Abstract
Published data on the association between three polymorphisms (Lys939Gln, Ala499Val, and PAT±) of Xeroderma Pigmentosum group C (XPC) and breast cancer risk are inconclusive. To derive a more precise estimation of the relationship, a meta-analysis was performed. Crude ORs with 95% CIs were used to assess the strength of association between them. A total of 11 studies including 5,090 cases and 5,214 controls were involved in this meta-analysis. For XPC Lys939Gln polymorphism, no obvious associations were found for all genetic models when all studies were pooled into the meta-analysis (Lys/Gln vs. Lys/Lys: OR = 1.00, 95% CI 0.92–1.10; Gln/Gln vs. Lys/Lys: OR = 0.96, 95% CI 0.84–1.09; dominant model: OR = 0.99, 95% CI 0.91–1.08; and recessive model: OR = 0.97, 95% CI 0.86–1.09). In the subgroup analysis by ethnicity or study design, still no obvious associations were found. For XPC Ala499Val polymorphism, also no obvious associations were found for all genetic models when all studies were pooled into the meta-analysis (Val/Ala vs. Ala/Ala: OR = 0.91, 95% CI 0.79–1.05; Val/Val vs. Ala/Ala: OR = 1.07, 95% CI 0.80–1.44; dominant model: OR = 0.93, 95% CI 0.81–1.06; and recessive model: OR = 1.11, 95% CI 0.84–1.48). For XPC PAT± polymorphism, obvious associations were found for recessive model when all studies were pooled into the meta-analysis (OR = 1.41, 95% CI 1.05–1.89). In conclusion, this meta-analysis suggests that the XPC PAT± polymorphism allele may be a low-penetrant risk factor for developing breast cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Breast cancer is currently the most frequently occurring cancer and one of the leading causes of cancer-related death in the world, which has become a major public health challenge [1]. The mechanism of breast carcinogenesis is still not fully understood. It has been suggested that low-penetrance susceptibility genes combining with environmental factors may be important in the development of cancer [2]. In recent years, several common low-penetrant genes have been identified as potential breast cancer susceptibility genes [3–6]. An important one is Xeroderma Pigmentosum group C (XPC), which is localized at 3p25 and encodes a protein of 940 amino acids that in vivo form a supramolecular complex including HR23B [7]. XPC plays an important role in DNA repair [8]. In XPC gene, three common polymorphisms are widely studied: (a) a substitution of alanine for valine in codon 499 (Ala499Val), in the interaction domain of XPC with hHRAD23; (b) an A to C transversion in exon 15 resulting in a lysine-to-glutamine transition at position 939 (Lys939Gln), located in the interaction domain with TFIIH; and (c) a poly AT region on intron 9 [9]. These polymorphisms to breast cancer risk have been a research focus in scientific community and have drawn increasing attention. Several original studies have reported the role of XPC polymorphisms in breast cancer risk [10–17], but the results are inconclusive, partially because of the possible small effect of the polymorphism on breast cancer risk and the relatively small sample size in each of published studies. Therefore, we performed this meta-analysis to derive a more precise estimate of these associations.
Methods
Publication search
Medline, PubMed, Embase, and Web of Science were searched (last search was updated on Sep 10, 2010, using the search terms: “XPC”, “polymorphism”, and “breast”). All searched studies were retrieved, and their bibliographies were checked for other relevant publications. Review articles and bibliographies of other relevant studies identified were hand-searched to find additional eligible studies. Only published studies with full text articles were included. When more than one of the same patient population was included in several publications, only the most recent or complete study was used in this meta-analysis.
Inclusion criteria
The inclusion criteria were: (a) evaluation of the XPC polymorphisms and breast cancer risk, (b) case–control studies, and (c) sufficient published data for estimating an odds ratio (OR) with 95% confidence interval (CI).
Data extraction
Information was carefully extracted from all eligible publications independently by two of the authors according to the inclusion criteria listed above. Disagreement was resolved by discussion between the two authors. If these two authors could not reach a consensus, another author was consulted to resolve the dispute and a final decision was made by the majority of the votes. The following data were collected from each study: first author’s name, publication date, ethnicity, study design, total number of cases and controls, and numbers of cases and controls with the XPC genotypes, respectively. Different ethnicities were categorized as Caucasian, Asian, African, and mixed. Study design was stratified to population-based studies or hospital-based studies. We did not define any minimum number of patients to include in our meta-analysis.
Statistical methods
Crude ORs with 95% CIs were used to assess the strength of association between the XPC polymorphisms and breast cancer risk. The pooled ORs were performed for co-dominant model, dominant model, and recessive model, respectively. Heterogeneity assumption was checked by the chi-square-based Q-test [18]. A P value greater than 0.10 for the Q-test indicates a lack of heterogeneity among studies, so the pooled OR estimate of the each study was calculated by the fixed-effects model (the Mantel–Haenszel method) [19]. Otherwise, the random-effects model (the DerSimonian and Laird method) was used [20]. Subgroup analyses were performed by ethnicity and study design. Sensitivity analysis was performed to assess the stability of the results. A single study involved in the meta-analysis was deleted each time to reflect the influence of the individual data set to the pooled ORs. The results (95% CIs) changed from statistically significant to not significant were considered as significant change, vice versa [21]. An estimate of potential publication bias was carried out by the funnel plot, in which the standard error of log (OR) of each study was plotted against its log (OR). An asymmetric plot suggests a possible publication bias. Funnel plot asymmetry was assessed by the method of Egger’s linear regression test, a linear regression approach to measure funnel plot asymmetry on the natural logarithm scale of the OR. The significance of the intercept was determined by the t-test suggested by Egger (P < 0.05 was considered representative of statistically significant publication bias) [22]. If publication bias existed, the Duval and Tweedie non-parametric “trim and fill” method was used to adjust for it [23]. All the statistical tests were performed with STATA version 10.0 (Stata Corporation, College Station, TX).
Results
Study characteristics
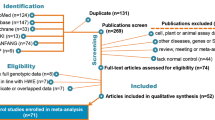
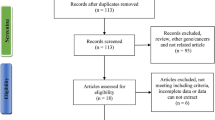
A total of eight publications met the inclusion criteria [10–17]. In the studies of Mechanic et al., Forsti et al., and Smith et al. [10, 13, 15], the ORs were presented separately according to different ethnicity. Therefore, each group in one study was considered separately for pooling subgroup analyses by ethnicity. Hence, a total of 11 studies including 5,090 cases and 5,214 controls were used in the meta-analysis. Table 1 lists the studies identified and their main characteristics.
Main results
Table 2 lists the main results of this meta-analysis. For XPC Lys939Gln polymorphism, no obvious associations were found for all genetic models when all studies were pooled into the meta-analysis (Lys/Gln vs. Lys/Lys: OR = 1.00, 95% CI 0.92–1.10; Gln/Gln vs. Lys/Lys: OR = 0.96, 95% CI 0.84–1.09; dominant model: OR = 0.99, 95% CI 0.91–1.08; and recessive model: OR = 0.97, 95% CI 0.86–1.09). In the subgroup analysis by ethnicity or study design, still no obvious associations were found. For XPC Ala499Val polymorphism, also no obvious associations were found for all genetic models when all studies were pooled into the meta-analysis (Val/Ala vs. Ala/Ala: OR = 0.91, 95% CI 0.79–1.05; Val/Val vs. Ala/Ala: OR = 1.07, 95% CI 0.80–1.44; dominant model: OR = 0.93, 95% CI 0.81–1.06; and recessive model: OR = 1.11, 95% CI 0.84–1.48). For XPC PAT± polymorphism, obvious associations were found for recessive model when all studies were pooled into the meta-analysis (OR = 1.41, 95% CI 1.05–1.89).
Sensitivity analysis
A single study involved in the meta-analysis was deleted each time to reflect the influence of the individual data set to the pooled ORs, and the corresponding pooled ORs were not materially altered (data not shown), indicating that our results were statistically robust.
Publication bias
Begg’s funnel plot and Egger’s test were performed to access the publication bias of literatures for XPC Lys939Gln polymorphism. The shape of the funnel plot revealed obvious asymmetry except for recessive model (figures not shown). Then, the Egger’s test was used to provide statistical evidence of funnel plot asymmetry. The results still suggested no obvious publication bias except for recessive model (P = 0.061 for Lys/Gln vs. Lys/Lys; P = 0.195 for Gln/Gln vs. Lys/Lys; P = 0.192 for dominant model; and P = 0.034 for recessive model). The Duval and Tweedie non-parametric “trim and fill” method was used to adjust for publication bias. Meta-analysis with and without “trim and fill” method did not draw different conclusion (data not shown), indicating that our results were statistically robust.
Discussion
To demonstrate the relationship between three polymorphisms (Lys939Gln, Ala499Val, and PAT±) of XPC and breast cancer risk, a meta-analysis involving 5,090 cases and 5,214 controls was performed. For XPC Lys939Gln and Ala499Val polymorphisms, no obvious associations were found for all genetic models. For XPC PAT± polymorphism, obvious associations were found for recessive model. It suggests that the XPC PAT± polymorphism allele may be a low-penetrant risk factor for developing breast cancer. Since the PAT± polymorphism occurs in an intronic sequence and may be linkage disequilibrium with another XPC polymorphism. The polymorphism located at the −5 position of intron 11 is a possible candidate since it is at a splice acceptor site which causes a deletion and skipping of exon 12 and is in strong linkage disequilibrium with the PAT± polymorphism [24–26]. It is suggested that the homozygous PAT+/+ genotype shows decreased DNA NER capacity and increased risk of cancer compared to the other genotypes [27].
Some limitations of this meta-analysis should be acknowledged. Firstly, in the subgroup analyses, the number of Asians and Africans were relatively small, not having enough statistical power to explore the real association. Secondly, our results were based on unadjusted estimates, while a more precise analysis should be conducted if individual data were available, which would allow for the adjustment by other co-variants including age, ethnicity, menopausal status, smoking status, drinking status, obesity, environmental factors, and other lifestyle [28].
In conclusion, this meta-analysis suggests that the XPC PAT± polymorphism allele may be a low-penetrant risk factor for developing breast cancer. However, it is necessary to conduct large sample studies using standardized unbiased genotyping methods, homogeneous breast cancer patients, and well-matched controls. Moreover, gene–gene and gene–environment interactions should also be considered in the analysis. Such studies taking these factors into account may eventually lead to our better, comprehensive understanding of the association between the XPC polymorphisms and breast cancer risk.
References
Parkin DM, Bray F, Ferlay J, Pisani P (2005) Global cancer statistics 2002. CA Cancer J Clin 55:74–108
Pharoah PD, Dunning AM, Ponder BA, Easton DF (2004) Association studies for finding cancer-susceptibility genetic variants. Nat Rev Cancer 4:850–860
Qiu LX, Zhang J, Zhu XD, Zheng CL, Sun S, Wang ZH (2010) The p21 Ser31Arg polymorphism and breast cancer risk: a meta-analysis involving 51, 236 subjects. Breast Cancer Res Treat 124:475–479
Qiu LX, Yao L, Zhang J, Zhu XD, Zhao XM, Xue K (2010) XPD Lys751Gln polymorphism and breast cancer susceptibility: a meta-analysis involving 28, 709 subjects. Breast Cancer Res Treat 124:229–235
Qiu LX, Yao L, Mao C, Chen B, Zhan P, Xue K (2010) TGFB1 L10P polymorphism is associated with breast cancer susceptibility: evidence from a meta-analysis involving 47, 817 subjects. Breast Cancer Res Treat 123:563–567
Qiu LX, Yao L, Xue K, Zhang J, Mao C, Chen B (2010) BRCA2 N372H polymorphism and breast cancer susceptibility: a meta-analysis involving 44, 903 subjects. Breast Cancer Res Treat 123:487–490
Araki M, Masutani C, Takemura M (2001) Centrosome protein centrin 2/caltractin 1 is part of the xeroderma pigmentosum group C complex that initiates global genome nucleotide excision repair. J Biol Chem 276:18665–18672
Sugasawa K, Shimizu Y, Iwai S, Hanaoka F (2002) A molecular mechanism for DNA damage recognition by the xeroderma pigmentosum group C protein complex. DNA Repair 1:95–107
Khan SG, Metter EJ, Tarone RE (2000) A new xeroderma pigmentosum group C poly(AT) insertion/deletion polymorphism. Carcinogenesis 21:1821–1825
Smith TR, Levine EA, Freimanis RI, Akman SA, Allen GO, Hoang KN (2008) Polygenic model of DNA repair genetic polymorphisms in human breast cancer risk. Carcinogenesis 29:2132–2138
Shen J, Gammon MD, Terry MB, Teitelbaum SL, Eng SM, Neugut AI (2008) Xeroderma pigmentosum complementation group C genotypes/diplotypes play no independent or interaction role with polycyclic aromatic hydrocarbons-DNA adducts for breast cancer risk. Eur J Cancer 44:710–717
Jorgensen TJ, Visvanathan K, Ruczinski I, Thuita L, Hoffman S, Helzlsouer KJ (2007) Breast cancer risk is not associated with polymorphic forms of xeroderma pigmentosum genes in a cohort of women from Washington County, Maryland. Breast Cancer Res Treat 101:65–71
Mechanic LE, Millikan RC, Player J (2006) Polymorphisms in nucleotide excision repair genes, smoking and breast cancer in African Americans and whites: a population-based case-control study. Carcinogenesis 27:1377–1385
Zhang L, Zhang Z, Yan W (2005) Single nucleotide polymorphisms for DNA repair genes in breast cancer patients. Clin Chim Acta 359:150–155
Forsti A, Angelini S, Festa F (2004) Single nucleotide polymorphisms in breast cancer. Oncol Rep 11:917–922
Shen J, Desai M, Agrawal M (2006) Polymorphisms in nucleotide excision repair genes and DNA repair capacity phenotype in sisters discordant for breast cancer. Cancer Epidemiol Biomarkers Prev 15:1614–1619
Shore RE, Zeleniuch-Jacquotte A, Currie D, Mohrenweiser H, Afanasyeva Y, Koenig KL (2008) Polymorphisms in XPC and ERCC2 genes, smoking and breast cancer risk. Int J Cancer 122:2101–2105
Cochran WG (1954) The combination of estimates from different experiments. Biometrics 10:101–129
Mantel N, Haenszel W (1959) Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 22:719–748
DerSimonian R, Laird N (1986) Meta-analysis in clinical trials. Control Clin Trials 7:177–188
Tobias A (1999) Assessing the influence of a single study in the meta-analysis estimate. Stata Tech Bull 8:15–17
Egger M, Davey Smith G, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315:629–634
Taylor SJ, Tweedie RI (1998) Practical estimates of the effect of publication bias in meta- analysis. Australas Epidemiologist 5:14–17
Khan SG, Muniz-Medina V, Shahlavi T (2002) The human XPC DNA repair gene: arrangement, splice site information content and influence of a single nucleotide polymorphism in a splice acceptor site on alternative splicing and function. Nucleic Acids Res 30:3624–3631
Marin MS, Lopez-Cima MF, Garcia-Castro L, Pascual T, Marron MG, Tardon A (2004) Poly (AT) polymorphism in intron 11 of the XPC DNA repair gene enhances the risk of lung cancer. Cancer Epidemiol Biomarkers Prev 13:1788–1793
Sak SC, Barrett JH, Paul AB, Bishop DT, Kiltie AE (2005) The polyAT, intronic IVSII-6 and Lys939Gln XPC polymorphisms are not associated with transitional cell carcinoma of the bladder. Br J Cancer 92:2262–2265
Qiao Y, Spitz MR, Shen H, Guo Z, Shete S, Hedayati M (2002) Modulation of repair of ultraviolet damage damage in the host-cell reactivation assay by polymorphic XPC and XPD/ERCC2 genotypes. Carcinogenesis 23:295–299
Hirschhorn JN, Lohmueller K, Byrne E (2002) A comprehensive review of genetic association studies. Genet Med 4:45–61
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zheng, W., Cong, XF., Cai, WH. et al. Current evidences on XPC polymorphisms and breast cancer susceptibility: a meta-analysis. Breast Cancer Res Treat 128, 811–815 (2011). https://doi.org/10.1007/s10549-011-1369-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-011-1369-6