Abstract
Published data on the association between BRCA2 N372H polymorphism and breast cancer risk are inconclusive. To derive a more precise estimation of the relationship, a meta-analysis was performed. Crude ORs with 95% CIs were used to assess the strength of association between them. A total of 22 studies including 22,515 cases and 22,388 controls were involved in this meta-analysis. Overall, no significant associations were found between BRCA2 N372H polymorphism and breast cancer risk when all studies pooled into the meta-analysis (NH versus NN: OR = 1.01, 95% CI = 0.97–1.05; HH versus NN: OR = 1.05, 95% CI = 0.97–1.13; dominant model: OR = 1.01, 95% CI = 0.98–1.05; and recessive model: OR = 1.05, 95% CI = 0.98–1.13). In the subgroup analysis by ethnicity, still no significant associations were found for Caucasians, Asians, or Africans. When stratified by study design, statistically significantly elevated risk was found for 372H allele based on population-based studies (HH versus NN: OR = 1.11, 95% CI = 1.01–1.21; dominant model: OR = 1.05, 95% CI = 1.00–1.10; recessive model: OR = 1.09, 95% CI = 1.00–1.18). In conclusion, this meta-analysis suggests that the BRCA2 372H allele may be a low-penetrant risk factor for developing breast cancer. However, large sample and representative population-based studies with homogeneous breast cancer patients and well matched controls are warranted to confirm this finding.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Breast cancer is currently the most frequently occurring cancer and one of the leading causes of cancer-related death in the world, which has become a major public health challenge [1]. The mechanism of breast carcinogenesis is still not fully understood. It has been suggested that low-penetrance susceptibility genes combining with environmental factors may be important in the development of cancer [2]. In recent years, several common low-penetrant genes have been identified as potential breast cancer susceptibility genes. An important one is BRCA2. Carriers of rare, mostly protein-truncating mutations in the BRCA2 gene are strongly predisposed to developing breast cancer [3]. The only common non-synonymous polymorphism in BRCA2 is N372H polymorphism (rs144848) with the A to C change resulting in a asparagine (Asn, N) to histidine (His, H) transition in exon 10 [4]. This polymorphism to breast cancer risk has been a research focus in scientific community and has drawn increasing attention. Several original studies have reported the role of BRCA2 N372H polymorphism in breast cancer risk [5–13], but the results are inconclusive, partially because of the possible small effect of the polymorphism on breast cancer risk and the relatively small sample size in each of published studies. Therefore, we performed this meta-analysis to derive a more precise estimation of these associations.
Methods
Publication search
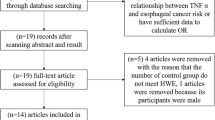
Medline, PubMed, Embase, and Web of Science were searched (last search was updated on Dec 30, 2009, using the search terms: “BRCA2”, “polymorphism” and “breast”). All searched studies were retrieved, and their bibliographies were checked for other relevant publications. Review articles and bibliographies of other relevant studies identified were hand-searched to find additional eligible studies. Only published studies with full text articles were included. When more than one of the same patient population was included in several publications, only the most recent or complete study was used in this meta-analysis.
Inclusion criteria
The inclusion criteria were: (a) evaluation of the BRCA2 N372H polymorphism and breast cancer risk, (b) case–control studies, and (c) sufficient published data for estimating an odds ratio (OR) with 95% confidence interval (CI).
Data extraction
Information was carefully extracted from all eligible publications independently by two of the authors according to the inclusion criteria listed above. Disagreement was resolved by discussion between the two authors. If these two authors could not reach a consensus, another author was consulted to resolve the dispute and a final decision was made by the majority of the votes. The following data were collected from each study: first author’s name, publication date, ethnicity, study design, total number of cases and controls, and numbers of cases and controls with the BRCA2 N372H genotypes, respectively. Different ethnicities were categorized as Caucasian, Asian, African, and mixed. Study design was stratified to population-based studies or hospital-based studies. We did not define any minimum number of patients to include in our meta-analysis.
Statistical methods
Crude ORs with 95% CIs were used to assess the strength of association between the BRCA2 N372H polymorphism and breast cancer risk. The pooled ORs were performed for co-dominant model (NH versus NN, HH versus NN), dominant model (NH + HH versus NN), and recessive model (HH versus NN + NH), respectively. Heterogeneity assumption was checked by the chi-square-based Q-test [14]. A P-value greater than 0.10 for the Q-test indicates a lack of heterogeneity among studies, so the pooled OR estimate of the each study was calculated by the fixed-effects model (the Mantel–Haenszel method) [15]. Otherwise, the random-effects model (the DerSimonian and Laird method) was used [16]. Subgroup analyses were performed by ethnicity and study design. Sensitivity analysis was performed to assess the stability of the results. A single study involved in the meta-analysis was deleted each time to reflect the influence of the individual data-set to the pooled ORs [17]. An estimate of potential publication bias was carried out by the funnel plot, in which the standard error of log (OR) of each study was plotted against its log (OR). An asymmetric plot suggests a possible publication bias. Funnel plot asymmetry was assessed by the method of Egger’s linear regression test, a linear regression approach to measure funnel plot asymmetry on the natural logarithm scale of the OR. The significance of the intercept was determined by the t-test suggested by Egger (P < 0.05 was considered representative of statistically significant publication bias) [18]. All the statistical tests were performed with STATA version 10.0 (Stata Corporation, College Station, TX).
Results
Study characteristics
A total of nine publications met the inclusion criteria [5–13]. In five of these publications, the ORs were presented separately according to the different subgroup. Therefore, each group in one publication was considered separately for subgroup analysis. Hence, a total of 22 studies including 22,515 cases and 22,388 controls were involved in this meta-analysis. Table 1 lists the studies identified and their main characteristics. Of the 22 studies, sample sizes ranged from 246 to 8,991. There were 19 studies of Caucasians, one study of Asians, one study of Africans, and one study of mixed populations. Almost all of the cases were pathologically confirmed. Controls were mainly healthy populations and matched for age. Among these studies, 14 were population-based and five were hospital-based.
Main results
Table 2 lists the main results of this meta-analysis. Overall, no significant associations were found between BRCA2 N372H polymorphism and breast cancer risk when all studies pooled into the meta-analysis (NH versus NN: OR = 1.01, 95% CI = 0.97–1.05; HH versus NN: OR = 1.05, 95% CI = 0.97–1.13; dominant model: OR = 1.01, 95% CI = 0.98–1.05; and recessive model: OR = 1.05, 95% CI = 0.98–1.13). In the subgroup analysis by ethnicity, still no significant associations were found for Caucasians, Asians, or Africans. When stratified by study design, statistically significantly elevated risk was found for 372H allele based on population-based studies (HH versus NN: OR = 1.11, 95% CI = 1.01–1.21; dominant model: OR = 1.05, 95% CI = 1.00–1.10; recessive model: OR = 1.09, 95% CI = 1.00–1.18). Surprisingly, statistically significantly reduced risk was found based on hospital-based study (NH versus NN: OR = 0.89, 95% CI = 0.80–0.99; dominant model: OR = 0.89, 95% CI = 0.80–0.99).
Sensitivity analysis
A single study involved in the meta-analysis was deleted each time to reflect the influence of the individual data-set to the pooled ORs, and the corresponding pooled ORs were not materially altered (data not shown), indicating that our results were statistically robust.
Publication bias
Begg’s funnel plot and Egger’s test were performed to access the publication bias of literatures. The shape of the funnel plot did not reveal any evidence of obvious asymmetry (figures not shown). Then, the Egger’s test was used to provide statistical evidence of funnel plot symmetry. The results still did not suggest any evidence of publication bias (P = 0.22 for NH versus NN; P = 0.43 for HH versus NN; P = 0.43 for dominant model; and P = 0.31 for recessive model).
Discussion
The present meta-analysis, including 22,515 cases and 22,388 controls, explored the association between the BRCA2 N372H polymorphism and breast cancer risk. Our results indicated that significantly increased breast cancer risk in BRCA2 372H allele carriers were found based on the population-based studies but not on hospital-based studies. This reason may be that the hospital-based studies have some biases because such controls may just represent a sample of ill-defined reference population, and may not be representative of the general population very well, particularly when the genotypes under investigation were associated with the disease conditions that the hospital-based controls may have. Therefore, using a proper and representative population-based study is very important to reduce biases in such genetic association studies.
Some limitations of this meta-analysis should be acknowledged. First, the controls were not uniformly defined. Although most of the controls were selected mainly from healthy populations, some had benign disease. Therefore, non-differential misclassification bias was possible because these studies may have included the control groups who have different risks of developing breast cancer. Second, in the subgroup analyses, the number of Asians and Africans were relatively small, not having enough statistical power to explore the real association. Third, our results were based on unadjusted estimates, while a more precise analysis should be conducted if individual data were available, which would allow for the adjustment by other co-variants including age, ethnicity, menopausal status, smoking status, drinking status, obesity, environmental factors, and other lifestyle.
In conclusion, this meta-analysis suggests that the BRCA2 372H allele may be a low-penetrant risk factor for developing breast cancer based on the population-based studies. However, it is necessary to conduct large sample studies using standardized unbiased genotyping methods, homogeneous breast cancer patients, and well matched controls. Moreover, gene–gene and gene–environment interactions should also be considered in the analysis. Such studies taking these factors into account may eventually lead to our better, comprehensive understanding of the association between the BRCA2 N372H polymorphism and breast cancer risk.
References
Parkin DM, Bray F, Ferlay J, Pisani P (2005) Global cancer statistics 2002. CA Cancer J Clin 55:74–108
Lichtenstein P, Holm NV, Verkasalo PK (2000) Environmental and heritable factors in the causation of cancer. N Engl J Med 343:78–85
Wooster R, Bignell G, Lancaster J (1995) Identification of the breast cancer susceptibility gene BRCA2. Nature 378:789–792
Pharoah PD, Antoniou A, Bobrow M (2002) Polygenic susceptibility to breast cancer and implications for prevention. Nat Genet 31:33–36
Healey CS, Dunning AM, Teare MD (2000) A common variant in BRCA2 is associated with both breast cancer risk and prenatal viability. Nat Genet 26:362–364
Spurdle AB, Hopper JL, Chen X (2002) The BRCA2 372 HH genotype is associated with risk of breast cancer in Australian women under age 60 years. Cancer Epidemiol Biomarkers Prev 11:413–416
Cox DG, Hankinson SE, Hunter DJ (2005) No association between BRCA2 N372H and breast cancer risk. Cancer Epidemiol Biomarkers Prev 14:1353–1354
Garcia-Closas M, Egan KM, Newcomb PA, Brinton LA, Titus-Ernstoff L, Chanock S (2006) Polymorphisms in DNA double-strand break repair genes and risk of breast cancer: two population-based studies in USA and Poland, and meta-analyses. Hum Genet 119:376–388
Pharoah P for The Breast Cancer Association Consortium (2006) Commonly studied single-nucleotide polymorphisms and breast cancer: results from the breast cancer association consortium. J Natl Cancer Inst 98:1382–1396
Seymour IJ, Casadei S, Zampiga V (2008) Disease family history and modification of breast cancer risk in common BRCA2 variants. Oncol Rep 19:783–786
Ishitobi M, Miyoshi Y, Ando A (2003) Association of BRCA2 polymorphism at codon 784 (Met/Val) with breast cancer risk and prognosis. Clin Cancer Res 9:1376–1380
Menzel HJ, Sarmanova J, Soucek P (2004) Association of NQO1 polymorphism with spontaneous breast cancer in two independent populations. Br J Cancer 90:1989–1994
Millikan RC, Player JS, Decotret AR, Tse CK, Keku T (2005) Polymorphisms in DNA repair genes, medical exposure to ionizing radiation, and breast cancer risk. Cancer Epidemiol Biomarkers Prev 14:2326–2334
Cochran WG (1954) The combination of estimates from different experiments. Biometrics 10:101–129
Mantel N, Haenszel W (1959) Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 22:719–748
DerSimonian R, Laird N (1986) Meta-analysis in clinical trials. Control Clin Trials 7:177–188
Tobias A (1999) Assessing the influence of a single study in the meta-analysis estimate. Stata Tech Bull 8:15–17
Egger M, Davey Smith G, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315:629–634
Author information
Authors and Affiliations
Corresponding author
Additional information
Li-Xin Qiu, Lei Yao, Kai Xue contributed equally to this work and should be considered as co-first authors.
Rights and permissions
About this article
Cite this article
Qiu, LX., Yao, L., Xue, K. et al. BRCA2 N372H polymorphism and breast cancer susceptibility: a meta-analysis involving 44,903 subjects. Breast Cancer Res Treat 123, 487–490 (2010). https://doi.org/10.1007/s10549-010-0767-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-010-0767-5