Abstract
Styela clava, a solitary ascidian native to the NW Pacific, has become a conspicuous member of fouling communities in NW European waters. As its natural dispersal appears to be limited, the wide distribution of S. clava along coasts within its introduced range may be attributed to secondary spread assisted by human activities. Here, we used six microsatellite loci to examine the genetic diversity and extent of gene flow among S. clava populations in its European introduced range. Samples were collected from 21 populations within Europe (N = 808), 4 populations within the USA and two populations within the native range (Japan). Large variation in genetic diversity was observed among the European populations but were not explained either by the geographic distance from the first introduction area (i.e. Plymouth, UK) nor by the time elapsed since the introduction. No founder effect was observed in the introduced populations, except possibly in Puget Sound (USA). At least two different introductions occurred in Europe, identified as distinct genetic clusters: northern Danish populations (resembling one Japanese population), and the rest of Europe; a sample from Shoreham (England) possibly represents a third introduction. In North America, the population from the Atlantic was genetically similar to the majority of European populations, suggesting a European origin for populations on this seaboard, while populations from the Pacific coast were genetically similar to the same Japanese population as the Danish populations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Introduction of non-indigenous marine species to new geographical areas is a wide-spread phenomenon (Carlton 1996; Vermeij 1996; Ruiz et al. 1997; Castilla et al. 2004) which is often mediated by transport on ships’ hulls and in ballast water (Carlton and Geller 1993). Marinas and harbours are thus repeatedly entrance gates for new invasions of exotic fauna and flora (Zibrowius 1991; Goulletquer et al. 2002) and ascidians in particular (Carlisle 1954; Holmes 1968; Brunetti 1979; Monniot 1981; Castilla et al. 2002; Lambert and Lambert 2003; Lambert 2004). Ascidians are now recognized as important invaders, dominating fouling communities and interfering with bivalve culture (Osman and Whitlatch 1999; Lambert 2007). Among colonial ascidians, Didemnum vexillum is an aggressive invader with rapidly expanding populations on the east and west coasts of North America that reach high abundance in newly colonized areas and affect aquaculture habitats by overgrowing shellfish and infrastructure (Valentine et al. 2007). Among solitary species, Styela clava is considered to be a serious pest into Prince Edward Island, Canada where it has become a nuisance to mussel culture activities (Bourque et al. 2007 and references therein).
Invasions may constitute rapid evolutionary events in which populations are subjected to founder effects during colonization, followed by a rapid expansion (e.g. Sakai et al. 2001). Introduced populations are thus more likely than native populations to be out of mutation/migration-drift equilibrium (Eckert et al. 1996). In particular, colonization events are predicted to result in reduced genetic diversity, a loss of rare alleles and an increase in population differentiation owing primarily to founder effects and genetic drift (Wright 1943; Nei et al. 1975; Cornuet and Luikart 1996). However, human transport vectors may promote high genetic diversity within introduced populations when independent introductions from different regions of the species’ native range merge (Simon-Bouhet et al. 2006). These opposing scenarios have important implications for the adaptation, and therefore the successful establishment, spread, and proliferation of exotic species.
Styela clava, a solitary ascidian native to the coasts of Japan, Korea, China and Siberia (Herdman 1882; Abbott and Johnson 1972), has become a conspicuous member of fouling communities in various parts of the world. First reported outside its native range in California in the late 1920s, this large sea squirt has subsequently also colonized NW Europe, eastern and western USA, southern Australia, eastern and western Canada New Zealand and one site in the Mediterranean basin (Abbott and Johnson 1972; Lambert and Lambert 1998; Lützen 1999; Davis and Davis 2006, 2008; Locke et al. 2007). Since its initial discovery in Plymouth (UK) in 1953 (Carlisle 1954), S. clava has spread around Britain and also to the Channel Islands, Ireland, Scotland and along the coast of Europe from Denmark to Portugal (Davis and Davis 2004a). This species is mostly confined to sheltered localities free of strong wave action, such as inlets, bays, harbours and marinas. The total time spent as planktonic egg and larva is approximately 24–43 h (see references in Dupont et al. 2009). Adults might also disperse occasionally attached to drifting wood or weed (e.g. Sargassum muticum, Lützen 1999; Davis and Davis 2004a). However, as natural dispersal appears to have a limited range, the widespread occurrence of Styela clava within its introduced range may be attributed to secondary transportation by human activity. It has been suggested that dispersal could occur as settled juveniles attached to commercial oysters that have been transported and re-laid (Christiansen and Thomsen 1981; Minchin and Duggan 1988), as mature adults attached to the hulls of ships (Millar 1960; Buizer 1980) or within sea-chests and, finally, as eggs and larvae carried in ballast water (Davis and Davis 2004a).
A recent population genetic study using microsatellites at small and intermediate geographical scales highlighted the importance of human-mediated dispersal in range expansion and occupancy by S. clava in south-west England, while the limited dispersal ability of this ascidian resulted in fine-scale population structure in several marinas (Dupont et al. 2009). In addition, Dupont et al. (2009) showed that enclosed marinas might function as reservoirs of propagules for subsequent spread whereas others might be sinks for migrants.
Here, we used microsatellite nuclear markers to examine the overall connectivity, i.e. genetic relationships, among S. clava populations located in its Northern European introduced range. We carried out extensive sampling of S. clava along European coasts, with emphasis on English and French coasts, in order to test the hypothesis that Plymouth might have been the source for secondary introductions in other localities in Europe. Specifically we ask: (1) is there any evidence for founder effects or bottlenecks in the introduced populations? (2) is there evidence for multiple invasion sources or multiple introductions through human-mediated processes? and (3) can the pattern of genetic structure among European populations be used to infer the vectors of introduction and/or pathways of spread?
Methods
Sampling and DNA isolation
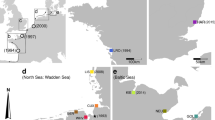
Samples of Styela clava were collected from 21 populations within Europe (Fig. 1, details in Table 1) between January 2004 and September 2005. In order to compare genetic diversity indices with populations introduced outside Europe, 4 American populations were added to the dataset. Most of the introduced populations were collected in sites where the date of first observation of S. clava was recorded in the literature. Extensive sampling was undertaken along the Channel coasts around Plymouth, the first site where S. clava was recorded in Europe. In addition, two populations within the native range (Japan) were obtained for preliminary comparison of genetic diversity between introduced and native populations. A portion of the branchial sac tissue of each individual was dissected and preserved in 96% ethanol for DNA analysis. Total genomic DNA was extracted using a CTAB method (described in Sambrook and Russell 2001, p. 6.61). Extractions were visually confirmed using 1% agarose gels stained with ethidium bromide.
Location of the study populations. Two clusters identified by BAPS (see text and Fig. 3) were composed of one population (SHO and TSU), one cluster was composed of the populations indicated by italic font and the fourth cluster was composed of all other populations
Microsatellite genotyping
For the samples from Plymouth, Mission Bay and Otsuchi, genotypes were obtained from Dupont et al. (2006) and for Falmouth population, genotypes were obtained from Dupont et al. (2009) with additional genotyping completed for the locus Sc3e1. For the 23 other populations, individuals were genotyped at six microsatellite loci (Sc1b3, Sc1c8, Sc1h1, Sc2b12, Sc2h9 and Sc3e1) following protocols detailed in Dupont et al. (2006). Only loci presenting no or negligible heterozygote deficiency in Dupont et al. (2006) were chosen in order to avoid null-allele biases. In order to limit genotyping errors, all samples presenting new or rare alleles were amplified twice. PCR products were screened on a 6.5% polyacrylamide gel using a Li-Cor NEN Global IR2 DNA sequencer system.
Genetic diversity analyses
For each population, the genetic diversity was analyzed by computing allele frequencies, number of alleles (N all) and expected heterozygozity (H e) using GENETIX v 4.04 (Belkhir et al. 2004). To take into account variation in sample size, allelic richness (A r; El Mousadik and Petit 1996) was estimated using FSTAT v.2.9.3 (Goudet 1999). The null hypothesis of independence between loci was tested from statistical genotypic disequilibrium analysis using exact tests implemented in the GENEPOP v 3.4 program (Raymond and Rousset 1995).
Analysis of genetic structure within populations
Distribution of the genetic diversity of introduced species can depart from equilibrium models for several reasons, including (1) recent demographic expansion of a local and isolated founding population or (2) recent mixing within a population of a set of genetically differentiated immigrants. We tested for deviation from mutation-drift equilibrium in the study populations using the approach proposed by Cornuet and Luikart (1996) and implemented in their software BOTTLENECK. Using a Wilcoxon test, the observed heterozygosity is compared with the heterozygosity expected under equilibrium considering a two-phase mutation model (TPM) recommended for microsatellite data (Di Rienzo et al. 1994). Recently founded populations were expected to display a transient excess of heterozygosity whereas expanding populations (e.g. recovering from a bottleneck) or populations resulting from immigration from differentiated sources should exhibit the opposite (Cornuet and Luikart 1996).
We used population heterozygote deficiencies to investigate the occurrence of cryptic population structure within populations (i.e. Wahlund effect, see Hartl and Clark 1997). Such a pattern may appear in invasive species because of their regular transportation from one site to another through human-mediated activities (e.g. fouling), generating mixtures between genetically differentiated pools of individuals. We quantified the Walhund effect within each population by calculating the Weir and Cockerham’s (1984) \( \hat{f} \), a monolocus estimator of the fixation index F IS, with GENEPOP v 3.4. Conformity to Hardy–Weinberg equilibrium (HWE) was assessed with exact tests implemented in the GENEPOP v 3.4 program (Raymond and Rousset 1995) with specified Markov chain parameters of 10,000 dememorization steps, followed by 1,000 batches of 10,000 iterations per batch.
Analysis of genetic structure among populations
In order to investigate the importance of genetic exchanges among populations, exact tests of allelic differentiation were carried out between populations using GENEPOP v.3.4 software. To adjust for multiple comparisons, sequential Bonferroni correction was used. We also used a traditional population differentiation approach based on F ST analysis. Weir and Cockerham’s (1984) estimator of the fixation index F ST (\( \hat{\theta } \)) was calculated with GENEPOP v.3.4. Multidimensional scaling (MDS) plots of the localities were produced from the matrix of pairwise F ST estimates using STATISTICA v.6 software (StatSoft, Inc. www.statsoft.com.).
We used the program BAPS v.4.14 (Corander et al. 2003, 2008) to detect clusters of genetically similar populations in the Northern Hemisphere introduced range of S. clava and to estimate individual coefficients of ancestry with regard to the detected clusters. BAPS uses a stochastic optimization to infer the posterior mode of genetic structure which greatly improves the speed of the analysis compared to traditional MCMC-based algorithms (Corander and Marttinen 2006). When testing for population clusters, we ran 5 replicates for k = 5, k = 10, k = 15, k = 20, k = 25 and k = 30, where k is the maximum number of genetically divergent groups (populations). When estimating individual ancestry coefficients via admixture analysis we used recommended values of (1) the number of iterations used to estimate the admixture coefficients for the individuals [100], (2) the number of reference individuals from each population [200] and (3) the number of iterations used to estimate the admixture coefficients for the reference individuals [20].
Results
Genetic diversity
Values of genetic diversity indices were similar when comparing European and American populations (Table 1). For example, H e ranged from 0.449 to 0.589 (mean = 0.536) in American populations and from 0.477 to 0.622 (mean = 0.574) in European populations. Values computed in the two Japanese (native) populations were in the same range with H e estimates of 0.556 and 0.575 (OTS and TSU respectively; mean = 0.566). Among populations in the introduced range, allelic richness and gene diversity were highly variable (Table 1). PUG presented the lowest level of genetic diversity (A r = 2.93; H e = 0.449). In Europe, the two Danish populations (JEG and DOV) presented the lowest levels of genetic diversity (A r = 4.00; H e = 0.480 and A r = 3.91; H e = 0.477 respectively) while two French populations (BRE and LEZ) had high levels of allelic richness (≥5). These large variations of genetic diversity among European populations were not explained by the time since the approximate date of introduction (i.e. first report in the literature). For instance, H e was not correlated with this time since introduction (r 2 = −0.129, P = 0.673). Under the hypothesis that English populations originated in Plymouth (i.e. the first recorded European population, 1953; Carlisle 1954) and spread progressively from there along the coasts, undergoing successive bottlenecks, we expect a decrease of genetic diversity in English populations with increasing distance from Plymouth, but no significant correlation was observed for H e (r 2 = −0.287, P = 0.640) and Ar (r 2 = −0.506, P = 0.385).
Genetic structure within populations
When testing for deviation from mutation-drift equilibrium in BOTTLENECK, a significant genetic diversity deficit (Wilcoxon test P < 0.05) was detected in 62% of European populations and in both Japanese populations. Under a two-phase mutation model (TPM) and in the absence of sub-structure within a population, which is the case for COR, FLE, GOS, RAM, LEZ, CON and RIA and both native populations (See values of \( \hat{f} \) in Table 1), such a result supports the hypothesis of population expansion. Alternatively, as genetic structure within a population mimics the effect of demographic expansion (Cornuet and Luikart 1996), the deviation from mutation-drift equilibrium observed in FEN, CAI, DVR, GUE, PER and BRE might be due to sub-structure: significant departures from Hardy–Weinberg equilibrium were indeed observed in these sites (Table 1) (as well as in ROC, SAN and WOO) suggesting sub-structure (Walhund effect) within these localities.
Genetic structure among populations
FST analysis showed a significant genetic structure at the level of the whole study (27 populations, FST = 0.031, P < 0.001) and when considering only the introduced populations (American and European populations, FST = 0.030, P < 0.001) suggesting limited gene exchange among populations and a non-unique source (in time or space) for the introduced populations.
When considering each population pair, 209 values of F ST out of 351 (59%) were associated with a significant exact test after Bonferroni correction (Table 2). When considering only pairs of European populations, 42% of F ST values were significant. These pairwise F ST estimates were pictured with MDS plots which clearly illustrated the genetic isolation of the Danish, Japanese and western American populations from the remaining European populations (Fig. 2a). In agreement with this picture, it is noteworthy that the percentage of significant pairwise F ST values fell to 30% of pairwise comparisons if the Danish populations were excluded from the analysis. In particular DOV was found to be different from every other European population except JEG, the other Danish population studied. Figure 2b illustrates the central position of GUE, FAL, PLY and BRE and the genetic differentiation of STM and SHO from the others among the European populations. This explains the significant F ST value computed among the 21 European sampled sites (F ST = 0.019, P < 0.001; without Danish populations F ST = 0.015, P < 0.001), The American introduced populations were also genetically different (F ST = 0.047, P < 0.001) even when only Pacific populations were considered (i.e. without WOO population F ST = 0.033, P < 0.001). Moreover, the group of populations from the Pacific Ocean (American and Japanese) were genetically different from the group of populations from the Atlantic (American and European)(F ST = 0.029, P < 0.001). Our study did not aim focus on native populations. However, it is interesting to note that the two sampled Japanese populations also showed significant genetic differences between them (F ST = 0.026, P < 0.001).
Multidimensional scaling plots constructed using pairwise F ST estimates among a all populations (stress = 0.09) and b all European populations except Danish ones (stress = 0.15). The same colour code as in Fig. 1 was used
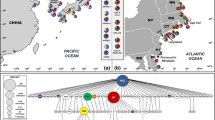
Main results from F ST computations were confirmed with Bayesian analyses. Analyses using BAPS identified 4 genetic clusters overall (P = 1; Fig. 3a, b): two clusters were composed of only one population (SHO and TSU), one cluster was composed of all the non-Danish European populations except SHO plus the American population from the Atlantic (WOO), and the fourth cluster was composed of the Danish populations, the American populations from the Pacific coast and one Japanese population (OTS).
Discussion
None of the studied introduced populations of Styela clava in Europe and America displayed evidence of founder effects. Genetic diversity within introduced populations was generally high and similar whatever their geographic origin. In addition, the two samples from Japan, within the native range of S. clava, showed genetic diversity of the same level. Altogether, there was little evidence of population bottlenecks with attendant loss of genetic diversity during introduction and secondary spread. Only the sample from Puget Sound (PUG) showed lower, but still appreciable, diversity. No general link was demonstrated between the date of first occurrence of S. clava at a locality and the genetic diversity there, although Puget Sound, with relatively low diversity, had also been colonised relatively recently.
The relative genetic distinctness of the two Japanese samples, taken on opposite sides of the main island Honshu, suggests that populations of S. clava are genetically differentiated within its native range, although much more extensive sampling is clearly necessary to confirm this. If so, and given sufficiently strong differentiation within the north-west Pacific, independent introductions derived from different parts of the native range would be genetically distinguishable even in the absence of bottlenecking and related founder effects. Pairwise F ST, MDS and BAPS analyses all pointed out the genetic isolation of two Danish populations (JEG and DOV). Our data thus suggest that (at least) two separate introductions have occurred in Europe, represented by the samples from Denmark and the rest of Europe with the possible exception of Shoreham (SHO). According to the BAPS analysis, the majority of samples from Europe belongs to a single genetic cluster, but differs from the Japanese populations on either side of Honshu. It has been suggested that the introduction of S. clava in the early 1950s was linked to the return of warships from the Korean War (1950–1953) to naval bases in southern England (Plymouth, the site of first occurrence, and perhaps also Portsmouth) (Coughlan 1969; Minchin and Duggan 1988), presumably involving hull fouling. The genetic distinctness of the bulk of European populations from the Japanese samples is in keeping with an origin elsewhere in the species’ native range, and thus with the hypothesis of a Korean derivation, but more extensive sampling of the native range would again be necessary to test this theory properly.
The Limfjord, a shallow sound in northern Denmark where JEG and DOV populations were collected, has high salinity and high summer temperatures compared to other areas of the region. As a result it holds several southern faunal elements, and has been a recipient of several species of immigrants that originate from widely different regions (Knudsen 1989). Lützen (1999) noted that the first record of S. clava in the Limfjord was in an oyster bed to which spat from the English Channel had been transferred as part of commercial operations. He also suggested that many French and some Dutch populations of S. clava could be attributed to importation of the Pacific Oyster (Crassostrea gigas) from Japan. Our molecular data suggest the reverse relationships: (1) they link the Northern Danish populations with Japan (and/or the west coast of North America), rather than England, thus potentially implicating the importation of oysters from a Pacific site, and (2) indicate a common origin for French and English populations, with the chronology of first occurrences suggesting spread from England to France. The first definite Danish record was in 1980, concerning individuals believed to represent the 1978 year-class (Christiansen and Thomsen 1981), but Lützen and Sørensen (1993) and Lützen (1999) note that the species ‘had presumably been present from the mid-1960 s’. The basis for this statement is that a fisherman interviewed by J. Lützen recalled seeing S. clava for the first time in the mid-1960 s while operating close to oyster beds in the northern part of Nissum Bredning (Lützen and Sørensen 1993 and J. Lützen in litt. to JDDB in 2006); in contrast to the 1978 date, this timing would place the arrival of S. clava before the recorded commercial importation of C. gigas to Europe had begun. However, the molecular data and a first occurrence in the late 1970 s are in keeping with the suggestion of Christiansen and Thomsen (1981) that the repeated importation of commercial oysters from California to the western Limfjord during the 1970 s may explain the origin of the populations of S. clava in northern Denmark.
The existence of a genetically distinct population of S. clava at Shoreham (SHO) in SE England was not anticipated, and no particular explanation can be offered. Lady Bee Marina, with berths for 120 vessels, lies within Shoreham Harbour, a nationally significant commercial and fishing port. Of possible relevance are the facts that (1) the sampled site is in the inner part of the harbour in the lock-gated Shoreham Harbour Canal, and thus relatively limited exchange with other populations would be expected, and (2) the harbour was heated by cooling water discharge for most of the twentieth century until the coal-powered Brighton ‘B’ Power Station closed in 1987. This may have explain why some warm-water species have been recorded in Shoreham Harbour (e.g. the barnacle Amphibalanus amphitrite (Bishop 1947) and the polychaete Hydroides elegans (Monro 1938, as H. incrustans). The new gas-powered Shoreham Power Station, was commissioned in 2000, and uses the same cooling water outfall structures in the Shoreham Harbour Canal as Brighton ‘B’; Lady Bee Marina is nearby, on the opposite bank of the canal. The genetic differentiation of the SHO population was supported by four of the six supposedly neutral markers (i.e. microsatellites developed from a conventional enriched library, Dupont et al. 2006) used in this study. Indeed, between 60 and 100% of significant tests were obtained for the loci Sc1b3, Sc1h1, Sc2h9 and Sc3e1 when monolocus exact tests of allelic differentiation were carried out between SHO and the other European population (data not shown). Thus, a hypothesis of selection of some alleles by temperature may be ruled out. The relatively high genetic diversity of the Shoreham population of S. clava, comparable to that of the surrounding sites, and the presence of two unique alleles, albeit as single copies in our sample, suggest that this sample may represent a third independent European introduction, reflected in its recognition as a separate BAPS cluster.
The clustering of the sample from Woods Hole (WOO), Massachusetts, with the most widespread European grouping of populations suggests a European origin for the initial colonisation of the eastern coast of North America. The timing of appearance of S. clava on the E. coast of the USA is in keeping with this suggestion: the species was first noted off Massachusetts in 1970 (Berman et al. 1992), by which time it had become widespread and locally abundant on the S coast of England and had spread to Wales and mainland Europe. Concerning the western coast of North America, S. clava is believed to have been introduced to Californian waters in the 1920s via ships previously docked in Asian ports (Abbott and Johnson 1972). Our findings are in keeping with this hypothesis: the populations from the west coast of the USA are genetically similar to the Otsuchi population (OTS) from Japan.
Conclusion
The six microsatellite markers used in this study all showed a relatively high degree of polymorphism in the introduced European and North American populations, indicating a lack of strong founder effects. This result adds to the long list of marine invaders not showing the expected founder effects associated with introduction processes (see Roman and Darling 2007 for a review). Similar results have been recorded in other introduced ascidians, namely Microcosmus squamiger and, to a lesser extent, Botryllus schlosseri (Table 3). These findings corroborate the study by Silva and Smith (2008) using AFLPs that showed that invasive populations of ten different ascidian species (including S. clava) are highly polymorphic. In contrast, extremely low genetic diversity due to a combination of founder effects and selfing was observed in introduced populations of the tunicate Corella eumyota (Dupont et al. 2007, Table 3).
A more complete survey on the whole distribution range would be necessary to fully understand the pattern of invasion of Styela clava in the Northern Hemisphere as well as the pattern of spread of the species at a worldwide level. However, the study dataset helped to substantiate two independent routes of introduction of S. clava into Europe by different vectors, in accordance with previous suggestions based on circumstantial evidence. Because of the multiplicity of introduction pathways and vectors (see for example Voisin et al. 2005), cryptic introductions appear to be common in marine biological invasions.
References
Abbott PD, Johnson JV (1972) The ascidians Styela barnharti, S. plicata, S. clava and S. montereyensis in Californian waters. Bull South Calif Acad Sci 71:95–105
Belkhir K, Borsa P, Goudet J, Chikhi L, Bonhomme F (2004) GENETIX 4.05, logiciel sous Windows pour la génétique des populations. Laboratoire Génome, Population, Interactions, CNRS UMR 5000, Université Montpellier II, Montpellier
Ben-Schlomo R, Paz G, Rinkevich B (2006) Postglacial-period and recent invasions shape the population genetics of botryllid ascidians along European Atlantic coasts. Ecosystems 9:1118–1127
Ben-Shlomo R, Douek J, Rinkevich B (2001) Heterozygote deficiency and chimerism in remote populations of a colonial ascidian from New Zealand. Mar Ecol Prog Ser 209:109–117
Berman J, Harris L, Lambert W, Buttrick M, Dufresne M (1992) Recent invasions of the Gulf of Maine—3 contrasting ecological histories. Conserv Biol 6:435–441
Bishop MWH (1947) Establishment of an immigrant barnacle in British coastal waters. Nature 159:501–502
Bourque D, Davidson J, MacNair NG, Arsenault G, LeBlanc AR, Landry T, Miron G (2007) Reproduction and early life history of an invasive ascidian Styela clava Herdman in Prince Edward Island, Canada. J Exp Mar Biol Ecol 342:78–84
Breton G, Dupont W (1973) Styela clava Herdmann, ascidie nouvelle pour les côtes de la baie de Seine abonde dans le port du Havre (76). Bull Trimest Soc Géol Normandie 65:32
Brunetti R (1979) Polyandrocarpa zorritensis (Van Name, 1931) a colonial ascidian new to Mediterranean record. Vie Milieu 29:647–652
Buizer DAG (1980) Explosive development of Styela clava Herdman 1882, in the Netherlands after its introduction (Tunicata ascidiacea). Bull Zool Mus Univ Amsterdam 7:181–187
Carlisle DB (1954) Styela mammiculata N.SP., a new species of ascidian from the Plymouth area. J Mar Biol Ass UK 33:329–334
Carlton JT (1996) Pattern, process, and prediction in marine invasion ecology. Biol Conserv 78:97–106
Carlton JT, Geller JB (1993) Ecological roulette: the global transport of nonindigenous marine organisms. Science 261:78–82
Castilla JC, Collins AG, Meyer CP, Guinez R, Lindberg DR (2002) Recent introduction of the dominant tunicate, Pyura praeputialis (Urochordata, Pyuridae) to Antofagasta, Chile. Mol Ecol 11:1579–1584
Castilla JC, Guinez R, Caro AU, Ortiz V (2004) Invasion of a rocky intertidal shore by the tunicate Pyura praeputialis in the Bay of Antofagasta, Chile. Proc Natl Acad Sci USA 101:8517–8524
Christiansen J, Thomsen JC (1981) Styela clava Herdman 1882, a species new to the Danish fauna (Tunicata, Ascidiacea). Steenstrupia 7:15–24
Cohen AN, Carlton JT (1995) Nonindigenous aquatic in a United States estuary: a case of the biological invasions of the San Francisco Bay and Delta. A report for the United States Fish and Wildlife service, Washington DC and the National Sea Grant College Program, Connecticut State Grant (NOAA Grant Number NA36RG0467)
Cohen AN, Mills C, Berry H, Wonham M, Bingham B, Bookheim B, Carlton JT, Chapman J, Cordell J, Harris L, Klinger T, Kohn A, Lambert CC, Lambert G, Li K, Secord D, Toft J (1998) Report of the Puget Sound Expedition September 8–16, 1998. A rapid assessment survey of non-indigenous species in the shallow waters of Puget Sound, Washington State Department of Natural Resources, Olympia, WA. United States Fish and Wildlife Service, Lacey
Corander J, Marttinen P (2006) Bayesian identification of admixture events using multilocus molecular markers. Mol Ecol 15:2833–2843
Corander J, Waldmann P, Sillanpaa MJ (2003) Bayesian analysis of genetic differentiation between populations. Genetics 163:367–374
Corander J, Siren J, Arjas E (2008) Bayesian spatial modeling of genetic population structure. Comput Stat 23:111–129
Cornuet J-M, Luikart G (1996) Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 144:2001–2014
Coughlan J (1969) The leathery sea-squirt—a new ascidian from Milford Haven. Nat Wales 11:192–193
Coughlan J (1985) Occurrence of the immigrant ascidian Styela clava Herdman in Heysham harbour, Lancashire. Porc Mar Nat Hist Soc Newsl 3:85–87
Davis MH, Davis ME (2004a) The distribution limits of Styela clava (Tunicata, Ascidiacea) in European waters. Porc Mar Nat Hist Soc Newsl 15:35–43
Davis MH, Davis ME (2004b) New records of Styela clava Herdman, 1882 (Tunicata, Ascidiacea) in Europe. Porc Mar Nat Hist Soc Newsl 14:24–28
Davis MH, Davis ME (2006) Styela clava (Tunicata, Ascidiacea)—a new addition to the fauna of New Zealand. Porc Mar Nat Hist Soc Newsl 20:19–22
Davis MH, Davis ME (2008) First record of Styela clava (Tunicata, Ascidiacea) in the Mediterranean region. Aquat Invasions 3:125–132
Di Rienzo A, Peterson AC, Garcza JC, Valdes AM, Slatkin M, Freimer NB (1994) Mutational processes of simple-sequence repeat loci in human populations. Proc Natl Acad Sci USA 91:3166–3170
Dupont L, Viard F, Bishop JDD (2006) Isolation and characterization of twelve polymorphic microsatellite markers for the invasive ascidian Styela clava (Tunicata). Mol Ecol Notes 6:101–103
Dupont L, Viard F, David P, Bishop JDD (2007) Combined effects of bottlenecks and selfing in populations of Corella eumyota, a recently introduced sea squirt in the English Channel. Diversity Distrib 13:808–817
Dupont L, Viard F, Dowell MJ, Wood C, Bishop JDD (2009) Fine- and regional-scale genetic structure of the exotic ascidian Styela clava (Tunicata) in south-west England, 50 years after its introduction. Mol Ecol 18:442–453
Eckert CG, Manicacci D, Barrett SCH (1996) Genetic drift and founder effect in native versus introduced populations of an invading plant, Lythrum salicaria (Lythraceae). Evolution 50:1512–1519
El Mousadik A, Petit RJ (1996) High level of genetic differentiation for allelic richness among populations of the argan tree Argania spinosa (L.) Skeels endemic to Morocco. Theorl Appl Genet 92:832–839
Goudet J (1999) FSTAT vers. 2.8, updated from Goudet (1995). Fstat: a computer program to calculate F-statistics. J Hered 86:485–486
Goulletquer P, Bachelet G, Sauriau PG, Noel P (2002) Open Atlantic coast of Europe—a century of introduced species into French waters. In: Leppäkoski E, Gollash S, Olenin S (eds) Invasive aquatic species of Europe. Kluwer, Dordrecht
Guiry GM, Guiry MD (1973) Spread of an introduced ascidian to Ireland. Mar Poll Bull 4:127
Hartl DL, Clark AG (1997) Principles of population genetics. Sinauer Associates, Sunderland
Herdman WA (1882) Report on the Tunicata collected during the voyage of H.M.S. Challenger during the years 1873–76. Part I.-Ascidiæ simplices. Zool Chall Exped 6:1–296
Holmes NJ (1968) Aspects of the biology of Styela clava Herdman. PhD thesis, University of Southampton
Huwae PHM, Lavaleye MSS (1975) Styela clava Herdman, 1882 (Tunicata, Ascidiacea) nieuw voor Nederland. Zool Bijdr 17:79–81
Knudsen J (1989) Immigration of marine invertebrates to the Limfjord (Denmark) and the North Sea—Baltic transition area. In: Steinberger Y, Luria M, Spanier E (eds) Environmental quality and ecosystem stability. ISEEQ, Jerusalem, pp 135–145
Lambert G (2004) The south temperate and Antarctic ascidian Corella eumyota reported in two harbours in north-western France. J Mar Biol Assoc UK 84:239–241
Lambert G (2007) Invasive sea squirts: a growing global problem. J Exp Mar Biol Ecol 342:3–4
Lambert CC, Lambert G (1998) Non-indigenous ascidians in southern California harbors and marinas. Mar Biol 130:675–688
Lambert CC, Lambert G (2003) Persistence and differential distribution of nonindigenous ascidians in harbors of the Southern California Bight. Mar Ecol Prog Ser 259:145–161
Locke A, Hanson JM, Ellis KM, Thompson J, Rochette R (2007) Invasion of the southern Gulf of St. Lawrence by the clubbed tunicate (Styela clava Herdman): potential mechanisms for invasions of Prince Edward Island estuaries. J Exp Mar Biol Ecol 342:69–77
Lopez-Legentil S, Turon X, Planes S (2006) Genetic structure of the star sea squirt, Botryllus schlosseri, introduced in southern European harbours. Mol Ecol 15:3957–3967
Lützen J (1999) Styela clava Herdman (Urochordata, Ascidiacea), a successful immigrant to North West Europe: ecology, propagation and chronology of spread. Helgol Meeresunters 52:383–391
Lützen J, Sørensen V (1993) Ecology, reproduction and further spread of the immigrant East-Asiatic ascidian Styela clava Herdman in Danish waters. Flora Fauna 2:75–79
Millar RH (1960) The identity of the ascidians Styela mammiculata Carlisle and S. clava, Herdman. J Mar Biol Assoc UK 39:509–511
Minchin D, Duggan CB (1988) The distribution of the exotic ascidian, Styela clava, Herdman, in Cork harbour. Irish Nat J 22:388–393
Monniot C (1981) Apparition de l’ascidie Microcosmus exasperatus dans les ports méditerrannéens. Tethys 10:59–62
Monro CCA (1938) On a new species of serpulid polychaete from the Shoreham Harbour Canal, Sussec. Ann Mag Nat Hist 1:73–78
Nei M, Maruyama T, Chakraborty R (1975) The bottleneck effect and genetic variability in populations. Evolution 29:1–10
Osman RW, Whitlatch RB (1999) Ecological interactions of invading ascidians within epifaunal communities of southern New England. In: Pederson J (ed) Marine bioinvasions: proceedings of conference January 24–27, 1999. Massachusetts Institute of Technology, Cambridge, Massachusetts, pp 164–174
Paz G, Douek J, Mo CQ, Goren M, Rinkevich B (2003) Genetic structure of Botryllus schlosseri (Tunicata) populations from the Mediterranean coast of Israel. Mar Ecol Prog Ser 250:153–162
Raymond M, Rousset F (1995) GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Hered 86:248–249
Rinkevich B, Paz G, Douek J, Ben-Schlomo R (2001) Allorecognition and microsatellite allele polymorphism of Botryllus schlosseri from the Adriatic Sea. In: Sawada H, Yokosawa H, Lambert CC (eds) The biology of Ascidians. Springer, Tokyo, pp 426–435
Rius M, Pascual M, Turon X (2008) Phylogeography of the widespread marine invader Microcosmus squamiger (Ascidiacea) reveals high genetic diversity of introduced populations and non-independent colonizations. Divers Distrib 14:818–828
Roman J, Darling JA (2007) Paradox lost: genetic diversity and the success of aquatic invasions. Trends Ecol Evol 22:454–464
Ruiz GM, Carlton JT, Grosholz ED, Hines AH (1997) Global invasions of marine and estuarine habitats by non-indigenous species: mechanisms, extent, and consequences. Am Zool 37:621–632
Sakai AK, Allendorf FW, Holt JS, Lodge DM, Molofsky J, With KA, Baughman S, Cabin RJ, Cohen JE, Ellstrand NC, McCauley DE, O’Neil P, Parker IM, Thompson JN, Weller SG (2001) The population biology of invasive species. Ann Rev Ecol Syst 32:305–332
Sambrook J, Russell DW (2001) Molecular clonning. A laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, New York
Silva N, Smith WC (2008) Inverse correlation of population similarity and introduction date for invasive ascidians. PLoS One 3:e2552
Simon-Bouhet B, Garcia-Meunier P, Viard F (2006) Multiple introductions promote range expansion of the mollusc Cyclope neritea (Nassariidae) in France: evidence from mitochondrial sequence data. Mol Ecol 15:1699–1711
Stoner DS, Ben-Schlomo R, Rinkevich B, Weissman IL (2002) Genetic variability of Botryllus schlosseri invasions to the east and west coasts of the USA. Mar Ecol Prog Ser 243:93–100
Valentine PC, Collie JS, Reid RN, Asch RG, Guida VG, Blackwood DS (2007) The occurrence of the colonial ascidian Didemnum sp on Georges Bank gravel habitat—ecological observations and potential effects on groundfish and scallop fisheries. J Exp Mar Biol Ecol 342:179–181
Vázquez E, Urgorri V (1992) Ascidians of the fouling in the Ría de Ferrol. Nova Acta Científ Compostel 3:161–167
Vermeij GJ (1996) An agenda for invasion biology. Biol Conserv 78:3–9
Voisin M, Engel CR, Viard F (2005) Differential shuffling of native genetic diversity across introduced regions in a brown alga: Aquaculture vs. maritime traffic effects. Proc Natl Acad Sci USA 102:5432–5437
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
Wright S (1943) Isolation by distance. Genetics 28:114–138
Zibrowius H (1991) Ongoing modification of the Mediterranean marine fauna and flora by the establishment of exotic species. Mésogée 51:83–107
Acknowledgments
This research was supported by the European Network of Excellence “Marine Genomics Europe” (contract no. 505403) and the NERC (UK) Oceans 2025 programme. FV and JB are the beneficiaries of a financial contribution from the AXA Research Fund (AXA Marine Aliens and Climate Change Project). We thank G. & C. Lambert, J. Lützen, D. Minchin and X. Turon for collecting and sending specimens. The authors are also grateful to two anonymous referees for points that improved the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Dupont, L., Viard, F., Davis, M.H. et al. Pathways of spread of the introduced ascidian Styela clava (Tunicata) in Northern Europe, as revealed by microsatellite markers. Biol Invasions 12, 2707–2721 (2010). https://doi.org/10.1007/s10530-009-9676-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10530-009-9676-0