Abstract
2,4,6-Trinitrotoluene (TNT) is the most widely used nitroaromatic compound and is highly resistant to degradation. Most aerobic microorganisms reduce TNT to amino derivatives via formation of nitroso- and hydroxylamine intermediates. Although pathways of TNT degradation are well studied, proteomic analysis of TNT-degrading bacteria was done only for some individual Gram-negative strains. Here, we isolated a Gram-positive strain from TNT-contaminated soil, identified it as Bacillus pumilus using 16S rRNA sequencing, analyzed its growth, the level of TNT transformation, ROS production, and revealed for the first time the bacillary proteome changes at toxic concentration of TNT. The transformation of TNT at all studied concentrations (20–200 mg/L) followed the path of nitro groups reduction with the formation of 4-amino-2,6-dinitrotoluene. Hydrogen peroxide production was detected during TNT transformation. Comparative proteomic analysis of B. pumilus showed that TNT (200 mg/L) inhibited expression of 46 and induced expression of 24 proteins. Among TNT upregulated proteins are those which are responsible for the reductive pathway of xenobiotic transformation, removal of oxidative stress, DNA repair, degradation of RNA and cellular proteins. The production of ribosomal proteins, some important metabolic proteins and proteins involved in cell division are downregulated by this xenobiotic.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
2,4,6-trinitrotoluene (TNT) is one of the most common nitroaromatic explosives in the world. TNT is toxic to pro- and eukaryotes, potential mammalian carcinogen and resistant to biodegradation (Alothman et al. 2020; Esteve-Nuñez et al. 2001; Smets et al. 2007). Every year 1000 tons of TNT are produced globally, and nearly 2 million L of TNT-contaminated wastewater, along with other nitro aromatic compounds are discarded into the environment every day (Whitacre 2012). Contamination with this xenobiotic occurs in places where ammunition is manufactured, at training grounds, and in areas of former and current hostilities (Khan et al. 2015). The most promising method of soil purification is in situ bioremediation based on the ability of soil microorganisms to decompose these pollutants. The main problems of bioremediation are associated with difficulties of microbial adaptation to toxic pollutants and with a high probability of pollutants migration to adjacent environments.
The toxic effect of TNT in relation to bacteria is manifested in the inhibition of culture growth, changes in the morphology and physical properties of cells, decrease of carbohydrates utilization rate and the level of reduced pyridine and oxidized flavin cofactors, as well as in suppression of respiration and decrease in the transmembrane potential (Kurinenko et al. 2003, 2007; Cherepnev et al. 2007; Konovalova et al. 2013). The TNT toxicity is associated not only with the products of its microbial transformation, but also with the formation of reactive oxygen species (ROS) during the transformation (Ziganshin et al. 2015).
Bacteria of genera Pseudomonas, Enterobacter, Rhodococcus, Mycobacterium, Clostridium and Desulfovibrio are capable of degrading TNT (Serrano-González et al. 2018). Bacilli also were shown to transform several nitroaromatic compounds including TNT (Khan et al. 2015; Konovalova et al. 2013). Previous studies have shown the possibility of TNT transformation by strains B. subtilis SK1 (Kurinenko et al. 2003), B. cereus (Mercimek et al. 2013), B. mycoides (Lin et al. 2013), Bacillus VT-8 (Solyanikova et al. 2014). TNT transformation pathways by microorganisms are quite well studied, the formation of 2,4- and 2,6-dinitrotoluenes, nitrites, hydroxylamine derivatives, ammonium, etc. has been proven (Wijker et al. 2013; Ziganshin and Gerlach 2014). Several strains harbor dual pathways: nitro reduction (reduction of the nitro group in TNT to a hydroxyl amino and/or amino group) and denitration (reduction of the aromatic ring of TNT to Meisenheimer complexes with the release of nitrites). TNT can serve as a nitrogen source for some strains, and the postulated mechanism involves ammonia release from hydroxyl amino intermediates (Esteve-Nuñez et al. 2001; Smets et al. 2007; Mercimek et al. 2013; Yin et al. 2005).
However, there is still insufficient information on the changes in the protein profile of microorganisms exposed to TNT. To date, TNT-induced changes of protein profile were demonstrated in yeast Yarrovia lipolytica (Khilyas et al. 2017) and only in several Gram-negative bacteria, namely Stenotrophomonas sp. OK-5 (Ho et al. 2004), Pseudomonas sp. HK-6 (Cho et al. 2009), Citrobacter (Liao et al. 2018), and Buttiauxella (Xu et al. 2021).
The aim of this work was to study changes in the proteome of Gram-positive bacterium, which we isolated from TNT-contaminated soil and identified as B. pumilus using 16S rRNA sequencing during its growth on high TNT concentrations. Herein, we analyzed the growth of this strain, the level of TNT transformation, ROS production, and revealed for the first time the proteome changes of B. pumilus at toxic concentration of TNT.
Materials and methods
Soil sample and bacteria cultivation
The Bacillus strain was isolated from an urban leached black soil of a park adjacent to the territory of the Federal Enterprise “Kazan Gunpowder Plant”. This soil has never been purposefully exposed to TNT, nitroaromatic compounds or pesticides. However, we found negligible concentrations of TNT in this soil using extraction method followed by HPLC method followed by HPLC analysis (Williford and Mark Bricka 1999). TNT concentration was ~ 5 mg/kg of soil, when extracted with acetonitrile. No TNT was found in the aqueous extract.
The strain was grown in flasks on LB medium at 30 °C for 16 h. Inoculate was added of a final concentration of 107 cells/mL to synthetic medium (g/L: MgSO4 × 7H2O—0.25, (NH4)2SO4—0.5, NaCl—0.5, glucose—3, supplemented by 4% (v/v) 0,2 M phosphate buffer pH 7.0). TNT was added at the final concentrations of 20–200 mg/L. The cultivation was carried out in 250 mL flasks in a shaker-incubator (KS 4000, IKA, China) at 120 rpm and a temperature of 30 °C.
Taxonomic identification of isolated strain
DNA was isolated from bacterial biomass using DNA Extraction Kit (BIO-RAD). The purity of the isolated DNA was assessed by agarose gel electrophoresis, and the DNA concentration was determined using the Qubit dsDNA BR assay kit and Qubit 2.0 Fluorometer (Invitrogen, Carlsbad, CA, USA). The 16S rRNA gene was amplified by PCR using primers (Forward primer 27F 5’-AGAGTTTGATCMTGGCTCAG-3’; Reverse primer 1492R 5’-TACGGYTACCTTGTTACGACTT-3’) (Wu et al. 2014) and pyrosequenced using the GS Junior System (454 Life Sciences, Roche). PCR products were then purified using QIAquick PCR purification kit (Qiagen) and analyzed using 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) according to the manufacturer’s instructions. Identification of the isolate was made using BLAST algorithm against GENBANK database. Phylogenetic analysis of the microorganism was performed using MEGA 6.0 software with the 16S rRNA sequences of 18 Bacillus species and a strain of Lactobacillus delbrueckii as an outgroup. Further, these sequences were aligned using the ClustalW software; incomplete sites at the 5 ‘and 3’ ends of the 16S rRNA gene sequences were excluded from the alignment. The resulting aligned sequences were selected for further analysis. The phylogenetic tree was built using the maximum likelihood (ML) algorithm with 1000 bootstrap iterations. The sequence of B. pumilus GY was deposited in NCBI database (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA822692/). The sequence of B. pumilus GY is identical to the reference strain B. pumilus Meyer and Gottheil registered in All-Russian Collection of Microorganisms (VKM) as B-508 (http://www.vkm.ru/rus/Catalogue.htm ) and in ATCC as Bacillus pumilus ATCC 7061 (https://www.atcc.org/products/7061 ).
High-performance liquid chromatography detection of TNT
TNT and its metabolites were analyzed in cell-free cultural fluids using Thermo Scientific Ultimate 3000 chromatograph (USA) equipped with an automatic sampler, injector, fraction collector, diode detector, SupelcosilLC-8 column (250 × 4.6 mm; 5 μm) and a wavelength detector (254 nm). For gradient system, two mobile phases were used: methanol and phosphate buffer (0.025 M, pH 7). The flow rate was 1 mL/min. The identification of metabolites was carried out by comparing the release times of markers including 4-amino-2,6-dinitrotoluene, (4-ADNT, 19.8), 2-amino-4,6-dinitrotoluene (2-ADNT, 10.8), 2,6-diamino-4-nitrotoluene (2,6-DANT, 20.0), TNT (17.5); the time of the chromatographic peak emergence are indicated in parentheses. Data collection and processing was carried out using the control system Chromeleon 6.80.
Nitrite ions analysis using ion chromatography
Nitrite ions were analyzed using Thermo Scientific Dionex AS 22 ion chromatograph (USA) equipped with a GP40 gradient pump, a CD20 conductivity detector, an AS40 autosampler, an IonPac AG9-HC guard column (4 × 50 mm), and an IonPac AS9-HC separation column (4 × 250 mm). A solution of 4.5 mM Na2CO3 and 1.4 mM NaHCO3 was used as an eluent. Elution was performed at a rate of 1.0 mL/min. A commercial Thermo Scientific Dionex anion solution (USA) which included (mg/L): chlorides—30, nitrites—100, bromides—100 mg, nitrates—100 mg, phosphates—150, sulfates—150, was used as a standard for plotting calibration curves.
ROS measurement
H2O2 formed from the O2− alongside nitro groups reduction pathway was measured using chemiluminometer Lum-100 (DISoft LLC, Russia). To detect the formation of hydrogen peroxide in a short time, we increased the concentration of bacterial cells in the cultivation medium by a factor of 10 compared to the growth conditions and added 100 µg/L and 200 µg/L of TNT to the suspension. After incubation time of 40, 80 and 120 min, samples of cultural fluid were treated with 2 mL of 1 M trichloroacetic acid and centrifuged for 20 min (5000 g, 4 °C), the resulting supernatant was titrated with 10 M KOH to pH 7. Then, 1 mL of a luminol-peroxidase mixture (1 µM peroxidase and 0.2 mM luminol) was added to 50 µL of each sample and luminescence was registered. Using a calibration curve built on the basis of known H2O2 concentrations (0.9–10 µM), its concentration in the samples was determined.
Isolation of bacillary proteins
To obtain a fraction of B. pumilus intracellular proteins, the bacterial culture grown for 24 h was harvested by centrifugation. 1 µL of a proteinase inhibitor cocktail (P8340, Sigma Aldrich), 1 µL of DNase (1 mg/mL, P07-01, BioloT), 10 µL of a solution of 22% Chaps + 16% NP40 (Sigma Aldrich), 50 µL 10% Triton-X 100 (Helicon) and 5 µL of lysozyme (10 µg/µL) were added to the pellet. The suspension was incubated for 20 min at 37 °C with shaking at 200 rpm. For mechanical destruction, glass beads (0.5 mm, 0.1 mm, BioSpec Products; FastPrep®Systems, MP Biomedicals) were used. Then the mixture was kept on ice for 10 min and subjected to ultrasonic destruction for 10 min. After ultrasound, 50 µL of cell lysate (urea 0.48 g, Tris-HCl 1.2 mg, 133 µL Chaps + NP40 in 1 mL, pH = 8.5) was added and centrifuged for 10 min at 10,000×g. The supernatant was collected and protein concentration was measured using a 2-D Quant kit (GE Healthcare, USA).
Two-dimensional electrophoresis
Rehydration buffer (30 mM DTT, 4% CHAPS, 20 mM Tris, 2% IPG buffer, 1% bromphenol blue, pH = 8.0) was added to the protein solution so that the final volume of each sample was 300 µL. The procedure for isoelectric focusing of this fraction of samples was carried out using strips (Bio-Rad # 163–2032, 18 cm, pH 3–10) and a PowerPacHV protein isoelectric focusing device (Bio-Rad, USA) according to the manufacturer’s instructions. To separate proteins in the second direction, 9–16% vertical gradient gels with marker # 26,610 prepared according to the standard protocol were used. Densitometric analysis was performed using PDQuest software (Bio-Rad) after Coomassie staining. The gels were fixed in a solution of 10% acetic acid and 20% isopropanol.
Trypsinolysis of proteins
Protein spots were excised from Coomassie stained SDS-polyacrylamide gel, transferred to non-siliconized tubes, and incubated in water (ddiH2O) for 15 min. After water was removed, 50µL of a 1:1 mixture of acetonitrile and 200 mM aqueous solution of ammonium bicarbonate was added to the protein spots. The tubes were incubated at 50 °C on a shaker for 20–30 min, after which the solution was removed. For dehydration, 100 µL of 100% acetonitrile was added and incubated for 20 min. After drying, 5 µL of trypsin solution (12.5 ng/µL) was added to each excised spot. The tubes were incubated in ice at 4 °C for one hour. Next, they were transferred to a thermostat at 37 °C and incubated for 12 h. To extract proteins, 20 µL of a 0.1–0.5% trifluoroacetic acid (TFA) solution was added to each sample. Samples were placed in an ultrasonic bath for 10 min and incubated for 40 min at room temperature. The solution was moved a new tube, and the spots were subjected to additional extraction with a solution of 100% acetonitrile and 0.5% TFA in a 1:1 ratio. The solution was evaporated on a rotary evaporator (Eppendorf, Concentratorplus) at 45 °C for 1 h. The dried extract was dissolved in 5 µL of 0.1% TFA and used for the analysis on a mass spectrometer.
Mass spectrometry
Samples were applied to plates with dehydroxybenzoic acid as a matrix in the ratio of 4: 1. Analysis was performed on MALDI using flexControl-ultraflexTOF/TOF software. Calibration was performed against a standard peptide solution. Ionizing irradiation was applied to the samples to obtain 104 peaks. The resulting peaks were saved and transferred for analysis to the flex Analysis software. An internal calibration was performed to remove peaks of keratin, trypsin, and contaminants found in empty SDS-polyacrylamide gel pieces.
Bioinformatic analysis of proteins
Identification of proteins was carried out using the BioTools software using the Mascot search engine (http://www.matrixscience.com) and Bacillus pumilus Uniprot database (20,180,131; 35,840 sequences; 10,623,105 residues). For the false discovery rate (FDR) statistical significance p ≤ 0.05 was used. Alkylation of cysteine by carbamidomethylation was set as a fixed modification and oxidation of methionine was applied as a variable modification. Information about functions of proteins was obtained from UniProt and NCBI (PubMed) databases; mapping of protein-protein interactions (interactome) was performed using the STRING online program (https://string-db.org/).
Statistics
Mathematical data processing was carried out using Microsoft Office Excel 7.0. All experiments were carried out in at least three replicates. A group of data was considered homogeneous if the standard deviation σ in it did not exceed 13%. The difference between the groups was considered significant if the probability criterion was P < 0.05.
Results
Taxonomy of Bacillus strain
To clarify the taxonomic position of the isolated Bacillus strain, 16S rRNA gene was sequenced. According to the screening performed in the GenBank database on the basis of fragment sequence, the closest sequence to the obtained sequence was 16S rRNA of B. pumilus NBRC 12092 strain. The 16S rRNA gene sequence of B. pumilus GY was most closely related to the reference strain with 100% identity. Phylogenetic tree showing links with other members of the genus Bacillus and with other B. pumilus strains is presented in Fig. 1. The isolated strain was designated as B. pumilus GY. The optimal tree with the sum of branch length = 0.21147444 is shown. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree.
TNT transformation
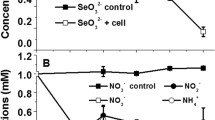
The ability of B. pumilus GY to grow on a synthetic medium in the presence of various concentrations of TNT (20–200 mg/L) was assessed. Bacterial growth was observed at all TNT concentrations, decreasing to the minimum at 200 mg/L (Fig. 2 A). Despite the growth suppression of B. pumilus GY, it was able to eliminates 20–50 mg/mL TNT and reduce higher concentrations by this amount (Fig. 2B). In medium with an initial TNT concentration of 20 mg/L, the xenobiotic disappeared after 4 h of cultivation. Afterwards, the strain began to grow rapidly. The TNT concentration of 50 mg/L was eliminated after 24 h of cultivation. Other TNT concentrations were transformed at 24 h of growth by 40, 24, and 18% from an initial TNT concentration of 100, 150, and 200 mg/L, respectively.
Identification of transformation products of high TNT concentrations (100–200 mg/L) at 24 h of cultivation revealed 4-amino-2,6-dinitrotoluene (4-ADNT) (Fig. 3A), at the same time, there were no nitrite ions in the cultivation medium. This suggests that the transformation of xenobiotic follows the pathway of nitro group reduction with the formation of monoamine derivatives of TNT. The first step in nitro group reduction can be achieved through one-electron transfer (solid line) or two-electron transfer (dashed line) (Fig. 3B, according to Esteve-Nuñez et al. 2001).
The first mechanism produces a nitro anion radical that could react with oxygen to form a superoxide radical and the original nitroaromatic compound through a futile cycle (dotted line). If the mechanism occurs via the transfer of two electrons, the nitroso derivative formed is the first putative intermediate; following two consecutive electron transfers, a hydroxylamine and an aromatic amine are produced. The superoxide radical is capable of further dismutation, which leads to the formation of hydrogen peroxide H2O2 (Ziganshin et al. 2015; Naumenko et al. 2013, 2017; Khilyas et al. 2013).
ROS detection
The level of extracellular hydrogen peroxide, which is formed upon dismutation of the superoxide radical anion during TNT transformation by the reduction of nitro groups was measured in reaction mixtures where the concentration of cells was about 10 times higher than it was under growth conditions. In 2 h, B. pumilus GY completely transformed TNT at concentration of 100 mg/L and reduced by 80% the concentration of 200 mg/L (Fig. 3A). The highest Н2О2 level which exceeded of the control one’s by 3 times was detected at TNT concentration of 200 mg/L (Fig. 4). The permanent Н2О2 baseline level is due to ROS normally generated as a subproduct of cellular metabolism, and the decrease in the peroxide level after 120 min of incubation is related to the adaptive antioxidant response of the bacteria.
Proteome profiling
To determine the up- and down-regulation of proteins under the influence of TNT, densitometric analysis was carried out using the PDQuest software, during which the images of acrylamide gels containing proteins of the control and experimental groups were compared. All identified proteins correspond to the reference ones from reference strain Bacillus pumilus NBRC 12,092. Under the action of 200 mg/L TNT, we visually observed the downregulation of 46 proteins; in addition to these proteins, 24 upregulated proteins were detected in the presence of TNT (Supplementary Fig. 1). According to the databases, out of 24 upregulated proteins 23 were identified (Table 1), and out of 46 downregulated ones, 41 were identified (Table 2). Briefly summarizing the results of proteomic analysis, we can conclude that TNT upregulated proteins responsible for the reduction pathway of this xenobiotic transformation, removal of oxidative stress, for DNA repair, degradation of RNA and proteins, have been detected. TNT downregulated the production of ribosomal proteins, some important metabolic proteins and proteins involved in the cell division.
Using STRING software, we estimated the percentage of proteins involved in various metabolic processes of the cell in control (without TNT) and experimental (200 mg/L TNT) groups (Fig. 5). When proteins are grouped according to their biological functions, it can be seen that control cells perform a wider range of functions compared to TNT-exposed bacterial cells, that is manifested in 11 functional groups (Fig. 5B) compared to 7 ones (Fig. 5A). Among the proteins of the experimental group, the predominant enzymes are those responsible for maintaining the redox state of the cell, the metabolism of the nitrogen-containing compound, and the degradation of RNA indicating a response to TNT-induced stress.
Proteins of B. pumilus GY growing 24 h by TNT concentration of 200 mg/L (A) and without TNT (B) participating in various metabolic processes. All identified proteins are taken as 100%. Proteins, the concentration of which is less than 1%, are classified in the section “other”. Proteins were sorted by functional groups using the STRING online program
We also constructed a scheme of protein-protein interactions in experimental (Supplementary, Fig. 2A) and control group (Supplementary, Fig. 2B), which shows the changes in interteractome of B. pumilus GY with and without TNT.
Consequently, in the process of TNT transformation by B. pumilus GY, upregulated proteins are responsible for the reduction pathway of xenobiotic transformation, removal of oxidative stress and DNA repair, as well as RNA and proteins degradation. TNT inhibited the production of ribosomal proteins and proteins involved in in basic microbial metabolism.
Discussion
The huge potential of microorganisms capable of transforming and destructing nitroaromatic compounds attracts close attention of scientists involved in the development of methods for combating anthropogenic environmental pollution.
Even those microorganisms that were unlikely to be exposed to TNT in nature are capable to transform it. Bacteria that are part of the intestinal tract microbiota, such as E. coli, Citrobacter sp., have proven to be successful TNT destructors (Kurinenko et al. 2007; Liao et al. 2018). Degradation rate of 100 mg/L TNT reached 100% within 30 h after inoculation with Klebsiella variicola originally identified as a benign endosymbiont in plants (Yang et al. 2021). TNT-degrading Pseudomonas strain was isolated from Antarctic environmental samples of Deception Island (Cabrera et al. 2020). These facts indicate the wide prevalence of microbial oxidoreductases, in particular FMN-dependent NADH-nitro reductase (EC1.5.1.34), capable to nitroreduction. In studied strain B. pumilus GY this enzyme—FMN-dependent NADH-azoreductase was upregulated by TNT (Table 1) supporting that biotransformation occurs along the nitro reduction pathway with formation of monoamine derivatives (Fig. 3). Among the proteins upregulated by TNT nitrate reductase, which also able to reduce TNT nitro groups to amino derivatives, was identified. Obviously, both proteins took part in the formation of 4-ADNT which we detected at 24 h of cultivation (Fig. 3 A). Our data is consistent with upregulation of aldehyde dehydrogenase and monooxygenase, which are key enzymes of the TNT degradation pathway in Buttiauxella sp. (Xu et al. 2021) and upregulation of assimilatory nitrate reductase in Pseudomonas sp. HK-6 (Cho et al. 2009), indicating the similarity of initial stages of TNT transformation by Gram-negative and Gram-positive bacteria.
A natural ecotope where B. pumilus GY existed was not directly exposed to TNT, however it contained insignificant TNT amounts due to long-term migration from the contaminated area. It is unlikely that the isolated strain effectively transforms TNT directly in the soil, where this compound is tightly adsorbed on soil particles. However, in laboratory conditions it was shown that B. pumilus GY eliminates 20 mg/L of TNT already after 4 h, and 50 mg/L of TNT after 24 h (Fig. 2B). The higher concentrations of 100, 150 and 200 mg/L were transformed after 24 h by 40, 24 and 18%, respectively. This means that with a gradual transition of low concentrations of TNT into the aqueous phase, soil bacteria will be able to degrade this xenobiotic.
It is not TNT biotransformation per se that deserves special attention, but elucidating response of B. pumilus GY to TNT-induced toxic stress. The adaptive response system of microorganisms combines repair mechanisms, induction of protective proteins, the action of which is aimed at neutralizing the harmful effects, and dissociative processes of splitting a homogeneous population into variants. It could be assumed that B. pumilus GY was already partially adapted to TNT in soil; therefore even a highly toxic concentration of 200 mg/L TNT was transformed by this strain by about 10% after 8 h and did not completely suppress biomass growth (Fig. 2). In response to stress conditions, the elimination of oxidized RNA by degradosomes containing polynucleotide phosphorylase, enolase, RNA helicase and RNase E is a well-known mechanism (Miczak et al. 1996). We also identified enolase among the TNT-upregulated proteins. Bacterial Clp proteases are involved in the degradation of proteins that are aggregated and incorrectly assembled during folding, as well as of proteins damaged by oxidative stress. So it’s quite clear that upregulation of Clp protease together with chaperonin which carries out the assembly of both newly synthesized and denatured proteins are detected under stress (Table 1). According results obtained by Ho et al. (2004), under TNT-mediated stress in Stenotrophomonas sp. OK-5 chaperone protein DnaK also was identified.
Proteome analysis revealed upregulation of proteins which counteract oxidative stress caused by TNT. We confirmed the existence of TNT-induced oxidative stress in B. pumilus GY by detecting hydrogen peroxide which is formed upon dismutation of the superoxide radical anion (Fig. 4). ROS production during TNT nitro reduction established for B. pumilus GY (Fig. 4) is characteristic for all organisms, including human, mouse and rat cells (Naumenko et al. 2017) as well as plants (Naumenko et al. 2013) and yeasts (Ziganshin et al. 2015; Khilyas et al. 2013, 2017). Protein profiling of B. pumilus GY revealed upregulated catalase, which promotes the removal of TNT-induced ROS. It is known that H2O2 affects the expression of the mdh2 gene encoding malate dehydrogenase, which increases catalase activity by NADH production during conversion of malate to oxaloacetate (Heyno et al. 2014). We also detected increasing level of NAD-dependent malate dehydrogenase together with components of the antioxidant system Trx/TrxR. Upregulation of these proteins is in line with the data of literature and indicates powerful counteraction of B. pumilus GY to oxidative stress caused by TNT.
Among the proteins downregulated by TNT were many enzymes participating in basic microbial metabolism including glycolysis, pentose phosphate pathway, pyruvate metabolism and citrate cycle indicating the general decrease in metabolic activity of bacterium (Table 2). For example, NADH: quinone oxidoreductase involved in the first step of the electron transport chain was downregulated. Additionally, TNT alters the metabolism of amino acids downregulating alanine dehydrogenase and ornithine aminotransferase. Decreased levels of L3 protein of 50 S ribosomal subunit and of S1 protein of 30 S subunit were identified. Apparently, TNT-induced downregulation of ribosomal proteins may indicate reduced biosynthesis of all proteins that leads to the essential decrease of bacterial biomass (Fig. 1A).
In the study, we focused not on the well-studied pathways of TNT complete destruction, but on the adaptive potential of the strain, which quickly changed the level of synthesized proteins. This result is of great ecological importance and indicates a high adaptability of soil bacteria to grow in the presence of nitroaromatic pollutants. Microbial response to TNT-induced stress and its impact on biodegradation need further investigation in order to have a better understanding of ways and methods to counteract environmental pollution.
References
Alothman ZA, Bahkali AH, Elgorban AM, Al-Otaibi MS, Ghfar AA, Gabr SA, et al. (2020) Bioremediation of explosive TNT by Trichoderma viride. Molecules 25(6):1393. https://doi.org/10.3390/molecules25061393
Cabrera MÁ, Márquez SL, Quezada CP, Osorio MI, Castro-Nallar E, González-Nilo FD, Pérez-Donoso JM (2020) Biotransformation of 2,4,6-trinitrotoluene by Pseudomonas sp. TNT3 isolated from Deception island, Antarctica. Environ Pollut 262:113922. https://doi.org/10.1016/j.envpol.2020.113922
Cherepnev GV, Velizhinskaya TA, Yakovleva GYu, Denivarova NA, Kurinenko BM (2007) Assessing the toxic effect of 2,4,6-trinitrotoluene on cells of Escherichia coli K12 by flow cytofluorometry. Microbiology 76(3): 331–335. https://doi.org/10.1134/S0026261707030101
Cho Y-S, Lee B-U, Kahng H-Y, Oh K-H (2009) Comparative analysis of 2,4,6-trinitrotoluene (TNT)-induced cellular responses and proteomes in Pseudomonas sp. HK-6 in two types of media. J Microbiol 47(2):220–224. https://doi.org/10.1007/s12275-008-0108-0
Esteve-Nuñez A, Caballero A, Ramos JL (2001) Biological degradation of 2,4,6-trinitrotoluene. Microbiol Mol Biol Rev 65(3):333–352. https://doi.org/10.1128/MMBR.65.3.335-352.2001
Heyno E, Innocenti G, Lemaire SD, Issakidis-Bourguet E, Krieger-Liszkay A (2014) Putative role of the malate valve enzyme NADP-malate dehydrogenase in H2O2 signaling in Arabidopsis. Philos Trans R Soc Lond B Biol Sci 369(1640):20130228. https://doi.org/10.1098/rstb.2013.0228
Ho E-M, Chang H-W, Kim S-I, Kahng H-Y, Oh K-H (2004) Analysis of TNT (2,4,6-trinitrotoluene)-inducible cellular responses and stress shock proteome in Stenotrophomonas sp. OK-5. Curr Microbiol 49(5):346–352. https://doi.org/10.1007/s00284-004-4322-7
Khan MI, Lee J, Yoo K, Kim S, Park J (2015) Improved TNT detoxification by starch addition in a nitrogen-fixing Methylophilus-dominant aerobic microbial consortium. J Hazard Mater 300:873–881. https://doi.org/10.1016/j.jhazmat.2015.08.032
Khilyas IV, Ziganshin AM, Pannier AJ, Gerlach R (2013) Effect of ferrihydrite on 2,4,6-trinitrotoluene biotransformation by an aerobic yeast. Biodeg 24(5):631–644. https://doi.org/10.1007/s10532-012-9611-4
Khilyas IV, Lochnit G, Ilinskaya ON (2017) Proteomic analysis of 2,4,6-trinitrotoluene degrading yeast Yarrowia lipolytica. Front Microbiol 8:A.2600. https://doi.org/10.3389/fmicb.2017.02600
Konovalova OA, Yakovleva GY, Steryakov OV, Trushin MV (2013) Scanning probe microscopy in the study of morphometric changes and physical parameters of Escherichia coli bacteria under the action of 2,4,6 - trinitrotoluene. World Appl Sci J 23(4):507–509. https://doi.org/10.5829/idosi.wasj.2013.23.04.13077
Kurinenko BM, Yakovleva GY, Denivarova NA, Abreimova YV (2003) Specific toxic effects of 2,4,6-trinitrotoluene on Bacillus subtilis SK1. Appl Biochem Microbiol 39(3):275–8
Kurinenko BM, Denivarova NA, Davydov RE, Yakovleva GYu (2007) Features of the toxic action of 2,4,6-trinitrotoluene on Escherichia coli K12. Appl Biochem Microbiol 43(1):52–56. https://doi.org/10.1134/S0003683807010097
Liao H-Y, Chien C-C, Tang P, Chen C-C, Chen C-Y, Chen S-C (2018) The integrated analysis of transcriptome and proteome for exploring the biodegradation mechanism of 2,4,6-trinitrotoluene by Citrobacter sp. J Hazard Mater 349:79–90. https://doi.org/10.1016/j.jhazmat.2018.01.039
Lin H, Chena Z, Megharajc M, Naidu R (2013) Biodegradation of TNT using Bacillus mycoides immobilized in PVA-sodium alginate-kaolin. Appl Clay Sci 83:336–342. https://doi.org/10.1016/j.clay.2013.08.004
Mercimek HA, Dincer S, Guzeldag G, Ozsavli A, Matyar F (2013) Aerobic biodegradation of 2,4,6-trinitrotoluene (TNT) by Bacillus cereus isolated from contaminated soil. Microb Ecol. 66(3):512–521. https://doi.org/10.1007/s00248-013-0248-6
Miczak A, Kaberdin VR, Wei CL, Lin-Chao S. (1996) Proteins associated with RNase E in a multicomponent ribonucleolytic complex. Proc Natl Acad Sci USA 93(9):3865–3869. https://doi.org/10.1073/pnas.93.9.3865
Naumenko EA, Sibgatullina GV, Mukhitov AR, Rodionov AA, Ilinskaya ON, Naumova RP (2013) 2,4,6-Trinitrotoluene as a trigger of oxidative stress in Fagopyrum tataricum callus cells. Rus J Plant Physiol 60(3):404–410. https://doi.org/10.1134/S1021443713030102
Naumenko EA, Ahlemeyer B, Baumgart-Vogt E (2017) Species-specific differences in peroxisome proliferation, catalase, and SOD2 upregulation as well as toxicity in human, mouse, and rat hepatoma cells induced by the explosive and environmental pollutant 2,4,6-trinitrotoluene. Environ Toxicol 32(3):989–1006. https://doi.org/10.1002/tox.22299
Serrano-González MY, Chandra R, Castillo-Zacarias C, Robledo-Padilla F, Rostro-Alanis M, Parra-Saldivar R (2018) Biotransformation and degradation of 2,4,6-trinitrotoluene by microbial metabolism and their interaction. Defence Technol 14(2):151–164. https://doi.org/10.1016/j.dt.2018.01.004
Smets BF, Yin H, Esteve-Nuñez A (2007) TNT biotransformation: when chemistry confronts mineralization. Appl Microbiol Biotechnol 76(2):267–277. https://doi.org/10.1007/s00253-007-1008-7
Solyanikova P, Robota IV, Mazur DM, Lebedev AT, Golovleva LA (2014) Application of Bacillus sp. strain VT-8 for decontamination of TNT-polluted sites. Microbiology 83(5):577–584. https://doi.org/10.1134/S0026261714050257
Whitacre DM (2012) Reviews of environmental contamination and toxicology. Springer, New York. http://www.springer.com/gp/book/9781461414629.
Wijker RS, Bolotin J, Nishino SF, Spain JC, Hofstetter TB (2013) Using compound-specific isotope analysis to assess biodegradation of nitroaromatic explosives in the subsurface. Environ Sci Technol 47(13):6872–6883. https://doi.org/10.1021/es3051845
Williford CW Jr, Mark Bricka R (1999) Extraction of TNT from aggregate soil fractions. J Hazard Mater 66(1–2):1–13. https://doi.org/10.1016/S0304-3894(98)00214-3
Wu CF, Xu XM, Zhu Q, Deng MC, Feng L, Peng J, et al. (2014)An effective method for the detoxification of cyanide-rich wastewater by Bacillus sp. CN-22. Appl Microbiol Biotechnol 98(8):3801–7. https://doi.org/10.1007/s00253-013-5433-5
Xu M, Liu D, Sun P, Li Y, Wu M, Liu W, et al. (2021) Degradation of 2,4,6-trinitrotoluene (TNT): involvement of protocatechuate 3,4-dioxygenase (P34O) in Buttiauxella sp. S19-1. Toxics 9(10):231. https://doi.org/10.3390/toxics9100231
Yang X, Lai JL, Li J, Zhang Y, Luo XG, Li ZG (2021) Biodegradation and physiological response mechanism of a bacterial strain to 2,4,6-trinitrotoluene contamination. Chemosphere 270:129280. https://doi.org/10.1016/j.chemosphere.2020.129280
Yin H, Wood TK, Smets BF (2005) Reductive transformation of TNT by Escherichia coli: pathway description. Appl Microbiol Biotechnol 67(3):397–404. https://doi.org/10.1007/s00253-004-1736-x
Ziganshin AM, Gerlach R (2014) Pathways of 2,4,6-trinitrotoluene transformation by aerobic yeasts. In: Singh SN (ed) Biological remediation of explosive residues, environmental science and engineering. Springer, Cham, pp 301–311
Ziganshin AM, Ziganshina EE, Byrne J, Gerlach R, Struve E, Biktagirov T, et al. (2015) Fe(III) mineral reduction followed by partial dissolution and reactive oxygen species generation during 2,4,6-trinitrotoluene transformation by the aerobic yeast Yarrowia lipolytica. AMB Express 5:1–12. https://doi.org/10.1186/s13568-014-0094-z
Acknowledgements
This work was carried out under the Program of Kazan Federal University Strategic Academic Leadership and supported by Russian Science Foundation Grant No 22-24-00036.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yakovleva, G., Kurdy, W., Gorbunova, A. et al. Bacillus pumilus proteome changes in response to 2,4,6-trinitrotoluene-induced stress. Biodegradation 33, 593–607 (2022). https://doi.org/10.1007/s10532-022-09997-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10532-022-09997-8