Abstract
Purpose
As a primary risk factor and modifier of breast cancer incidence and prognosis, obesity may contribute to race disparities in breast cancer outcomes. This study examined association between obesity and DNA methylation in non-Hispanic Black and White women diagnosed with breast cancer.
Methods
Genome-wide DNA methylation was measured in the breast cancer tumor tissue of 96 women using the EPIC array. To examine the association between obesity and tumor methylation, linear regression models were used—regressing methylation β value for each cytosine and guanine (CpG) site on body mass index adjusting for covariates. Significance was set at false discovery rate (FDR) < 0.05. In the top 20 CpG sites, we explored the interactions with race and estrogen receptor (ER) status. We used multivariable Cox-proportional hazard models to examine whether methylation in the top 20 sites was associated with all-cause mortality.
Results
While none of the CpG sites passed the FDR threshold for significance, among the top 20 CpG sites, we observed interactions with race (TOMM20) and ER status (PSMB1, QSOX1 and PHF1). The same CpG sites in TOMM20, PSMB1, and QSOX1 were associated with all-cause mortality.
Conclusions
We identified novel interactions between obesity-associated methylation and both race and ER status in genes that have been associated with tumor regulation. Our data suggest that dysregulation in two sites may associate with all-cause mortality.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Breast cancer is the leading cause of cancer death in women; however, substantial heterogeneity exists in terms of prognosis by race [1], estrogen receptor (ER) status [2], and body size [3]. As obesity is a potentially modifiable prognostic factor, it is critical to understand its role in disparate breast cancer outcomes. Studies have reported conflicting associations between obesity and breast cancer, depending on various factors including hormone receptor status, race, menopausal status, and hormone therapy use [3]. For example, among pre-menopausal women, obesity has been associated with reduced risk of breast cancer in some, but not all, studies [3,4,5,6], whereas among post-menopausal women, obesity is consistently associated with an increased risk of breast cancer [7,8,9,10], although the association may vary by intrinsic subtype. Following a diagnosis of breast cancer, obesity is associated with poor outcomes irrespective of menopausal status or tumor characteristics [11,12,13,14] which may suggest that multiple mechanisms are involved across the carcinogenic pathway. Given the rising rates of obesity in cancer patients [3, 15], and disproportionate burden of obesity among minority women [16, 17], it is important to identify the underlying mechanisms driving the association between body size and breast cancer prognosis.
Epigenetic mechanisms are functionally relevant changes to the genome that do not involve a change to the underlying DNA sequence and are a promising biomarker that can comprehensively capture the result of both genetic and environmental influences. DNA methylation has become a promising target for assessing the etiology and progression of cancers due its malleability following environmental and lifestyle exposures and influence on gene expression [18]. Cancer cells are known to display aberrant methylation patterns leading to the silencing of tumor suppressor genes and increased expression of oncogenes [18, 19]. As a potential modulator of exposure-outcome relationships, uncovering obesity-related perturbations in tumor tissue may shed light on the pathophysiology of cancer progression and outcomes, informing behavioral and pharmacologic targets for intervention.
While the relationship between obesity and breast cancer prognosis has been examined using candidate gene approaches, no study to date has used a genome-wide approach to assess obesity-associated methylation signatures and outcomes in the tumor tissue of women diagnosed with a first-primary breast cancer. Thus, the purpose of this study was to conduct an epigenome-wide association study (EWAS) within breast tumor tissue to identify unknown CpG sites associated with obesity. We further explored modification by race, ER status, and downstream associations with breast cancer prognosis.
Methods
Study population
The Glenn Family Breast Satellite Tissue Bank at the Winship Cancer Institute of Emory University has a history of collecting clinical data and storing fresh tumor specimens from patients receiving surgery at three local hospitals in the metro-Atlanta area (Emory University Hospital, Emory University Hospital Midtown, and Grady Memorial Hospital). We used stratified sampling to identify 99 non-Hispanic Black (NHB) and non-Hispanic White (NHW) women diagnosed between 2008 and 2017 who were ideal weight, overweight, or obese. This sampling strategy was implemented to enhance our ability to detect obesity-associated tumor methylation and identify potential modification by race. Women were eligible for inclusion if they were 21 years of age or older, NHB or NHW by self-report, diagnosed with a first-primary stage I–III breast cancer, and received surgery at one of the participating hospitals. Women with a previous diagnosis of breast cancer or without an available fresh tissue specimen were excluded.
Data collection
Anthropometric and covariate data were obtained from the clinical records of women undergoing surgery. The primary exposure, body mass index (BMI, kg/m2), was derived from body weight and height, obtained at the time of the diagnosis. Age at diagnosis, family history of breast cancer, race, educational attainment, history of pregnancy, menarche and breast feeding, hormone replacement therapy use, and self-reported smoking status (current/former/never) were similarly abstracted from the clinical record. Clinical characteristics obtained from the record included: ER, progesterone receptor (PR), and human epidermal growth factor 2 (HER2)-status; tumor grade; receipt of chemotherapy, endocrine therapy, and radiation; comorbidities; as well as breast cancer and all-cause mortality. Poor prognosis was defined as all-cause mortality (including mortality from breast cancer). Given the short follow-up period (10 years), we anticipate that any mortality would be driven, in part, by underlying breast cancer [20].
Methylation data
DNA methylation was measured in 99 breast tumor tissue samples using the Illumina Infinium MethylationEPIC Beadchip (Illumina, San Diego, CA, USA). Methylation assays were performed in accordance with the Infinium HD Methylation Assay protocol. The protocol uses bisulfite treatment of the DNA to convert unmethylated cytosine to uracil, allowing identification of methylated vs. unmethylated loci. Following bisulfite treatment, two site-specific probes bind to sequences flanking methylated and unmethylated loci. The fluorescent signal from the methylated probe relative to the total signal for both methylated and unmethylated probes represents the proportions of DNA strands that are methylated at the CpG site [21]. This is described by the β-value, the proportion of probes methylated (M) divided by the combined methylated (M) and unmethylated (U) probes (β = M/[M + U]). The β-value is measured on a scale from 0–1, where 1 indicates 100% of the cells were methylated at the CpG site. Three samples were removed during pre-processing due to poor performance.
Quality control (QC) was conducted on the resulting data using the CpGassoc package [22]. Data points with low signal or detection p-values > 0.001 were set to missing, and 2869 CpG sites with missing values in > 10% of the samples were removed. All final samples had non-missing data for ≥ 95% of CpG sites, so no samples were removed. We additionally filtered out probes per general masking recommendations from Zhou et al. including probes with SNPs at a minor allele frequency > 1%, non-unique hybridization, probes with inconsistent mapping quality, probes with non-unique sub-sequence, and probes with a SNP that causes a color channel switch from the official annotation [23]. After QC, 759,156 CpG sites were evaluated.
Statistical analysis
Analyses were carried out using R (www.r-project.org/). Demographic characteristics were reported as frequencies or means. Mean methylation, as defined by the individual combined methylation β-value across the genome, was examined using linear regression models to assess whether individual mean β-values differed according to BMI, race, ER status, or poor prognosis, adjusting for model-specific covariates based on a-priori knowledge of the literature and causal graphical analyses [24, 25].
For the EWAS, the CpGassoc package was used to fit a linear regression model for each CpG site. Each regression modeled methylation and BMI as the primary predictor, adjusting for age, race, smoking status, and chip position. To account for potential chip-to-chip differences in measurement and to adjust for batch effects, a fixed effect for each BeadChip was included in all models. Significance was defined as a false discovery rate (FDR) q-value < 0.05. Since the β-values can display non-normal distribution, violating our assumption for linear regression, we examined the residuals of a select few CpG sites identified in the discovery and interaction analyses described below.
To assess whether the relationship between BMI and tumor methylation was modified by race or ER status (positive or negative), the top 20 CpG sites identified in the primary analysis were tested for interaction. For each of these CpG sites, we regressed the β-values on BMI with an interaction between BMI and race or ER status, respectively. We were underpowered to further explore intrinsic subtypes of breast cancer (ER, PR, HER2 status). All interaction analyses were adjusted for age, race (only in the ER interaction model), and chip position. Statistical significance was set at p < 0.05.
To examine whether BMI-associated methylation was associated with mortality, multivariable Cox-proportional hazard ratios (HR) were used to examine associations between the top 20 CpG sites and all-cause mortality. Stage, ER status, and treatment characteristics were considered as potential covariates in the model but were ultimately excluded—as causal graphical analyses suggested that they were likely mediators of the exposure-outcome relationship. Thus, final models were adjusted for age only. In sites found to interact with race or ER status, we additionally presented stratified results.
Sensitivity analysis
We conducted post hoc sensitivity analyses, excluding outliers in interaction models of race and ER status which appeared to have been driven by one or two patients. In our race interaction models, we additionally restricted to women with BMI > 45 since NHW are not represented in this range. In the survival analysis, we examined models adjusting for (1) age and BMI, and (2) age, ER status, and stage. Finally, due to the potential for neoadjuvant therapy to affect the breast tumor epigenome, and known associations with breast cancer prognosis, we performed additional sensitivity analyses excluding women who underwent neoadjuvant systemic therapy.
Results
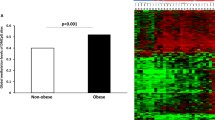
Demographic information is included in Table 1. NHB women in our study sample were older (mean age = 58 and 50 years, respectively) and had higher BMI than NHW women (BMI = 34.64 and 29.84 kg/m2, respectively, Supplemental Fig. 1). There were no differences by ER status, all-cause mortality, alcohol intake, and chemotherapy status between NHB and NHW women. When examining the participants mean methylation values collectively, we observed no differences by race, obesity status or breast cancer subtype, adjusting for covariates. However, overall DNA methylation was lower by 0.018 units (95% CI [− 0.032, − 0.005]) among women with 10-year mortality following breast cancer diagnosis compared to surviving patients adjusting for age, BMI, race, and smoking status.
For the primary analysis assessing the epigenome-wide association of DNA methylation with BMI, no sites passed the FDR threshold for significance. Given the exploratory nature of the analysis and the limited sample, we determined a priori to further investigate the top 20 CpG sites for interaction with race, ER status, and all-cause mortality. The top 20 sites are listed in Supplementary Table 1.
We first performed interaction analysis to investigate differences in the relationship between BMI and DNA methylation in NHB vs. NHW women. The relationship between BMI and DNA methylation varied by race at one site. In NHB women, as BMI increased by 1 kg/m2, methylation decreased by 0.003 at cg03731251 in the TOMM20 gene, whereas among NHW women methylation increased by 0.002 (p = 0.001; Fig. 1a). Following a sensitivity analysis excluding two extreme values, the relationship was no longer significant (p = 0.053; Supplemental Fig. 2a). An additional sensitivity analysis restricted to women with a BMI < 45 excluding 11 NHB women was no longer significant (p = 0.052; Supplemental Fig. 2b).
We performed similar tests of interaction to investigate the relationship between BMI and DNA methylation of the top 20 CpG sites by ER status. Three CpG sites, annotated to three genes: cg20174711 (PSMB1), cg08755040 (QSOX1), and cg23718418 (PHF1), were differentially associated by ER status. Among women who were ER-negative, as BMI increased by 1 kg/m2, methylation at cg20174711 in the PSMB1 gene decreased at 1.4 times the rate of methylation decrease compared to ER-positive patients (p = 0.00004; Fig. 1b). After a sensitivity analysis excluding one site, the interaction was no longer significant (p = 0.40; Supplemental Fig. 2c). Similarly, at the site cg08755040 in the QSOX1 gene with each incremental increase in BMI, methylation among women with ER-positive cancer increased at two times the rate of ER-negative cancer (p < 0.0001; Fig. 1c). Sensitivity analyses, removing a single outlier, remained significant (p = 0.0006, Supplemental Fig. 2d). Finally, at site cg23718418 (PHF1 gene), among women with ER-negative disease, methylation increased by 0.002 for every 1 kg/m2 increase in BMI, whereas the increase was less than 0.0005 for every 1 kg/m2 increase in BMI among women with ER-positive breast cancer (p = 0.002; Fig. 1d). After exclusion of one site, the interaction was no longer significant (p = 0.51; Supplemental Fig. 2e). We additionally examined the residuals of the four sites described above in the EWAS models to assess violations of a non-normal distribution (Supplemental Fig. 3). These plots appear to be consistent with a normal distribution.
We identified three sites that were associated with poor prognosis (Table 2). In the TOMM20 gene, every 1% increase in methylation in cg03731251 was associated with a 6% reduction in all-cause mortality (HR = 0.94; 95% CI [0.91, 0.99]). Our estimate was robust to subsequent adjustment by BMI (HR = 0.93; 95% CI [0.88, 0.99]), as well as ER status and stage (HR = 0.93; 95% CI [0.89, 0.97]). Upon removing women who had undergone neoadjuvant therapy (n = 22), the association remained significant (HR = 0.91; 95% CI [0.87, 0.95]). Given the interaction identified between cg03731251 and race, we additionally examined the stratified hazard ratios for each model in NHW and NHB women. The effects seem to be primarily driven by NHB women (NHW HR [95% CI] 1.98 [0.97, 4.04]; NHB HR [95% CI] 0.46 [0.22, 0.94]). In the PSMB1 gene, we also observed an inverse association between methylation at the cg20174711 site and all-cause mortality, where an 9% reduction was observed for every 1% increase in methylation (HR = 0.91; 95% CI [0.83, 0.99]). Our estimate remained significant when adjusting for BMI (HR = 0.89; 95% CI [0.79, 1.00]), ER status and stage (HR = 0.91; 95% CI [0.84, 0.99]). When excluding women who had undergone neoadjuvant therapy, the association similarly strengthened (HR = 0.84; 95% CI [0.76, 0.92]). Given the observed interaction with ER status, we additionally examined the stratified models between ER-positive and ER-negative tumor types. While our multivariable models did not reach the threshold of statistical significance, we observed a modest risk reduction among women with ER-positive tumors (ER-positive HR [95% CI] 0.92 [0.84, 0.98]). The QSOX1 gene was not associated with poor prognosis overall. However, among women who were ER-negative, every 1% increase in methylation was associated with a 71% reduction in all-cause mortality adjusting for age (ER-negative HR [95% CI] 0.29 [0.11, 0.77]), although our estimate was imprecise [26, 27].
Discussion
This study is the first untargeted analysis to examine BMI-associated methylation in breast cancer tumor tissue using the EPIC array. While a greater number of women is required to detect robust associations between tumor DNA methylation and obesity, we found unique interactions between the top BMI-associated methylation sites and race and ER status. One site in the TOMM20 displayed differential associations with race. Three sites in PSMB1, QSOX1, and PHF1 genes showed differential associations by ER status. Finally, two sites near the TOMM20 and PSMB1 genes, respectively, were associated with mortality.
Obesity and differential DNA methylation have previously been examined in 935 CpG sites of cancer-associated genes in ER-positive breast tumor tissue. Hair et al. found that BMI was associated with differential methylation in 30 CpG sites primarily related to immune response and insulin-like growth factor [28]. While none of the top 20 sites identified in our study replicated from Hair et al., this is likely due to differences in arrays used (EPIC and Illumina GoldenGate Cancer I Panel, respectively). Hair et al. noted that only 7 probes of the 30 identified in their analysis matched directly with probes in the Illumina HumanMethylation 450 Beadchip panel; suggesting limited overlap with the EPIC array. Importantly, this study did not examine heterogeneity by race or ER status and did not assess downstream impacts on breast cancer outcomes. To our knowledge, only one previous investigation, limited to 13 genes, has considered interactions between obesity and methylation on mortality [29].
In our study, an interaction between BMI-associated methylation and race was found in the TOMM20 gene. TOMM20 is a protein coding gene which encodes for Tom20, a protein essential to the recognition and translocation of mitochondrial preproteins [30, 31]. Tom20 is highly expressed in breast cancer tissue [32] and may bind with Aryl-hydrocarbon receptor-interacting protein (AIP) to mediate uptake of Survivin, an anti-apoptotic protein [33]. In our study, NHB exhibited an inverse association between BMI and methylation in cg03731251 of the TOMM20 gene, whereas NHW women exhibited a positive association between BMI and methylation. While no longer significant after removal of outliers, we found that hypomethylation at this site also associated with poor prognosis, but only among NHB women. Given the potential anti-apoptotic role of TOMM20, these data could suggest a pathway through which obesity may differentially impact prognosis by race.
PSMB1 was found to interact with ER status and was associated with all-cause mortality. PSMB1 is a member of the Proteasome β subunit (PSMB) family and has been shown to play a role in promoting breast cancer cell growth and migration [34]. Previous gene expression analyses have found differential expression of the PSMB1 gene between ER-positive versus ER-negative tumor tissue. Graham et al. found a 2.33-fold increased expression of PSMB1 in ER-positive compared to ER-negative tumor tissue [35]. In our study, as BMI increased, methylation decreased in both groups—albeit more pronounced in ER-negative tumors. Hypomethylation in this CpG site also associated with poor prognosis. Taken together, our findings suggest that CpG methylation in PSMB1 could play an important role in breast cancer progression and prognosis, given that individuals with decreased methylation had the more aggressive form of breast cancer (ER-negative) and poorer outcomes.
One site in QSOX1 was associated with ER status. Quiescin Sulfhydryl Oxidase 1 is an enzyme which catalyzes disulfide bond formation during protein folding and may play a role in growth regulation [36]. QSOX1 expression has previously been examined related to breast cancer with divergent findings. Pernodet et al. found that higher expression of QSOX1 was associated with reduced tumorigenesis and better outcomes [37]. Whereas Katchman et al. found higher expression of QSOX1 was associated with ER-positive tumors, higher tumor grade and poorer survival in ER-positive tumors. However, expression was not associated with survival in ER-negative, HER2 or TNBC tumors [38]. Similarly, Soloviev et al. found elevated QSOX1 transcription primarily in higher grade tumors, with 67% of grade 3 tumors exceeding the normal range of transcription [39]. Knutsvik et al. also found higher expression of QSOX1 was associated with poorer prognosis including high tumor grade, hormone receptor negativity, HER2 positivity, and increased tumor proliferation [40]. cg08755040 is located in the body of QSOX1. Methylation in the gene body has primarily been cited as signs of an active gene [41]. In our study, as BMI increased we observed similar increases in methylation among women with ER-positive tumors, which may suggest a positive association with expression. Given previous studies suggesting a deleterious effect of QSOX1 when expressed, particularly in ER-positive tumors, our results shed light on a specific pathway through which increased BMI may be influencing outcomes in ER-positive tumor types.
One CpG site in the PHF1 gene also interacted with ER status. PHF1 has been previously identified as an important regulator of histone methylation and an activator of the tumor suppressor p53 pathway. Moreover, it has been shown to be down-regulated in breast cancer tissue [42]. We observed an interaction with ER status, with greater BMI-associated methylation in ER-negative tumors compared to ER-positive tumors. Given that cg23718418 is located at the transcription start site, an increase in methylation may suggest decreased expression. While this may represent a valid mechanism underpinning the poorer prognosis associated with ER-negative tumors, we observed no downstream associations with all-cause mortality.
Our study has several limitations. With the limited sample size, we were underpowered to detect a main effect between CpG site methylation and BMI after correcting for multiple comparisons. Additionally, we did not correct for multiple testing in our interaction and hazard analyses, so it should be noted that these are suggestive relationships. We also found that some interaction analyses were no longer significant after exclusion of just one or two samples. For our analyses examining differences by race, we had fewer NHW women (with a lower distribution of BMI) limiting our ability to make strong inferences about interactions between BMI and DNA methylation by race. There are also several known risk factors for breast cancer that were not accounted for in our primary analysis including breastfeeding, nulliparity, and hormone therapy use. However, it is unresolved whether these covariates associate with breast tumor methylation, thus age-adjusted models are likely appropriate.
With a limited sample of patients, many of our results did not hold following exclusion of one or two individuals. While these results should be interpreted with caution, these preliminary data shed light on plausible epigenetic drivers of the association between obesity and breast cancer prognosis. The primary strength of this study was our untargeted approach to examine epigenetic pathways associating BMI with breast cancer prognosis in a diverse population of women undergoing surgery for breast cancer. Additional research efforts with a larger, equally diverse, patient pool are needed to validate our preliminary findings and further interrogate the biologic mechanisms identified here.
Data availability
The datasets generated during and/or analyzed during the current study are not publicly available due to IRB protocol but are available from the corresponding author on reasonable request to study PI (lauren.mccullough@emory.edu).
References
Huo D, Hu H, Rhie SK, Gamazon ER, Cherniack AD, Liu J, Yoshimatsu TF, Pitt JJ, Hoadley KA, Troester M, Ru Y, Lichtenberg T, Sturtz LA, Shelley CS, Benz CC, Mills GB, Laird PW, Shriver CD, Perou CM, Olopade OI (2017) Comparison of breast cancer molecular features and survival by African and European ancestry in the cancer genome AtlasBreast Cancer Molecular Features and Survival by African and European AncestryBreast Cancer Molecular Features and Survival by African and European Ancestry. JAMA Oncol 3(12):1654–1662. https://doi.org/10.1001/jamaoncol.2017.0595
Howlader N, Cronin KA, Kurian AW, Andridge R (2018) Differences in breast cancer survival by molecular subtypes in the United States. Cancer Epidemiol Biomark Prev 27(6):619–626. https://doi.org/10.1158/1055-9965.Epi-17-0627
Picon-Ruiz M, Morata-Tarifa C, Valle-Goffin JJ, Friedman ER, Slingerland JM (2017) Obesity and adverse breast cancer risk and outcome: mechanistic insights and strategies for intervention. Cancer J Clin 67(5):378–397. https://doi.org/10.3322/caac.21405
van den Brandt PA, Spiegelman D, Yaun SS, Adami HO, Beeson L, Folsom AR, Fraser G, Goldbohm RA, Graham S, Kushi L, Marshall JR, Miller AB, Rohan T, Smith-Warner SA, Speizer FE, Willett WC, Wolk A, Hunter DJ (2000) Pooled analysis of prospective cohort studies on height, weight, and breast cancer risk. Am J Epidemiol 152(6):514–527
Yang XR, Sherman ME, Rimm DL, Lissowska J, Brinton LA, Peplonska B, Hewitt SM, Anderson WF, Szeszenia-Dabrowska N, Bardin-Mikolajczak A, Zatonski W, Cartun R, Mandich D, Rymkiewicz G, Ligaj M, Lukaszek S, Kordek R, Garcia-Closas M (2007) Differences in risk factors for breast cancer molecular subtypes in a population-based study. Cancer Epidemiol Biomarkers Prev 16(3):439–443. https://doi.org/10.1158/1055-9965.Epi-06-0806
Turkoz FP, Solak M, Petekkaya I, Keskin O, Kertmen N, Sarici F, Arik Z, Babacan T, Ozisik Y, Altundag K (2013) Association between common risk factors and molecular subtypes in breast cancer patients. Breast 22(3):344–350. https://doi.org/10.1016/j.breast.2012.08.005
Renehan AG, Tyson M, Egger M, Heller RF, Zwahlen M (2008) Body-mass index and incidence of cancer: a systematic review and meta-analysis of prospective observational studies. Lancet 371(9612):569–578. https://doi.org/10.1016/s0140-6736(08)60269-x
Cheraghi Z, Poorolajal J, Hashem T, Esmailnasab N, Doosti Irani A (2012) Effect of body mass index on breast cancer during premenopausal and postmenopausal periods: a meta-analysis. PLoS ONE 7(12):e51446. https://doi.org/10.1371/journal.pone.0051446
Nattenmüller CJ, Kriegsmann M, Sookthai D, Fortner RT, Steffen A, Walter B, Johnson T, Kneisel J, Katzke V, Bergmann M, Sinn HP, Schirmacher P, Herpel E, Boeing H, Kaaks R, Kühn T (2018) Obesity as risk factor for subtypes of breast cancer: results from a prospective cohort study. BMC Cancer 18(1):616–616. https://doi.org/10.1186/s12885-018-4548-6
Reeves GK, Pirie K, Beral V, Green J, Spencer E, Bull D (2007) Cancer incidence and mortality in relation to body mass index in the Million Women Study: cohort study. BMJ 335(7630):1134. https://doi.org/10.1136/bmj.39367.495995.AE
Protani M, Coory M, Martin JH (2010) Effect of obesity on survival of women with breast cancer: systematic review and meta-analysis. Breast Cancer Res Treat 123(3):627–635. https://doi.org/10.1007/s10549-010-0990-0
Jiralerspong S, Goodwin PJ (2016) Obesity and breast cancer prognosis: evidence, challenges, and opportunities. J Clin Oncol 34(35):4203–4216. https://doi.org/10.1200/jco.2016.68.4480
Cespedes Feliciano EM, Kwan ML, Kushi LH, Chen WY, Weltzien EK, Castillo AL, Sweeney C, Bernard PS, Caan BJ (2017) Body mass index, PAM50 subtype, recurrence, and survival among patients with nonmetastatic breast cancer. Cancer 123(13):2535–2542. https://doi.org/10.1002/cncr.30637
Chan DSM, Vieira AR, Aune D, Bandera EV, Greenwood DC, McTiernan A, Vieira AR, Aune D, Navarro Rosenblatt D, Greenwood DC, Chan DSM, Bandera EV, Thune I, Vieira R, Norat T (2014) Body mass index and survival in women with breast cancer—systematic literature review and meta-analysis of 82 follow-up studies. Ann Oncol 25(10):1901–1914. https://doi.org/10.1093/annonc/mdu042
Garcia-Estevez L, Moreno-Bueno G (2019) Updating the role of obesity and cholesterol in breast cancer. Breast Cancer Res 21(1):35. https://doi.org/10.1186/s13058-019-1124-1
Pratt CA, Loria CM, Arteaga SS, Nicastro HL, Lopez-Class M, de Jesus JM, Srinivas P, Maric-Bilkan C, Schwartz Longacre L, Boyington JEA, Wouhib A, Gavini N (2017) A systematic review of obesity disparities research. Am J Prev Med 53(1):113–122. https://doi.org/10.1016/j.amepre.2017.01.041
Bilger M, Kruger EJ, Finkelstein EA (2017) Measuring socioeconomic inequality in obesity: looking beyond the obesity threshold. Health Econ 26(8):1052–1066. https://doi.org/10.1002/hec.3383
Kulis M, Esteller M (2010) DNA methylation and cancer. Adv Genet 70:27–56. https://doi.org/10.1016/b978-0-12-380866-0.60002-2
Herman JG, Baylin SB (2003) Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med 349(21):2042–2054. https://doi.org/10.1056/NEJMra023075
He W, Sofie Lindström L, Hall P, Czene K (2017) Cause-specific mortality in women with breast cancer in situ. Int J Cancer 140(11):2414–2421. https://doi.org/10.1002/ijc.30413
Bock C (2012) Analysing and interpreting DNA methylation data. Nat Rev Genet 13(10):705–719. https://doi.org/10.1038/nrg3273
Barfield RT, Kilaru V, Smith AK, Conneely KN (2012) CpGassoc: an R function for analysis of DNA methylation microarray data. Bioinformatics (Oxford, England) 28(9):1280–1281. https://doi.org/10.1093/bioinformatics/bts124
Zhou W, Laird PW, Shen H (2016) Comprehensive characterization, annotation and innovative use of Infinium DNA methylation BeadChip probes. Nucleic Acids Res 45(4):e22–e22. https://doi.org/10.1093/nar/gkw967
Krieger N, Davey Smith G (2016) The tale wagged by the DAG: broadening the scope of causal inference and explanation for epidemiology. Int J Epidemiol 45(6):1787–1808. https://doi.org/10.1093/ije/dyw114
Howards PP, Schisterman EF, Poole C, Kaufman JS, Weinberg CR (2012) Toward a clearer definition of confounding revisited with directed acyclic graphs. Am J Epidemiol 176(6):506–511. https://doi.org/10.1093/aje/kws127
Poole C (2001) Low P-values or narrow confidence intervals: which are more durable? Epidemiology 12(3):291–294. https://doi.org/10.1097/00001648-200105000-00005
Naimi AI, Whitcomb BW (2020) Can confidence intervals be interpreted? Am J Epidemiol. https://doi.org/10.1093/aje/kwaa004
Hair BY, Troester MA, Edmiston SN, Parrish EA, Robinson WR, Wu MC, Olshan AF, Swift-Scanlan T, Conway K (2015) Body mass index is associated with gene methylation in estrogen receptor-positive breast tumors. Cancer Epidemiol Biomark Prev 24(3):580–586. https://doi.org/10.1158/1055-9965.EPI-14-1017
McCullough LE, Chen J, Cho YH, Khankari NK, Bradshaw PT, White AJ, Garbowski G, Teitelbaum SL, Terry MB, Neugut AI, Hibshoosh H, Santella RM, Gammon MD (2016) DNA methylation modifies the association between obesity and survival after breast cancer diagnosis. Breast Cancer Res Treat 156(1):183–194. https://doi.org/10.1007/s10549-016-3724-0
Stelzer G, Rosen N, Plaschkes I, Zimmerman S, Twik M, Fishilevich S, Stein TI, Nudel R, Lieder I, Mazor Y, Kaplan S, Dahary D, Warshawsky D, Guan-Golan Y, Kohn A, Rappaport N, Safran M, Lancet D (2016) The GeneCards suite: from gene data mining to disease genome sequence analyses. Curr Protoc Bioinform. https://doi.org/10.1002/cpbi.5
TOMM20 gene. https://www.genecards.org/cgi-bin/carddisp.pl?gene=TOMM20.
Madamba SM, Damri KN, Dejean LM, Peixoto PM (2015) Mitochondrial ion channels in cancer transformation. Front Oncol 5:120–120. https://doi.org/10.3389/fonc.2015.00120
Kang BH, Xia F, Pop R, Dohi T, Socolovsky M, Altieri DC (2011) Developmental control of apoptosis by the immunophilin aryl hydrocarbon receptor-interacting protein (AIP) involves mitochondrial import of the survivin protein. J Biol Chem 286(19):16758–16767. https://doi.org/10.1074/jbc.M110.210120
Wang CY, Li CY, Hsu HP, Cho CY, Yen MC, Weng TY, Chen WC, Hung YH, Lee KT, Hung JH, Chen YL, Lai MD (2017) PSMB5 plays a dual role in cancer development and immunosuppression. Am J Cancer Res 7(11):2103–2120
Graham K, Ge X, de Las MA, Tripathi A, Rosenberg CL (2011) Gene expression profiles of estrogen receptor-positive and estrogen receptor-negative breast cancers are detectable in histologically normal breast epithelium. Clin Cancer Res 17(2):236–246. https://doi.org/10.1158/1078-0432.Ccr-10-1369
QSOX1 gene GeneCards. https://www.genecards.org/cgi-bin/carddisp.pl?gene=QSOX1
Pernodet N, Hermetet F, Adami P, Vejux A, Descotes F, Borg C, Adams M, Pallandre JR, Viennet G, Esnard F, Jouvenot M, Despouy G (2012) High expression of QSOX1 reduces tumorogenesis, and is associated with a better outcome for breast cancer patients. Breast Cancer Res 14(5):R136. https://doi.org/10.1186/bcr3341
Katchman BA, Ocal IT, Cunliffe HE, Chang YH, Hostetter G, Watanabe A, LoBello J, Lake DF (2013) Expression of quiescin sulfhydryl oxidase 1 is associated with a highly invasive phenotype and correlates with a poor prognosis in Luminal B breast cancer. Breast Cancer Res 15(2):R28. https://doi.org/10.1186/bcr3407
Soloviev M, Esteves MP, Amiri F, Crompton MR, Rider CC (2013) Elevated transcription of the gene QSOX1 encoding quiescin Q6 sulfhydryl oxidase 1 in breast cancer. PLoS ONE 8(2):e57327. https://doi.org/10.1371/journal.pone.0057327
Knutsvik G, Collett K, Arnes J, Akslen LA, Stefansson IM (2016) QSOX1 expression is associated with aggressive tumor features and reduced survival in breast carcinomas. Mod Pathol 29(12):1485–1491. https://doi.org/10.1038/modpathol.2016.148
Jones PA (2012) Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 13(7):484–492. https://doi.org/10.1038/nrg3230
Yang Y, Wang C, Zhang P, Gao K, Wang D, Yu H, Zhang T, Jiang S, Hexige S, Hong Z, Yasui A, Liu JO, Huang H, Yu L (2013) Polycomb group protein PHF1 regulates p53-dependent cell growth arrest and apoptosis. J Biol Chem 288(1):529–539. https://doi.org/10.1074/jbc.M111.338996
Funding
This work was funded by Developmental Funds from the Winship Cancer Institute of Emory University through the Brenda Nease Breast Cancer Fund and the Glenn Family Breast Center (PI: LEM, Grant No. 00067187), as well as the AVON Foundation for Women (PI: SGM, Grant No. 01-2016-087). The research was supported, in part, by Emory's Integrated Genomics Core shared resource, a core supported by the Winship Cancer Institute of Emory University. WD was supported in part by the Nalini and Ravi Saligram Scholarship through the Rollins School of Public Health at Emory University.
Author information
Authors and Affiliations
Contributions
LM conceived the research and designed the study. LM, SGM, and KG secured funding for the research. OD, MT, and LM, curated clinical data for the research. UK and LM obtained and processed specimens. LM and KC drafted the analytic plan and supervised all the work. WD performed the statistical analysis. JMK aided in data interpretation. LM and WD drafted the manuscript. All authors contributed to the development of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
All authors declare no conflicts of interest to disclose.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. All procedures performed in this study were approved by the Institutional Review Board of Emory University (PI: Mylin Torres, #00018512).
Informed consent
Patients have signed an informed consent form for studying their tumor tissues and other bio-specimens for research purposes, as part of the Breast Cancer Satellite Bank.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Do, W.L., Conneely, K., Gabram-Mendola, S. et al. Obesity-associated methylation in breast tumors: a possible link to disparate outcomes?. Breast Cancer Res Treat 181, 135–144 (2020). https://doi.org/10.1007/s10549-020-05605-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-020-05605-6