graves JA (2003) Conservation of chromosome arrangement and position of the X in mammalian sperm suggests functional significance. Chromosome Res 11:503–512
Haaf T, Ward DC (1995) Higher order nuclear structure in mammalian sperm revealed by in situ hybridization and extended chromatin fibers. Exp Cell Res 219:604–611
Habermann FA, Cremer M, Walter J et al (2001) Arrangements of macro- and microchromosomes in chicken cells. Chromosome Res 9:569–584
Hammoud SS, Nix DA, Zhang H et al (2009) Distinctive chromatin in human sperm packages genes for embryo development. Nature 460:473–478
Hann MC, Lau PE, Tempest HG (2011) Meiotic recombination and male infertility: from basic science to clinical reality? Asian J Androl 13:212–218
Harton GL, Tempest HG (2012) Chromosomal disorders and male infertility. Asian J Androl 14:32–39
Hassold T, Hunt P (2001) To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet 2:280–291
Hazzouri M, Rousseaux S, Mongelard F et al (2000) Genome organization in the human sperm nucleus studied by FISH and confocal microscopy. Mol Reprod Dev 55:307–315
Ioannou D, Griffin DK (2011) Male fertility, chromosome abnormalities, and nuclear organization. Cytogenet Genome Res 133:269–279
Ioannou D, Meershoek EJ, Christopikou D et al (2011) Nuclear organisation of sperm remains remarkably unaffected in the presence of defective spermatogenesis. Chromosome Res 19:741–753
Ioannou D, Fonseka KG, Meershoek EJ et al (2012) Twenty-four chromosome FISH in human IVF embryos reveals patterns of post-zygotic chromosome segregation and nuclear organisation. Chromosome Res 20:447–460
Kuroda M, Tanabe H, Yoshida K et al (2004) Alteration of chromosome positioning during adipocyte differentiation. J Cell Sci 117:5897–5903
Luetjens CM, Payne C, Schatten G (1999) Non-random chromosome positioning in human sperm and sex chromosome anomalies following intracytoplasmic sperm injection. Lancet 353:1240
Lukasova E, Kozubek S, Kozubek M, Falk M, Amrichova J (2002) The 3D structure of human chromosomes in cell nuclei. Chromosome Res 10:535–548
Manuelidis L (1990) A view of interphase chromosomes. Science 250:1533–1540
Marella NV, Bhattacharya S, Mukherjee L, Xu J, Berezney R (2009) Cell type specific chromosome territory organization in the interphase nucleus of normal and cancer cells. J Cell Physiol 221:130–138
Martin C, Brochard V, Migne C et al (2006) Architectural reorganization of the nuclei upon transfer into oocytes accompanies genome reprogramming. Mol Reprod Dev 73:1102–1111
Meaburn KJ, Misteli T (2007) Cell biology: chromosome territories. Nature 445:379–781
Meaburn KJ, Newbold RF, Bridger JM (2008) Positioning of human chromosomes in murine cell hybrids according to synteny. Chromosoma 117(6):579–591
Meistrich ML, Mohapatra B, Shirley CR, Zhao M (2003) Roles of transition nuclear proteins in spermiogenesis. Chromosoma 111:483–488
Meyer-Ficca M, Muller-Navia J, Scherthan H (1998) Clustering of pericentromeres initiates in step 9 of spermiogenesis of the rat (Rattus norvegicus) and contributes to a well defined genome architecture in the sperm nucleus. J Cell Sci 111(Pt 10):1363–1370
Misteli T (2005) Concepts in nuclear architecture. Bioessays 27:477–487
Mora L, Sanchez I, Garcia M, Ponsa M (2006) Chromosome territory positioning of conserved homologous chromosomes in different primate species. Chromosoma 115:367–375
Moskovtsev SI, Willis J, White J, Mullen JB (2010) Disruption of telomere–telomere interactions associated with DNA damage in human spermatozoa. Syst Biol Reprod Med 56:407–412
Mudrak O, Tomilin N, Zalensky A (2005) Chromosome architecture in the decondensing human sperm nucleus. J Cell Sci 118:4541–4550
Olszewska M, Wiland E, Kurpisz M (2008) Positioning of chromosome 15, 18, X and Y centromeres in sperm cells of fertile individuals and infertile patients with increased level of aneuploidy. Chromosome Res 16:875–890
Parada L, Misteli T (2002) Chromosome positioning in the interphase nucleus. Trends Cell Biol 12:425–432
Petrova NV, Yakutenko II, Alexeevski AV et al (2007) Changes in chromosome positioning may contribute to the development of diseases related to X-chromosome aneuploidy. J Cell Physiol 213(1):278–283
Sbracia M, Baldi M, Cao D et al (2002) Preferential location of sex chromosomes, their aneuploidy in human sperm, and their role in determining sex chromosome aneuploidy in embryos after ICSI. Hum Reprod 17:320–324
Shah K, Sivapalan G, Gibbons N, Tempest H, Griffin DK (2003) The genetic basis of infertility. Reproduction 126:13–25
Sharbatoghli M, Valojerdi MR, Amanlou M, Khosravi F, Jafar-Abadi MA (2012) Relationship of sperm DNA fragmentation, apoptosis and dysfunction of mitochondrial membrane potential with semen parameters and ART outcome after intracytoplasmic sperm injection. Arch Gynecol Obstet 286(5):1315–1322
Skinner BM, Volker M, Ellis M, Griffin DK (2009) An appraisal of nuclear organisation in interphase embryonic fibroblasts of chicken, turkey and duck. Cytogenet Genome Res 126:156–164
Solovei I, Kreysing M, Lanctot C et al (2009) Nuclear architecture of rod photoreceptor cells adapts to vision in mammalian evolution. Cell 137:356–368
Solov’eva L, Svetlova M, Bodinski D, Zalensky AO (2004) Nature of telomere dimers and chromosome looping in human spermatozoa. Chromosome Res 12:817–823
Szczerbal I, Foster HA, Bridger JM (2009) The spatial repositioning of adipogenesis genes is correlated with their expression status in a porcine mesenchymal stem cell adipogenesis model system. Chromosoma 118:647–663
Takizawa T, Meaburn KJ, Misteli T (2008) The meaning of gene positioning. Cell 135:9–13
Tempest HG (2011) Meiotic recombination errors, the origin of sperm aneuploidy and clinical recommendations. Syst Biol Reprod Med 57:93–101
Tempest HG, Griffin DK (2004) The relationship between male infertility and increased levels of sperm disomy. Cytogenet Genome Res 107:83–94
Terada Y, Luetjens CM, Sutovsky P, Schatten G (2000) Atypical decondensation of the sperm nucleus, delayed replication of the male genome, and sex chromosome positioning following intracytoplasmic human sperm injection (ICSI) into golden hamster eggs: does ICSI itself introduce chromosomal anomalies? Fertil Steril 74:454–460
Tilgen N, Guttenbach M, Schmid M (2001) Heterochromatin is not an adequate explanation for close proximity of interphase chromosomes 1–Y, 9–Y, and 16–Y in human spermatozoa. Exp Cell Res 265:283–287
Ward WS, Coffey DS (1991) DNA packaging and organization in mammalian spermatozoa: comparison with somatic cells. Biol Reprod 44:569–574
World Health Organization (2010) WHO laboratory manual for the examination and processing of human semen. World Health Organization
Wykes SM, Krawetz SA (2003) The structural organization of sperm chromatin. J Biol Chem 278:29471–29477
Zalenskaya IA, Zalensky AO (2004) Non-random positioning of chromosomes in human sperm nuclei. Chromosome Res 12:163–173
Zalensky A, Zalenskaya I (2007) Organization of chromosomes in spermatozoa: an additional layer of epigenetic information? Biochem Soc Trans 35:609–611
Zalensky AO, Breneman JW, Zalenskaya IA, Brinkley BR, Bradbury EM (1993) Organization of centromeres in the decondensed nuclei of mature human sperm. Chromosoma 102:509–518
Zalensky AO, Allen MJ, Kobayashi A et al (1995) Well-defined genome architecture in the human sperm nucleus. Chromosoma 103:577–590
Acknowledgments
We would like to thank the following for their help in the success of this project: the patients who participated in this research study, IVF Florida Reproductive Associates staff, Michael Ellis (Digital Scientific UK), and Dr. Lakshmi Rao Kandukuri.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Conly Rieder.
Rights and permissions
About this article
Cite this article
Millan, N.M., Lau, P., Hann, M. et al. Hierarchical radial and polar organisation of chromosomes in human sperm. Chromosome Res 20, 875–887 (2012). https://doi.org/10.1007/s10577-012-9323-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10577-012-9323-y