Abstract
Rice (Oryza sativa L. ‘Nipponbare’) cDNA subtractive suppression hybridization (SSH) libraries constructed using cadmium (Cd)-treated seedling roots were screened to isolate Cd-responsive genes. A cDNA clone, encoding the rice homolog of Metal Tolerance Protein (OsMTP1), was induced by Cd treatment. Plant MTPs belong to cation diffusion facilitator (CDF) protein family, which are widespread in bacteria, fungi, plants, and animals. OsMTP1 heterologous expression in yeast mutants showed that OsMTP1 was able to complement the mutant strains’ hypersensitivity to Ni, Cd, and Zn, but not other metals including Co and Mn. OsMTP1 expression increased tolerance to Zn, Cd, and Ni in wild-type yeast BY4741 during the exponential growth phase. OsMTP1 fused to green fluorescent protein was localized in onion epidermal cell plasma membranes, consistent with an OsMTP1 function in heavy metal transporting. OsMTP1 dsRNAi mediated by transgenic assay in rice seedlings resulted in heavy metal sensitivity and changed the heavy metal accumulation in different organs of mature rice under low-concentration heavy metal stress. Taken together, our results show that OsMTP1 is a bivalent cation transporter localized in the cell membrane, which is necessary for efficient translocation of Zn, Cd and other heavy metals, and maintain ion homeostasis in plant.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Heavy metals such as Zn, Mn, Ni, Fe, Co, and Cu are essential trace elements for numerous physical processes in plants. In trace amounts, they can act as important cofactors for many enzymes and function in cellular physical processes (Williams and Mills 2005). In high quantities, they are toxic through biomolecular inactivation by blocking essential functional groups or displacement of essential metal ions (Goyer 1997). Other heavy metals such as Cd, Pb, and Hg are non-essential elements in plants and are generally considered toxic, because they can displace some essential elements or react with sulfur and nitrogen in amino acid side chains (Clemens 2001). Plants have developed a series of uptake, extrusion, chelation, trafficking, and storage mechanisms to maintain essential trace element homeostasis, exclude non-essential trace elements, and to alleviate heavy metal toxicity (Hall and Williams 2003).
Some characterized plant transporter proteins responsible for metal homeostasis are membrane proteins that mediate heavy metal movement through membranes. These transporters can be classified into channels, carriers, and pumps (Williams et al. 2000). Among these transporters, cation diffusion facilitator (CDF) family proteins are crucial for metal ion homeostasis. Members of the diverse CDF family have been cloned from bacteria, fungi, plants, and animals (Mäser et al. 2001). Most CDF proteins possess four to six putative transmembrane domains (TMDs) with cytoplasmic N and C termini, and the most conserved amphipathic TMDs I, II, V, and VI regions that are likely involved in metal transfer (Haney et al. 2005). According to protein sequence alignment and substrate specificities of some characterized transporters, CDF members from three kingdoms (Archaea, Eubacteria, Eukaryotes) were classified into three subgroups: Zn-CDF, Fe/Zn-CDF, and Mn-CDF (Montanini et al. 2007).
In plants, CDF proteins were also designated as metal tolerance proteins (MTPs). The first plant CDF protein identified was ZAT (zinc transporter of Arabidopsis thaliana). Because of its involvement in Arabidopsis heavy metal tolerance, ZAT was later renamed as AtMTP1 (Metal Tolerance Protein 1) (Van-Der-Zaal et al. 1999). AtMTP1 over-expression in Arabidopsis confers improved growth with supra-optimal Zn concentrations in the medium, and an AtMTP1 T-DNA insertion mutant line displays enhanced sensitivity to high Zn concentrations (Kobae et al. 2004), suggesting that AtMTP1 is involved in Zn tolerance. Heterologous expression of AtMTP3 in yeast double mutant zrc1Δ cot1Δ restores Zn and Co tolerance, and AtMTP3 ectopic over-expression increases Zn accumulation in Arabidopsis leaves, and enhances Zn tolerance, while AtMTP3 RNAi causes Zn hypersensitivity. Hence, AtMTP1 and AtMTP3 function similarly in responding to Zn. Unlike AtMTP1, however, AtMTP3 seems to be involved in metal transportation under Fe deficiency (Arrivault et al. 2006). Sequence analyses showed that AtMTP1 and AtMTP3 belonged to Zn-CDF, while AtMTP11 belonged to Mn-CDF (Montanini et al. 2007). Functional analysis showed that AtMTP11 can specifically complement the Mn-hypersensitive yeast mutant pmr1Δ (Delhaize et al. 2007), and might be involved in both Mn tolerance and homeostasis mechanisms in Arabidopsis. MTP genes have also been cloned from other plants, especially from some heavy metal hyperaccumulator plants such as Arabidopsis halleri (AhMTP1) (Dräger et al. 2004), Thlaspi goesingense (TgMTP1) (Kim et al. 2004), and Stylosanthes hamata L. (ShMTP1) (Delhaize et al. 2003). These genes can all response to Zn, Cd, or other bivalence heavy metal ions.
Rice is the most important cereal crop in China. It is also a model monocotyledon plant. Heavy metal contamination has become a serious environmental problem, and has greatly affected rice growth and quality. Heavy metal accumulation in water, soil, and air increasingly accompanies industrial pollution. Heavy metals accumulated in rice are toxic to growth and metabolism. Rice Metal Tolerance Protein1 (OsMTP1) gene expression is induced by Cd treatment, as indicated by our previous SSH study. Analysis of the rice genome has identified ten MTP genes (Montanini et al. 2007), and OsMTP1 belongs to the Zn-CDF subgroup. In this study, we mainly focus on OsMTP1 functional identification. Our results showed that OsMTP1 heterologous expression in a yeast mutant restored Ni, Zn, and Cd tolerance. OsMTP1 dsRNA interference can decrease markedly the rice seedling tolerance to heavy metals. Furthermore, down-regulation of OsMTP1 by double-stranded RNA interference (dsRNAi) can also change the heavy metal accumulation in different organs of rice. These results suggest that OsMTP1 is necessary for efficient translocation of Zn, Cd and other heavy metals as well as ion homeostasis in plant.
Materials and methods
Plant materials
Wild-type rice japonica variety Zhonghua 11 was used. Transgenic and wild-type plants were grown under a routine management regime in an experimental field at the South China Botanical Garden (Guangzhou, China). Rice seedlings were grown on one-half-strength MS medium with or without appropriate selection and heavy metals.
For heavy metal resistance assays of rice, homozygous T3 seeds of lines over-expressing OsMTP1 or transformed with dsRNAi OsMTP1 were sown on plates containing half-strength MS medium supplied with various concentrations of heavy metals. Wild-type plants were used as a control. After 2 weeks growth, rice seedlings were examined.
For measurements of Zn, Cd, and Ni contents in rice, wild-type rice and homozygous dsRNAi T3 rice seeds were grown hydroponically according to Yoshida et al. (1976). ZnSO4 (0.2 mM), CdCl2 (0.01 mM), and NiSO4 (0.05 mM) were added to the liquid culture, respectively.
Plasmid constructs and plant transformation
To study OsMTP1 function in plants, we performed transgenic over-expression and dsRNAi assays in rice. For OsMTP1 over-expression (OsMTP1-OE) in rice, the OsMTP1 open reading frame (ORF) was PCR amplified to obtain full-length cDNA fragments with AK100735 cDNA clone (KOME, http://cdna01.dna.affrc.go.jp/cDNA/) as a template using the following primers: pMTP1OEL: 5′-CGGGATCCATGGACAGCCATAACTCAGC-3′ and pMTP1OER: 5′-CGGGATCCCTACTCGCGCTCAATCTGAA-3′ (BamHI sites are underlined). PCR fragments were cloned into the BamHI site of pCU1301 (Chen et al. 2007) between the maize Ubi1 promoter and the nos site. For OsMTP1 dsRNAi in rice, the 5′ fragments of OsMTP1 (500 bp) were PCR amplified using the following primers: pMTP1RIL: 5′-CGGGATCC GAGCTCATGGACAGCCATAACTCAGC-3′ (BamHI and SacI sites are underlined), and pMTP1RIR: 5′-GGGGTACCACTAGT GCTGAGACAGCAAACATGAG-3′ (KpnI and SpeI sites are underlined). PCR products were first cloned into pMD18-T vector (TaKaRa, Japan) and confirmed by sequencing. OsMTP1 dsRNAi fragments were released by digesting with SacI and SpeI and subcloned into pTCK303 (SacI–SpeI sites) to generate the forward insertion. Further dsRNAi fragments were released by digesting with BamHI and KpnI and subcloned into the same pTCK303 (BamHI–KpnI sites) to generate the reverse insertion (Wang et al. 2004). The over-expression construct (OsMTP1/pCU1301) and dsRNAi construct (OsMTP1RI/pTCK303) were introduced into Agrobacterium strain EHA105 and transformed into wild-type Zhonghua 11 following a standard procedure (Hiei et al. 1994).
Bacterial and yeast strains, media, and transformation
Escherichia coli strain JM109 was used for molecular cloning. The metal tolerance assay was performed on wild-type Saccharomyces cerevisiae strain BY4741, the Zn-sensitive zrc1Δ (Y00829) strain, the Zn and Co sensitive cot1Δ (Y01613) strain, the Cd-sensitive ycf1Δ (Y04069) strain, the Mn-sensitive pmr1Δ (Y04534) strain, the Ni-sensitive smf1Δ (Y06272) strain, and vacuole acidification mutant strains vph2Δ (Y04969). Yeast mutants were derived from the parental strain BY4741 and were all obtained from Euroscarf (http://www.uni-frankfurt.de/fb15/mikro/euroscarf/). Growth was in yeast potato dextrose or in synthetic defined (SD) medium with 2% (w/v) galactose, supplemented as necessary.
The OsMTP1-coding region was inserted into the NcoI sites of pYES260 (from Euroscarf) behind the GAL1 promoter, yielding pYES260-OsMTP1. The plasmid was introduced into the yeast according to the method of Gietz and Schiestl (2007). Transformed cells were selected on solid SD medium (plus histidine, leucine, and methionine, without uracil) overnight and cultured in liquid SD medium overnight. The overnight cultures of transformed cells were each diluted to OD 1.0 at 600 nm. Diluted transformed cells (2 μl) were then spotted onto SD media plates with different heavy metals. For yeast growth curves, growth rates were measured according to OD 600 values.
RT-PCR and Northern blot analysis
Gene expression was first analyzed by semi-quantitative RT-PCR. Total RNAs were extracted from tissues using Trizol reagent (Invitrogen, USA) according to the instructions. Briefly, 5 μg total RNA was denatured at 65°C for 5 min followed by quick chilling on ice in a 14 μl reaction containing 1 μl oligo (dT)12–18 (50 μM) primer, and 1 μl of 40 μM dNTP mixture (10 mM each dATP, dGTP, dCTP, and dTTP at neutral pH). After addition of 4 μl 5× reaction buffer, the reaction was incubated at 37°C for 2 min, 1 μl (200 U) of M-MLV reverse transcriptase (Promega, Shanghai, China) was added to the reaction and incubated at 42°C for another 50 min. After terminating, the reaction was heated at 70°C for 15 min for inactivating. Thermocycling conditions were 94°C for 5 min followed by 30–35 cycles of 94°C for 1 min, 55–60°C for 30 s, and 72°C for 30 s. Due to the use of the OsMTP1 5′-terminal fragment (500 bp) to perform the dsRNAi, we detected the OsMTP1 3′-terminal (250 bp) to determine the efficiency of RNA-interfere with RT-PCR. The constitutively expressed OsActin7 gene was used as internal control. The primers used were as follows: 5′-GGTCCTGATGGAGAGCACGC-3′ and 5′-CTACTCGCGCTCAATCTGAA-3′ for OsMTP1, 5′-CGTGACCTTACCGACAACCT-3′ and 5′-GCACCTGAACCTTTCTGCTC-3′ for OsActin7.
Northern blotting was performed to identify OsMTP1 transcripts in transgenic rice and to test Cd-inducible OsMTP1 expression in rice roots and shoots. Total RNA from transgenic plant leaves and 2-week-old Cd-treated rice roots and shoots were isolated with Trizol as described by the manufacturer (Invitrogen, USA) and blotted onto a nylon membrane (Millipore, USA). Digoxigenin (DIG)-labeled specific probes of the OsMTP1 ORF were made using a PCR DIG probe synthesis kit (Roche, Germany) with primers pMTP1OEL and pMTP1OER. The membrane was hybridized with the DIG-labeled probe for 16 h at 65°C in DIG Easy Hyb (Roche, Germany), then were washed twice at 37°C in 2× SSC and 0.1% (w/v) SDS for 10 min, followed by washing twice at 65°C in 0.2× SSC and 0.1% (w/v) SDS for 30 min. The membranes were then subjected to immunological detection following the manufacturer’s instructions (Roche, Germany).
Construction of an OsMTP1–EGFP fusion expression vector and fluorescence microscopy
DNA fragments containing OsMTP1 ORFs were obtained by PCR with forward primer pM1GL (5′-CGGGATCCATGGACAGCCATAACTCAGC-3′) and reverse primer pM1GR (5′-CGGGATCCTCGCGCTCAATCTGAATGGT-3′) (BamHI sites are underlined). PCR products were cloned into pMD18 T-vector, digested, and further cloned into pUC/EGFP made by ligating the 35S promoter from pBIm (Hua et al. 2004) and GFP from pBI101-EGFP into pUC18 in frame to produce the C-terminal EGFP-tagged fusion protein. After sequence confirmation, these vectors were bombarded into onion epidermal cells. After incubation at 22°C for 24 h, GFP fluorescence was visualized using confocal laser scanning microscope (LSM, 510 META, Zeiss, Germany). GFP excitation was 488 nm, and emission was detected between 500 and 575 nm.
mRNA in situ hybridization
Tissue sectioning was as described previously (Zhang et al. 2005). Briefly, rice vegetative and reproductive tissues were fixed in 4% (w/v) paraformaldehyde, dehydrated in an ethanol series, cleared with xylene, and embedded in paraffin. Paraffin-embedded tissues were cut into 8 μm-thick sections and then attached to glass microscope slides coated with polylysine hydrobromide. The 3′ untranslated region (431 bp) of OsMTP1 full-length cDNA was chosen as a template to generate DIG-labeled probe, which was PCR-amplified with full-length cDNA as template, and the primers were as follows: pM1RHL: 5′-CATCAGCCATGTGACCATTC-3′ and pM1RHR: 5′-CCATGCCAGTGCAGATTTAG-3′. PCR products were then cloned into pGEM-T vector (Promega, Shanghai, China) to generate recombinant plasmid pGOsM1.
DIG-labeled antisense probe was transcribed from SphI-digested pGOsM1 using Sp6 RNA polymerase, and the sense RNA probe controls were transcribed from SacI-digested pGOsM1 using T7 RNA polymerase. The hybridization mixture consisted of 50% (v/v) deionized formamide, 10% (w/v) dextran sulfate, 0.3 M NaCl, 10 mM Tris–HCl (pH 6.8), 10 mM NaH2PO4 (pH 6.8), 5 mM EDTA, 2.2 mM dithiothreitol, 0.6 μg/μl yeast tRNA, 0.5 μg/μl yeast RNA, and about 500 ng denatured DIG-labeled RNA probe. About 100 μl mixture was mounted onto each slide and the slide was then covered with a clear coverslip. Hybridization was done at 65°C overnight. The slides were then washed twice in 2× SSC at room temperature, then in 1× SSC and 0.1× SSC at 55°C for 15 min each.
The DIG-labeled RNA was detected using anti-DIG alkaline phosphatase conjugate (anti-DIG-AP) according to the manufacturer (Roche, Germany). After the enzyme-catalyzed color reaction was done as previously described (Zhang et al. 2005), the slides were visualized with a microscope (Axioplan 2 imaging, Zeiss). The hybridization signal was visible as an insoluble blue precipitate.
Measurement of heavy metal content in plants
Plant tissues were harvested, dried and desorbed in 10 ml HNO3:H2O2 (4:1) at 100°C for 8 h. Then the HNO3 in mixture was subsequently released through volatilization at 100°C for 24 h. The rest extraction was settled to the volume of 20 ml with 0.2% HNO3. Zn, Cd, and Ni contents were analyzed by atomic absorption spectrophotometer (AAS, GBC3000).
Results
OsMTP1 is a member of rice CDF family
To isolate rice Cd-responsive genes, we constructed SSH libraries with Cd-treated or untreated rice roots. A cDNA fragment, designated as OsMTP1, was one of the Cd-induced genes in the SSH assay. OsMTP1 cDNA (Os05g03780; Accession number AK100735) encodes a protein of 419 amino acids (45.57 kD). Analysis of the cDNA predicted sequence showed 69 and 78% identity with AtMTP1 (Loci At2g46800; Mäser et al. 2001) at the nucleotide sequence and amino acid levels, respectively. On comparing with the rice genome sequence, it was found that OsMTP1 was located on chromosome 5.
Previously, Montanini et al. (2007) identified 10 MTP genes from rice and 12 from Arabidopsis (Supplement data 1). The phylogenetic analysis of 22 MTPs in rice and Arabidopsis indicates that MTPs are grouped into three branches according to their amino acid sequence and possible ion-transporting specificity (Montanini et al. 2007). In rice, there are five Mn-CDFs, three Zn-CDFs, one Fe/Zn-CDF, and one unclassified CDF. Among these, OsMTP1 has the highest homology with AtMTP1. OsMTP1 functional domains including six putative TM domains and His-rich regions are conserved in all of these three members.
OsMTP1 is constitutively expressed and up-regulated by Cd
To understand the possible function of OsMTP1 in rice, we examined its expression in different rice tissues by RT-PCR analysis (Fig. 1a). OsMTP1 was expressed in various organs, including roots, leaves, stems and flowers. The highest level of OsMTP1 transcript was detected in mature leaves and stems, which coincided with mRNA in situ hybridization results (Fig. 1b).
OsMTP1 expression patterns in wild-type Zhonghua 11 rice plants. a RT-PCR analysis of OsMTP1 expression. The amplification of the rice Actin7 gene was used as a control to show the approximately equal total RNA amounts used. b In situ hybridization of OsMTP1 mRNA in rice leaves. Blue precipitates indicate positive OsMTP1 signals. Transverse sections of a mature leaf (A; B shows enlargement of A); seedling leaf bud (C; D shows enlargement of C); and longitudinal sections of a seedling leaf bud (E); control shows seedling leaf bud (F). c OsMTP1 expression under Cd stress analyzed by Northern blots. OsMTP1 expression increased in rice seedlings treated with 100 μM CdCl2 for different periods
mRNA in situ hybridization of OsMTP1 was performed using a DIG-labeled RNA antisense probe against OsMTP1 cDNA 3’-untranslational region with control sense RNA. In rice leaves, OsMTP1 mRNA accumulated specifically near vascular tissue (Fig. 1b, A–D), and longitudinal leaf sections showed that the signals were distributed in specific sieve vessel cell files (Fig. 1b, E). No positive reaction above background levels was detected on sections hybridized with sense RNA (Fig. 1b, F).
We investigated OsMTP1 expression patterns following Cd treatment using RNA Northern blots (Fig. 1c). The 2-week-old rice seedlings were exposed to 100 μM CdCl2 and OsMTP1 gene expression was assayed with the OsMTP ORF DIG-labeled probe. OsMTP1 expression was induced in roots and shoots after a 10-h CdCl2 treatment.
Identification of OsMTP1 over-expressing and OsMTP1-dsRNAi transgenic plants
To further elucidate the OsMTP1 role in rice, we generated OsMTP1 over-expression and dsRNAi (OsMTP1-RI) transgenic plants. The OsMTP1 over-expression and dsRNAi transgenes were under the control of the maize (Zea mays) ubiquitin 1 (Ubi1) promoter (Fig. 2a). Transgenic plants were generated by introducing the constructs into the japonica rice variety Zhonghua 11 by Agrobacterium tumefaciens-mediated transformation (Hiei et al. 1994). Positive T1 lines segregating at or near 3:1 under hygromycin selection and GUS staining were selected for further identification.
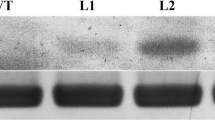
OsMTP1 expression patterns in transgenic rice. a Constructs used for rice transformation. The full-length cDNA encoding the OsMTP1 gene (1,257 bp) was inserted into the BamHI site of pCU1301 with sense orientation to construct the over-expression vector in rice. The 3′ terminal partial OsMTP1 cDNA (500 bp) was inserted into the SacI/SpeI site (behind the intron) in antisense orientation and BamHI/KpnI site (before the intron) in sense orientation to construct the dsRNAi vector in rice. b Northern blot analysis of OsMTP1 transcript levels in transgenic rice. Total RNA extracted from rice leaves at the heading stage was probed with full-length OsMTP1 cDNA. Loading of approximate amounts of RNA was confirmed by ethidium bromide staining. c RT-PCR analysis of OsMTP1 expression in wild type, over-expression, and dsRNAi rice plants. Amplification of the rice Actin7 gene was used as a control to show the approximately equal amounts of total RNA used in the RT-PCR analysis
To examine OsMTP1 expression in these transgenic lines, total RNA was isolated and RT-PCR was performed using specific primers. Northern blot results showed that in over-expression transgenic rice, the OsMTP1 transcript was highly expressed (Fig. 2b). In dsRNAi transgenic rice plants, the dsRNA (1.4 kb) could be detected (Fig. 2b) and was smaller than OsMTP1 transcripts. Endogenous OsMTP1 expression was relatively low in wild-type rice plants and cannot be detected at the same exposure time. OsMTP1 transcripts were further analyzed with semi-quantitative RT-PCR. As shown in Fig. 1a, OsMTP1 transcript was barely detected in the wild-type seedlings, but was greatly increased in the over-expression lines. In comparison, OsMTP1 transcript was not detectable in the dsRNAi lines, respectively (Fig. 2c).
OsMTP1 dsRNAi transgenic plants exhibit obviously decreased Zn, Cd, and Ni tolerance
We performed the transgenic assay in rice to identify OsMTP1 biological roles and elucidate the relevance between OsMTP1 transcripts and heavy metal resistance. Wild-type and transgenic T3 homologous seeds were chosen to detect Zn, Cd, and Ni tolerance. Over-expression or dsRNAi of OsMTP1 both decreased Zn, Cd, and Ni tolerance in rice, particularly when grown for 2 weeks in solid MS medium supplemented with different concentrations to determine at what levels the aberrant phenotypes became apparent (Fig. 3a). Seedling length (including root and shoot) was measured to monitor heavy metal tolerance (Fig. 3b). Under heavy metal (Zn, Cd, and Ni) stress at different concentrations, down-expression of OsMTP1 transgenic rice grew less than wild type, while the growth of over-expression of OsMTP1 rice was slightly suppressed by metal stress. This result indicated that the abnormal expression level of OsMTP1 may cause ionic disorder, and then affect the growth condition of rice transgenic plants.
Heavy metal tolerance test of OsMTP1 over-expression and dsRNAi rice plants. a Phenotype comparison of wild type, over-expression, and dsRNAi rice plants grown under different heavy metal ion levels. Rice seedlings were grown for 12 days on solid phytogel containing one-half-strength MS medium (control) or medium supplied with different Zn, Cd, and Ni concentrations. Images show two plants each of wild-type control (left), OsMTP1 over-expression (middle), and dsRNAi (right). White bars 3 cm. b Average rice seedling shoot and root length, averaged for 7–10 independent strains. Error bars represent the SE. *P < 0.05, and **P < 0.01
OsMTP1 dsRNAi transgenic plants have changed Zn, Cd, and Ni content
We measured the Zn, Cd and Ni levels in mature dsRNAi transgenic rice plants and wild-type rice plants that had been exposed to low-concentration of heavy metals during the whole growth period. We found that the metal content in rice roots, leaves and seeds showed obvious disparities between dsRNAi transgenic rice plants and wild-type rice plants (Fig. 4a–c). In rice roots, the part of a crop which is immersed in nutrient fluid supplied with heavy metal, dsRNAi transgenic plants had higher level accumulation of Zn, Cd and Ni contents than wild-type plants, while in rice aerial parts (leaves, stems, and seeds), the heavy metal concentrations were different from root parts. As a whole, in leaves and stems, the Zn and Cd accumulation levels were higher in wild type than in dsRNAi transgenic rice plants, while Ni accumulation level was similar between wild type and dsRNAi transgenic rice plants. The same results were also detected in the rice seeds. Therefore, these results support that OsMTP1 may be important in rice cation homeostasis mechanisms and down-regulation of OsMTP1 by dsRNAi can affect the heavy metal accumulation in different rice organs.
Heavy metal content analyses of wild type and dsRNAi rice plants. Potted rice plants grown in hydroponics supplied with ZnSO4 (0.2 mM), CdCl2 (0.01 mM), and NiSO4 (0.05 mM). After seed maturity, roots, leaves and stems, seeds were harvested for AAS (GBC3000) analyses. Mean values from four to six independent experiments are shown. Error bars represent the SE. *P < 0.05, and **P < 0.01. a Zn, Ni, and Cd accumulation in rice roots. b Zn, Ni, and Cd accumulation in rice leaves and stems. c Zn, Ni, and Cd accumulation in rice seeds
Expression of OsMTP1 in yeast confers tolerance to Ni, Zn, and Cd but not Mn
To characterize OsMTP1 specificity as a metal transporter, we performed the complementation assays in a series of yeast mutants, including zrc1Δ, cot1Δ, smf1Δ, ycf1Δ, pmr1Δ, and vph2Δ (Fig. 5a). OsMTP1 heterologous expression can recover some of these mutants’ metal tolerance. zrc1Δ yeast mutant is a Zn-sensitive strain, in which ZRC1 gene encoding transporter that sequesters Zn into the vacuole was deleted (Li and Kaplan 1998). OsMTP1 did not complement the zrc1Δ Zn-sensitive phenotype. cot1Δ is hypersensitive to both Zn and Co because COT1 mediates the efflux of both ions into the vacuole (Conklin et al. 1992; Li and Kaplan 1998; Lyons et al. 2000). Interestingly, OsMTP1 complemented the cot1Δ phenotype when grown on Zn but not Co. We further transformed the ycf1Δ, pmr1Δ, and smf1Δ mutant strains, which are unable to grow on Cd, Mn, and Ni, respectively. YCF1 is an ABC transporter that confers Cd tolerance through the Mn and Ca transport of Cd conjugates into the vacuole (Szczypka et al. 1994; Li et al. 1997). PMR1 is a yeast secretary pathway pump responsible for high-affinity transport into the golgi and confers Mn tolerance by effectively removing Mn from the cytoplasm (Ton et al. 2002). SMF1 functions in Ni and Mn cellular accumulation, and the smf1Δ mutant is Ni sensitive (Supek et al. 1996). To our surprise, OsMTP1 heterologous expression in smf1Δ can complement the Ni-sensitive phenotype, indicating that OsMTP1 can transport Ni in yeast. OsMTP1 can also partially complement the Cd-sensitive phenotype of ycf1Δ mutant, but did not increase Mn tolerance in pmr1Δ. OsMTP1 could not complement the Zn-sensitive phenotype of vph2Δ, which is unable to acidify the vacuole (Fig. 5a).
Complementation of yeast mutants on selective media. a S. cerevisiae wild type (WT) and mutant strains were transformed with the empty vector pYES260 or with pYES260-OsMTP1. Yeast cultures were adjusted to OD600 = 1.0, and 2 μl serial dilutions (from left to right in each panel) were spotted on SD medium without (control) or with different heavy metal concentrations. Plates were incubated for 6 days to 2 weeks at 30°C. b OsMTP1 confers Ni, Zn, and Cd tolerance when expressed in smf1Δ, cot1Δ, and ycf1Δ yeast mutants. Yeast mutant (smf1Δ, cot1Δ, and ycf1Δ), transformed with the empty vector pYES260 or pYES260-OsMTP1, were grown in liquid SD culture (control) or supplemented with Ni (0.5 mM), Zn (5 mM), and Cd (0.1 mM). The optical density at 600 nm (OD600) of the cultures was determined after different periods
To further confirm the OsMTP1’s contribution to heavy metal tolerance in yeast, OsMTP1 was heterologously expressed in yeast mutant strains smf1Δ, cot1Δ, and ycf1Δ and detected their Ni, Zn, and Cd tolerance (Fig. 5b). Yeast mutant strains transformed with empty vector pYES260 or pYES260-OsMTP1 had the same growth rate without heavy metal stresses, while OsMTP1 heterogonous expression in smf1Δ, cot1Δ, and ycf1Δ elevated their growth rate under 0.5 mM Ni, 5 mM Zn, and 0.1 mM Cd stress (Fig. 5b).
OsMTP1 is localized to the plasma membrane
To examine OsMTP1 localization, we fused OsMTP1 ORF to the enhanced green fluorescent protein (EGFP) gene under 35S promoter control. The recombinant and empty vectors were bombarded into onion cells and confocal microscopy showed that OsMTP1–EGFP was efficiently targeted to the cell periphery, suggesting OsMTP1–EGFP plasma membrane localization (Fig. 6a), while EGFP was distributed throughout the cell including the cytoplasm and nucleus (Fig. 6c). Moreover, we also performed the bioinformatics analysis of OsMTP1’s subcellular localization with three different programs (Supplement data 2). WoLF PSORT predictions showed that OsMTP1 localization at the plasma membrane, vacuolar membrane and endoplasmic reticulum, scored, respectively, 8.0, 3.0 and 3.0. This analysis showed that OsMTP1 may mainly distribute at the plasma membrane, which is coincided with our EGFP fusion protein assay results. The Target P predicted a relatively high OsMTP1 score value for a chloroplast transit peptide (0.675, cTP), and ARAMEMNON predicted OsMTP1 distributed primarily in chloroplast with a high absolute value (8.9). These two above predictions indicated that OsMTP1 might be involved in chloroplast ion metabolism, while the correlative experimental evidence needs further investigation. We also performed the transmembrane domain prediction by TMHMM2; and the result showed that OsMTP1, had six transmembrane regions (Supplement data 3), could be a membrane-integrated protein. These predictions, together with our experimental results of OsMTP1–EGFP fusion protein subcellular localization, suggest that OsMTP1 may be mainly plasma membrane localized, and probably less distributed in chloroplast and other organelle membrane, functioning in transmembrane transporting of metallic ions.
Discussion
Heavy metal pollution severely affected plant growth and development. As the most important cereal crop in China, rice and its growing space have been polluted by heavy metals, particularly Cd. Plants have developed a series of strategies to decrease metal ion toxicity, one of which is the cellular transport system. There are several groups of metal transport proteins identified in plants, including PIB-type ATPases (Eren and Argüello 2004), CDF proteins, natural resistance-associated macrophage proteins (NRAMP), and Zn-regulated transporter-/Fe-regulated transporter-like proteins (ZIP, Mäser et al. 2001). CDF proteins were later renamed as MTPs in plants, and are now identified as a group of important metal transporting proteins.
Plant MTPs were first characterized in Arabidopsis, and are now cloned from a wide variety of plants species, including Arabidopsis, poplar, T. goesingense, Brassica juncea, and tobacco. Most MTPs have four to six putative transmembrane domains and a signature N-terminal amino acid sequence. A His-rich region between TMD four and five may be specifically involved in metal binding. Here, we described a Cd-induced gene, OsMTP1 in rice. Bioinformatics analyses indicated that OsMTP1 encodes a 45.57 kD membrane protein with six TMDs. Homologous analysis revealed that OsMTP1 displayed many characteristic features of MTPs such as AtMTP1 and PtdMTP1 (Damien et al. 2003), implying a similar molecular mechanism and physiological function among them. In this study, RT-PCR analyses demonstrated that OsMTP1 had relatively low expression, while Cd treatment slightly induced it in roots and shoots. These results suggest that OsMTP1 responded to Cd stress.
We also performed transgenic experiments in rice to analyze whether OsMTP1 was involved in rice heavy metal tolerance. OsMTP1 over-expression and dsRNAi transgenic rice plants were generated. We found that OsMTP1 dsRNAi transgenic rice seedlings had obviously decreased heavy metal tolerance, while the growth of OsMTP1 over-expression seedlings was suppressed slightly by heavy metal. These results indicated that both up- and down-regulated OsMTP1 expression in rice may function in cation absorption and transferring balance, disrupting or unbalancing rice growth and development. The remaining challenge is to determine the specific contribution of OsMTP1 in maintaining ionic equilibrium in rice plants.
In rice, OsMTP gene family includes ten MTP members. Among them, OsMTP1 and other two MTPs (OsMTP5 and OsMTP12) belong to the Zn-CDF subgroup (Montanini et al. 2007; Gustin et al. 2011). It is possible that these three Zn-CDF members are functionally redundant. At low-concentration Zn, Cd and Ni stresses, dsRNAi transgenic plants had higher heavy metal contents than wild type in rice roots, but had lower heavy metal contents than wild type in leaves and seeds. We speculated that in dsRNAi transgenic rice plants, due to the gene functional redundancy, down-regulation of OsMTP1 could not affect the roots absorbing the heavy metals, but it could impair the translocation of heavy metal from rice roots to aerial parts. This indicated that OsMTP1 might play crucial roles in heavy metal transcellular transport or translocation.
In the yeast mutant complementation assay, OsMTP1 can totally suppress the Ni-sensitivity of the yeast smf1Δ mutant, and partially suppress the Zn, Cd sensitivity of the yeast cot1Δ and ycf1Δ. These data showed that OsMTP1 demonstrated obvious metal-specificity, possibly preferentially transporting Ni in yeast. While in rice plants, the metal-specificity was not obvious since the Zn, Cd and Ni content levels showed similar changed pattern in dsRNAi transgenic rice plants under low-concentration heavy metal stresses. Although the mechanism of metal selectivity has been discussed in previous reports (Damien et al. 2003; Montanini et al. 2007), and some PtdMTP1 site-mutated assays were performed (Montanini et al. 2007), the mechanism remains unclear and needs further investigation.
In summary, OsMTP1 contains all typical MTP domains and can complement the Ni-sensitive phenotype of yeast mutant smf1Δ. OsMTP1 can also partially complement the Zn-sensitive phenotype of yeast mutant cot1Δ and the Cd-sensitive phenotype of yeast mutant ycf1Δ. These results indicate that OsMTP1 can transport heavy metals and possessed cation-binding specificity. Although OsMTP1 was expressed in leaves, stems, roots, and flowers, its expression was relatively low and regulated spatially and temporally during rice development and also showed differential response to Cd stress. Transgenic assays in rice showed that OsMTP1 expression levels changed plant cation absorption and affected Zn, Ni, and Cd contents. Bioinformatic analyses indicated that OsMTP1 is a membrane-integrated protein, and subcellular localization of OsMTP1/EGFP fusion protein showed that OsMTP1 was mainly localized in plasmalemma. The identification and functional analysis of OsMTP1 in rice should be helpful in understanding its role in plant heavy metal transport. This is the first molecular identification of rice MTP and we propose that heavy metal (especially cadmium) transport through MTPs is indeed existent. Our observations may provide a key to the development of low-toxic heavy metal crops for food production, and are potentially of great importance for changed heavy metal remediation manners of other plants.
Abbreviations
- MTP:
-
Metal tolerance protein
- CDF:
-
Cation diffusion facilitator
- NRAMP:
-
Natural resistance-associated macrophage protein
- ZIP:
-
Zn-regulated transporter-/Fe-regulated transporter-like protein
- TMD:
-
Transmembrane domain
- ZAT:
-
Zinc transporter of Arabidopsis thaliana
- AAS:
-
Atomic absorption spectrometer
- dsRNAi:
-
Double-stranded RNA interference
- SD medium:
-
Synthetic defined medium
- RT-PCR:
-
Reverse transcription PCR
- DIG:
-
Digoxigenin
- ORF:
-
Open reading frame
- EGFP:
-
Enhanced green fluorescent protein
- SSH:
-
Subtractive suppression hybridization
- ISH:
-
In situ hybridization
References
Arrivault S, Senger T, Krämer U (2006) The Arabidopsis metal tolerance protein AtMTP3 maintains metal homeostasis by mediating Zn exclusion from the shoot under Fe deficiency and Zn oversupply. Plant J 46(5):861–879
Chen R, Zhao X, Shao Z, Wei Z, Wang Y, Zhu L, Zhao J, Sun M, He R, He G (2007) Rice UDP-glucose pyrophosphorylase1 is essential for pollen callose deposition and its cosuppression results in a new type of thermosensitive genic male sterility. Plant Cell 19(3):847–861
Clemens S (2001) Molecular mechanisms of plant metal tolerance and homeostasis. Planta 212:475–486
Conklin DS, McMaster JA, Culbertson MR, Kung C (1992) COT1, a gene involved in cobalt accumulation in Saccharomyces cerevisiae. Mol Cell Biol 12:3678–3688
Damien B, Annegret K, Francis DaleS, Michel C (2003) Poplar metal tolerance protein 1 confers zinc tolerance and is an oligomeric vacuolar zinc transporter with an essential leucine zipper motif. Plant Cell 15:2911–2928
Delhaize E, Kataoka T, Hebb DM, White RG, Ryan PR (2003) Genes encoding proteins of the cation diffusion facilitator family that confer manganese tolerance. Plant Cell 15(5):1131–1142
Delhaize E, Gruber BD, Pittman JK, White RG, Leung H, Miao Y, Jiang L, Ryan PR, Richardson AE (2007) A role for the AtMTP11 gene of Arabidopsis in manganese transport and tolerance. Plant J 51(2):198–210
Dräger DB, Desbrosses-Fonrouge AG, Krach C, Chardonnens AN, Meyer RC, Saumitou-Laprade P, Krämer U (2004) Two genes encoding Arabidopsis halleri MTP1 metal transport proteins co-segregate with zinc tolerance and account for high MTP1 transcript levels. Plant J 39(3):425–439
Eren E, Argüello JM (2004) Arabidopsis HMA2, a divalent heavy metal-transporting P(IB)-type ATPase, is involved in cytoplasmic Zn2+ homeostasis. Plant Physiol 136(3):3712–3723
Gietz RD, Schiestl RH (2007) High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat Protoc 2(1):31–34
Goyer RA (1997) Toxic and essential metal interactions. Annu Rev Nutr 17:37–50
Gustin JL, Zanis MJ, Salt DE (2011) Structure and evolution of the plant cation diffusion facilitator family of ion transporters. BMC Evol Biol 11:76
Hall JL, Williams LE (2003) Transition metal transporters in plants. J Exp Bot 54(393):2601–2613
Haney CJ, Grass G, Franke S, Rensing C (2005) New developments in the understanding of the cation diffusion facilitator family. J Ind Microbiol Biotechnol 32(6):215–226
Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6:271–282
Hua W, Zhang L, Liang S, Jones RL, Lu YT (2004) A tobacco calcium/calmodulin-binding protein kinase functions as a negative regulator of flowering. J Biol Chem 279:31483–31494
Kim D, Gustin JL, Lahner B, Persans MW, Baek D, Yun DJ, Salt DE (2004) The plant CDF family member TgMTP1 from the Ni/Zn hyperaccumulator Thlaspi goesingense acts to enhance efflux of Zn at the plasma membrane when expressed in Saccharomyces cerevisiae. Plant J 39(2):237–251
Kobae Y, Uemura T, Sato MH, Ohnishi M, Mimura T, Nakagawa T, Maeshima M (2004) Zinc transporter of Arabidopsis thaliana AtMTP1 is localized to vacuolar membranes and implicated in zinc homeostasis. Plant Cell Physiol 45:1749–1758
Li L, Kaplan J (1998) Defects in the yeast high affinity iron transport system result in increased metal sensitivity because of the increased expression of transporters with a broad transition metal specificity. J Biol Chem 273:22181–22187
Li ZS, Lu YP, Zhen RG, Szczypka M, Thiele DJ, Rea PA (1997) A new pathway for vacuolar cadmium sequestration in Saccharomyces cerevisiae: YCF1-catalyzed transport of bis (glutathionato) cadmium. Proc Natl Acad Sci USA 94:42–47
Lyons TJ, Gasch AP, Gaither LA, Botstein D, Brown PO, Eide DJ (2000) Genome-wide characterization of the Zap1p zinc-responsive regulon in yeast. Proc Natl Acad Sci USA 97:7957–7962
Mäser P, Thomine S, Schroeder JI, Ward JM, Hirschi K, Sze H, Talke IN, Amtmann A, Maathuis FJ, Sanders D, Harper JF, Tchieu J, Gribskov M, Persans MW, Salt DE, Kim SA, Guerinot ML (2001) Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol 126(4):1646–1667
Montanini B, Blaudez D, Jeandroz S, Sanders D, Chalot M (2007) Phylogenetic and functional analysis of the Cation Diffusion Facilitator (CDF) family: improved signature and prediction of substrate specificity. BMC Genomics 107:1471–2164
Supek F, Supekova L, Nelson H, Nelson N (1996) A yeast manganese transporter related to the macrophage protein involved in conferring resistance to mycobacteria. Proc Natl Acad Sci USA 93:5105–5110
Szczypka MS, Wemmie JA, Moye-Rowley WS, Thiele DJ (1994) A yeast metal resistance protein similar to human cystic fibrosis transmembrane conductance regulator (CFTR) and multidrug resistance-associated protein. J Biol Chem 269:22853–22857
Ton VK, Mandal D, Vahadji C, Rao R (2002) Functional expression in yeast of the human secretory pathway Ca2+/Mn2+ATPase defective in Hailey–Hailey disease. J Biol Chem 277:6422–6427
Van-Der-Zaal BJ, Neuteboom LW, Pinas JE, Chardonnens AN, Schat H, Verkleij JA, Hooykaas PJ (1999) Overexpression of a novel Arabidopsis gene related to putative zinc-transporter genes from animals can lead to enhanced zinc resistance and accumulation. Plant Physiol 119:1047–1055
Wang Z, Chen CB, Xu YY, Jiang RX, Han Y, Xu ZH, Chong K (2004) A practical vector for efficient knockdown of gene expression in rice (Oryza sativa L.). Plant Mol Biol Rep 22(4):409–417
Williams LE, Mills RF (2005) P1B-ATPases-an ancient family of transition metal pumps with diverse functions in plants. Trends Plant Sci 10:491–502
Williams LE, Pittman JK, Hall JL (2000) Emerging mechanisms for heavy metal transport in plants. Biochim Biophys Acta 1465:104–126
Yoshida S, Forno DA, Cock JH, Gomez KA (1976) Laboratory manual for physiological studies of rice, 3rd edn. The International Rice Research Institute, Manila, The Philippines
Zhang M, Liang SP, Lu YT (2005) Cloning and functional characterization of NtCPK4, a new tobacco calcium-dependent protein kinase. Biochim Biophys Acta 1729:174–185
Acknowledgments
We sincerely thank Wei Zhang for her help in the rice transformation assay. We are also very grateful to Prof. David Ow for his comments and constructive suggestions to improve this manuscript. This research was supported by the China National Natural Sciences Foundation (Grant no. 30900892), the International Foundation for Science (Grant no. C/4787) to Mei Zhang and the CAS ‘100 Talents’ Program to Keqiang Wu.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Y. Lu.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yuan, L., Yang, S., Liu, B. et al. Molecular characterization of a rice metal tolerance protein, OsMTP1 . Plant Cell Rep 31, 67–79 (2012). https://doi.org/10.1007/s00299-011-1140-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-011-1140-9