Abstract
Published data on the association between TGFB1 L10P polymorphism and breast cancer risk are inconclusive. In order to derive a more precise estimation of the relationship, a meta-analysis was performed. Crude ORs with 95% CIs were used to assess the strength of association between them. A total of 30 studies including 20,401 cases and 27,416 controls were involved in this meta-analysis. Overall, significantly elevated breast cancer risk was associated with TGFB1 10P allele when all studies were pooled into the meta-analysis (LP vs. LL: OR = 1.046, 95% CI = 1.003–1.090; dominant model: OR = 1.052, 95% CI = 1.012–1.095). In the subgroup analysis by ethnicity, statistically significantly elevated risk was found in Caucasians (dominant model: OR = 1.045, 95% CI = 1.001–1.091). When stratified by study design, statistically significantly elevated risk was found based on population-based studies (dominant model: OR = 1.076, 95% CI = 1.019–1.136). In conclusion, this meta-analysis suggests that the TGFB1 10P allele may be a low-penetrant risk factor for developing breast cancer. However, large sample and representative population-based studies with homogeneous breast cancer patients and well-matched controls are warranted to confirm this finding.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Breast cancer is currently the most frequently occurring cancer and one of the leading causes of cancer-related deaths in the world, which has become a major public health challenge [1]. The mechanism of breast carcinogenesis is still not fully understood. It has been suggested that low-penetrance susceptibility genes combining with environmental factors may be important in the development of cancer [2]. In recent years, several common low-penetrant genes have been identified as potential breast cancer susceptibility genes. An important one is transforming growth factor B1 (TGFB1), which is located at 19q13.1 [3]. Transgenic animal experiments indicate that increased expression of TGFB1 is protective against early cancer development, particularly in breast cancer [4]. Several polymorphisms have been identified in TGFB1. One of the most widely studied polymorphisms is TGFB1 L10P polymorphism (rs1982073), a T to C transition in the 29th nucleotide resulting in a leucine (L) to proline (P) substitution at the 10th amino acid [5]. This polymorphism to breast cancer risk has been a research focus in scientific community and has drawn increasing attention. Several original studies have reported the role of TGFB1 L10P polymorphism in breast cancer risk [6–20], but the results are inconclusive, partially because of the possibly insignificant effect of the polymorphism on breast cancer risk and the relatively small sample size in each of published studies. Therefore, we performed this meta-analysis to derive a more precise estimation of these associations.
Methods
Publication search
Medline, PubMed, Embase, and Web of Science were searched (last search was updated on December 30, 2009, using the search terms: “TGFB1,” “TGFβ1,” “polymorphism,” and “breast”). All searched studies were retrieved, and their bibliographies were checked for other relevant publications. Review articles and bibliographies of other relevant studies identified were hand-searched to find additional eligible studies. Only published studies with full text articles were included. When more than one of the same patient population was included in several publications, only the most recent or with complete study was used in this meta-analysis.
Inclusion criteria
The inclusion criteria were: (a) evaluation of the TGFB1 L10P polymorphism and breast cancer risk, (b) case–control studies, and (c) sufficient published data for estimating an odds ratio (OR) with 95% confidence interval (CI).
Data extraction
Information was carefully extracted from all eligible publications independently by two of the authors according to the inclusion criteria listed above. Disagreement was resolved by discussion between the two authors. If these two authors could not reach a consensus, another author was consulted to resolve the dispute, and a final decision was made by the majority of the votes. The following data were collected from each study: first author’s name, publication date, ethnicity, study design, total number of cases and controls, and numbers of cases and controls with the TGFB1 L10P genotypes, respectively. Different ethnicities were categorized as Caucasian, Asian, African, and mixed. Study design was stratified to population-based studies, hospital-based studies, or nested case–control studies. We did not define any minimum number of patients for inclusion in our meta-analysis.
Statistical methods
Crude ORs with 95% CIs were used to assess the strength of association between the TGFB1 L10P polymorphism and breast cancer risk. The pooled ORs were performed for co-dominant model (LP vs. LL, PP vs. LL), dominant model (LP + PP vs. LL), and recessive model (PP vs. LL + LP). Heterogeneity assumption was checked by the χ2-based Q-test [21]. A P value greater than 0.10 for the Q-test indicates a lack of heterogeneity among studies, and so the pooled OR estimate of the each study was calculated by the fixed-effects model (the Mantel–Haenszel method) [22]. Otherwise, the random-effects model (the DerSimonian and Laird method) was used [23]. Subgroup analyses were performed by ethnicity and study design. Sensitivity analysis was performed to assess the stability of the results. A single study involved in the meta-analysis was deleted each time to reflect the influence of the individual data set to the pooled ORs [24]. An estimate of potential publication bias was carried out by the funnel plot, in which the standard error of log (OR) of each study was plotted against its log (OR). An asymmetric plot suggests a possible publication bias. Funnel plot asymmetry was assessed by the method of Egger’s linear regression test, a linear regression approach to measure funnel plot asymmetry on the natural logarithm scale of the OR. The significance of the intercept was determined by the t-test suggested by Egger (P < 0.05 was considered representative of statistically significant publication bias) [25]. If publication bias existed, then the Duval and Tweedie nonparametric “trim and fill” method was used to adjust for it [26]. All the statistical tests were performed with STATA version 10.0 (Stata Corporation, College Station, TX).
Results
Study characteristics
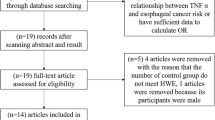
A total of 15 publications met the inclusion criteria [6–20]. In several publications, the ORs were presented separately according to the different subgroup. Therefore, each group in one publication was considered separately for subgroup analysis. Hence, a total of 30 studies including 20,401 cases and 27,416 controls were involved in this meta-analysis. Table 1 lists the studies identified and their main characteristics. Of the 30 studies, sample sizes ranged from 123 to 10,193. There were 20 studies of Caucasians, eight studies of Asians, one study of Africans, and one study of mixed populations. Almost all of the cases were pathologically confirmed. Controls were mainly healthy populations and matched for age. Among these studies, eight were population-based, 12 were hospital-based, and eight were nested case–control studies.
Main results
Table 2 lists the main results of this meta-analysis. Overall, significantly elevated breast cancer risk was associated with TGFB1 10P allele when all the studies were pooled into the meta-analysis (LP vs. LL: OR = 1.046, 95% CI = 1.003–1.090; dominant model: OR = 1.052, 95% CI = 1.012–1.095). In the subgroup analysis by ethnicity, statistically significantly elevated risk was found in Caucasians (dominant model: OR = 1.045, 95% CI = 1.001–1.091). When stratified by study design, statistically significantly elevated risk was found based on population-based studies (dominant model: OR = 1.076, 95% CI = 1.019–1.136).
Sensitivity analysis
A single study involved in the meta-analysis was deleted each time to reflect the influence of the individual data-set to the pooled ORs, and the corresponding pooled ORs were not materially altered (data not shown), indicating that our results were statistically robust.
Publication bias
Begg’s funnel plot and Egger’s test were performed to access the publication bias of literatures. The shape of the funnel plot did not reveal obvious asymmetry (figures not shown). Then, the Egger’s test was used to provide statistical evidence of funnel plot symmetry. The results still suggest no evidence of publication biases for LP vs. LL (P = 0.899) and dominant model (P = 0.675). However, modest publication biases were found for PP vs. LL (P = 0.047) and recessive model (P = 0.041). The Duval and Tweedie nonparametric “trim and fill” method was used to adjust for publication bias. Meta-analysis with and without “trim and fill” method did not draw different conclusions (data not shown), indicating that our results were statistically robust.
Discussion
The present meta-analysis, which included 20,401 cases and 27,416 controls, explored the association between the TGFB1 L10P polymorphism and breast cancer risk. Our results indicated that significantly increased breast cancer risk was found in TGFB1 10P allele carriers, which is contradictory with the biological function study. In vitro transfection experiments suggest that TGFB1 10P allele is associated with higher circulating levels of TGFB1 and increases TGFB1 secretion [7]. From a dual-role model for the action of TGFB1, TGFB1 is thought to inhibit the development of early benign tumors, but once somatic oncogenic mutations have destroyed the normal tumor suppressor action of TGFB1, it then promotes tumor invasion and metastasis [27, 28]. It is thought that the 10P allele would increase TGFB1 secretion and be associated with a reduced risk of in situ tumors but an increased risk of invasive cancer [27, 28]. Unfortunately, our study had insufficient information for subgroup analysis to detect whether there is a significant differential risk of ductal carcinoma in situ or invasive breast cancer. However, although the size of our meta-analysis is large, we cannot rule out the possibility that the association we found is a false positive one.
In the subgroup analysis based on ethnicities, significantly elevated risks were associated with the 10P allele in the Caucasians. However, no significant associations were found in the Asians and Africans. Actually, it might not be uncommon for the same polymorphism playing different roles in cancer susceptibility among different ethnic populations, because cancer is a complicated multi-genetic disease, and different genetic backgrounds may contribute to the discrepancy [29]. In Asians and Africans, the influence of the 10P allele might be masked by the presence of other as-yet unidentified causal genes involved in breast cancer development. In addition, the differences might arise by chance because studies with small sample size may have insufficient statistical power to explore a slight effect or may have generated a fluctuated risk estimate [30]. Considering the limited studies and limited numbers of Asians and Africans included in the meta-analysis, our results should be interpreted with caution.
Our results indicated that significantly increased breast cancer risk in TGFB1 10P allele carriers were found based on the population-based studies but not on hospital-based studies. This may be due to the fact that the hospital-based studies have some biases because such controls may just represent a sample of ill-defined reference population, and may not be a true representative of the general population, particularly when the genotypes under investigation were associated with the disease conditions that the hospital-based controls may have. Therefore, using a proper and representative population-based study is very important to reduce biases in such genetic association studies.
Some limitations of this meta-analysis should be acknowledged. First, the controls were not uniformly defined. Although most of the controls were selected mainly from healthy populations, some had benign disease. Therefore, non-differential misclassification bias was possible because these studies may have included the control groups who have different risks of developing breast cancer. Second, in the subgroup analyses, the number of Asians and Africans were relatively small, not having enough statistical power to explore the real association. Third, our results were based on unadjusted estimates, while a more precise analysis should be conducted if individual data were available, which would allow for the adjustment by other covariants including age, ethnicity, menopausal status, smoking status, drinking status, obesity, environmental factors, and other lifestyle habits.
In conclusion, this meta-analysis suggests that the TGFB1 10P allele may be a low-penetrant risk factor for developing breast cancer. However, it is necessary to conduct large sample studies using standardized unbiased genotyping methods, homogeneous breast cancer patients and well-matched controls. Moreover, gene–gene and gene–environment interactions should also be considered in the analysis. Such studies taking these factors into account may eventually lead to better, comprehensive understanding of the association between the TGFB1 L10P polymorphism and breast cancer risk by us.
References
Parkin DM, Bray F, Ferlay J, Pisani P (2005) Global cancer statistics 2002. CA Cancer J Clin 55:74–108
Lichtenstein P, Holm NV, Verkasalo PK (2000) Environmental and heritable factors in the causation of cancer. N Engl J Med 343:78–85
Fujii D, Brissenden JE, Derynck R, Francke U (1986) Transforming growth factor beta gene maps to human chromosome 19 long arm and to mouse chromosome 7. Somat Cell Mol Genet 12:281–288
Grainger DJ, Heathcote K, Chiano M, Snieder H, Kemp PR, Metcalfe JC (1999) Genetic control of the circulating concentration of transforming growth factor type beta1. Hum Mol Genet 8:93–97
Yokota M, Ichihara S, Lin TL, Nakashima N, Yamada Y (2000) Association of a T29/C polymorphism of the transforming growth factor-beta1 gene with genetic susceptibility to myocardial infarction in Japanese. Circulation 101:2783–2787
Ziv E, Cauley J, Morin PA, Saiz R, Browner WS (2001) Association between the T29 → C polymorphism in the transforming growth factor beta1 gene and breast cancer among elderly white women: the study of osteoporotic fractures. Jama 285:2859–2863
Dunning AM, Ellis PD, McBride S (2003) A transforming growth factor beta1 signal peptide variant increases secretion in vitro and is associated with increased incidence of invasive breast cancer. Cancer Res 63:2610–2615
Hishida A, Iwata H, Hamajima N (2003) Transforming growth factor B1 T29C polymorphism and breast cancer risk in Japanese women. Breast Cancer 10:63–69
Krippl P, Langsenlehner U, Renner W (2003) The L10P polymorphism of the transforming growth factor-beta 1 gene is not associated with breast cancer risk. Cancer Lett 201:181–184
Jin Q, Hemminki K, Grzybowska E (2004) Polymorphisms and haplotype structures in genes for transforming growth factor beta1 and its receptors in familial and unselected breast cancers. Int J Cancer 112:94–99
Le Marchand L, Haiman CA, van den Berg D, Wilkens LR, Kolonel LN, Henderson BE (2004) T29C polymorphism in the transforming growth factor beta1 gene and postmenopausal breast cancer risk: the Multiethnic Cohort Study. Cancer Epidemiol Biomarkers Prev 13:412–415
Saha A, Gupta V, Bairwa NK, Malhotra D, Bamezai R (2004) Transforming growth factor-beta1 genotype in sporadic breast cancer patients from India: status of enhancer, promoter, 5′-untranslated-region and exon-1 polymorphisms. Eur J Immunogenet 31:37–42
Kaklamani VG, Baddi L, Liu J (2005) Combined genetic assessment of transforming growth factor-beta signaling pathway variants may predict breast cancer risk. Cancer Res 65:3454–3461
Lee KM, Park SK, Hamajima N (2005) Genetic polymorphisms of TGF-beta1 and TNF-beta and breast cancer risk. Breast Cancer Res Treat 90:149–155
Shin A, Shu XO, Cai Q, Gao YT, Zheng W (2005) Genetic polymorphisms of the transforming growth factor-beta1 gene and breast cancer risk: a possible dual role at different cancer stages. Cancer Epidemiol Biomarkers Prev 14:1567–1570
Feigelson HS, Patel AV, Diver WR, Stevens VL, Thun MJ, Calle EE (2006) Transforming growth factor beta receptor type I and transforming growth factor beta1 polymorphisms are not associated with postmenopausal breast cancer. Cancer Epidemiol Biomarkers Prev 15:1236–1237
Scola L, Vaglica M, Crivello A (2006) Cytokine gene polymorphisms and breast cancer susceptibility. Ann N Y Acad Sci 1089:104–109
Cox DG, Penney K, Guo Q (2007) TGFB1 and TGFBR1 polymorphisms and breast cancer risk in the Nurses’ Health Study. BMC Cancer 7:175–181
Cox A, Dunning AM, Garcia-Closas M (2007) A common coding variant in CASP8 is associated with breast cancer risk. Nat Genet 39:352–358
Rajkumar T (2008) TGFb1 (Leu10Pro), p53 (Arg72Pro) can predict for increased risk for breast cancer in south Indian women and TGFb1 Pro (Leu10Pro) allele predicts response to neo-adjuvant chemo-radiotherapy. Breast Cancer Res Treat 112:81–87
Cochran WG (1954) The combination of estimates from different experiments. Biometrics 10:101–129
Mantel N, Haenszel W (1959) Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 22:719–748
DerSimonian R, Laird N (1986) Meta-analysis in clinical trials. Control Clin Trials 7:177–188
Tobias A (1999) Assessing the influence of a single study in the meta-analysis estimate. Stata Tech Bull 8:15–17
Egger M, Davey Smith G, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315:629–634
Taylor SJ, Tweedie RI (1998) Practical estimates of the effect of publication bias in meta- analysis. Australas Epidemiol 5:14–17
Bierie B, Moses HL (2006) Tumour microenvironment: TGFbeta: the molecular Jekyll and Hyde of cancer. Nat Rev Cancer 6:506–520
Derynck R, Akhurst RJ, Balmain A (2001) TGF-beta signaling in tumor suppression and cancer progression. Nat Genet 29:117–129
Hirschhorn JN, Lohmueller K, Byrne E (2002) A comprehensive review of genetic association studies. Genet Med 4:45–61
Wacholder S, Chanock S, Garcia-Closas M (2004) Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J Natl Cancer Inst 96:434–442
Author information
Authors and Affiliations
Corresponding author
Additional information
Li-Xin Qiu and Lei Yao have contributed equally to this work and should be considered as co-first authors.
Rights and permissions
About this article
Cite this article
Qiu, LX., Yao, L., Mao, C. et al. TGFB1 L10P polymorphism is associated with breast cancer susceptibility: evidence from a meta-analysis involving 47,817 subjects. Breast Cancer Res Treat 123, 563–567 (2010). https://doi.org/10.1007/s10549-010-0781-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-010-0781-7