Abstract
Objective
To strengthen NADH regeneration in the biosynthesis of l-2-aminobutyric acid (l-ABA).
Results
l-Threonine deaminase (l-TD) from Escherichia coli K12 was modified by directed evolution and rational design to improve its endurance to heat treatment. The half-life of mutant G323D/F510L/T344A at 42 °C increased from 10 to 210 min, a 20-fold increase compared to the wild-type l-TD, and the temperature at which the activity of the enzyme decreased by 50% in 15 min increased from 39 to 53 °C. The mutant together with thermostable l-leucine dehydrogenase from Bacillus sphaericus DSM730 and formate dehydrogenase from Candida boidinii constituted a one-pot system for l-ABA biosynthesis. Employing preheat treatment in the one-pot system, the biosynthesis of l-ABA and total turnover number of NAD+/NADH were 0.993 M and 16,469, in contrast to 0.635 M and 10,531 with wild-type l-TD, respectively.
Conclusions
By using the engineered l-TD during endured preheat treatment, the one-pot system has achieved a higher productivity of l-ABA and total turnover number of coenzyme.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
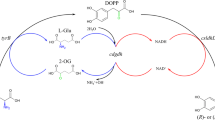
l-2-Aminobutyric acid (l-ABA) is an essential precursor in pharmaceutical manufacturing, such as in the production of the antiepileptic drug levetiracetam and the antituberculotic drug ethambutol (Shin and Kim 2009). Both chemical and enzymatic approaches for preparing l-ABA have been developed, yet enzymatic methods are currently preferred in consideration of the chirality and atomic economy issues (An et al. 2017). l-ABA can be biosynthesised by transamination using transaminase (Park et al. 2013) or by reductive amination using l-amino dehydrogenase (Galkin et al. 1997) from α-ketobutyric acid, which was deaminised from l-threonine by threonine deaminase (Fig. 1). Compared with the transamination process, the reductive amination process has a favourable reaction equilibrium and less or even no byproduct contamination and led to a yield of 97.3%, which was up-and-coming for l-ABA manufacture (Tao et al. 2014). However, in this reductive amination process, the complete bioconversion of l-threonine to l-ABA was obtained only if extra NAD+ was added to the mixture during the reaction in a timely fashion. As a result, the production of l-ABA still involves huge NAD+ costs despite depleted NADH being regenerated by formate dehydrogenase (FDH).
It was assumed that the main reason for the need of NAD+ re-addition was the irreversible consumption of NAD(H) by the NAD(H)-degrading activity of indigenous enzymes during the transformation (Heuser et al. 2007). One of the possible solutions is to use the recombinant mesophiles having a heterologous thermophilic enzyme (Krutsakorn et al. 2013). The preheat treatment of recombinant mesophiles could result in the denaturation of indigenous enzymes and minimisation of unwanted side reactions contained coenzyme.
In this study, a revised one-pot system consisting of l-threonine deaminase (l-TD), l-leucine dehydrogenase (l-LeuDH), and FDH was constructed to produce l-ABA from l-threonine. l-LeuDH from Bacillus sphaericus DSM730 and FDH from Candida boidinii were selected because of their favourable thermostability and specific activity. And l-TD from Escherichia coli K12 was chosen to catalyse the reaction of l-threonine to α-ketobutyric acid, owing to its high effective conversion (Eisenstein 1991). Nevertheless, l-TD would inactivate in the preheat treatment process; hence, we first tried to enhance its thermostability by directed evolution and rational design to match the one-pot system.
Materials and methods
Materials
DNA polymerase, restriction endonuclease and T4 DNA ligase were purchased from Transgen Biotech. The bacterial strains and plasmids used in this study are listed in Supplementary Table 1. All chemicals were of biochemical grade or the highest purity and used without further purification.
Gene cloning and construction of recombinants
Genes coding l-TD (GenBank: CQR83192.1) were amplified by PCR using E. coli K12 genomic DNA as a template. Genes coding l-LeuDH (GenBank: AB103386.1) were amplified from the chromosome of B. sphaericus DSM730. Genes coding FDH (GenBank: AJ011046.2) were amplified from the chromosome of C. boidinii. The PCR fragments were digested and then individually inserted into the corresponding sits of pET-28a(+) to generate pET28a-ilvA, pET28a-leudh and pET28a-fdh. The recombinant plasmids were transformed into E. coli BL21(DE3). Primer sequences are listed in Supplementary Table 2.
Directed evolution
Error prone PCR (ep PCR) was performed using the system according to Qiao et al. (2015), with some modifications. Primer sequences are listed in Supplementary Table 2. The mutant l-TD genes were digested by the restriction endonucleases BamH I and Hind III then inserted into the pET-28a(+) vector, and the plasmids were transformed into E. coli BL21(DE3) competent cells.
The cultivation of recombinant colonies was similar as our previous work (Qiao et al. 2015). Cell pellets were kept at 4 °C and 50 °C for 90 min, respectively. After the incubation period, 300 μL of 0.1 M l-threonine was deaminated by 100 μL of bacterial suspension at 30 °C for 30 min. The amount of α-ketobutyric acid was measured by mixing 50 μL of reaction solution, 200 μL of 2 mM 2,4-dinitrophenylhydrazine and 500 μL of 1 M NaOH (Hatfield and Umbarger 1970). The absorbance of the solution was gauged spectrophotometrically at 380 nm. By measuring the activities before and after heat treatment, stabilised mutants were isolated and sequenced.
Rational design
BLAST was performed on the NCBI database to search the homologous sequences of l-TD. Proteins characterised as thermophilic were chosen for sequence alignment. The Clustal Omega program (http://www.ebi.ac.uk) was used to perform multiple sequence alignments (MSAs). Simulated mutagenesis was performed on the crystallised structure of l-TD from E. coli (PDB ID: 1TDJ) (Gallagher et al. 1998) in Discovery Studio (DS) 3.0. The structures of modified l-TD were built on the homology-modelling program of DS 3.0 with 1TDJ as a template.
Site-directed and site-saturated mutagenesis
Site-directed mutagenesis and site-saturated mutagenesis were performed using the Fast Mutagenesis System kit (Transgen Biotech) following the manufacturer’s protocol. Primer sequences are listed in Supplementary Table 2. The mutation of site-directed mutagenesis was confirmed by DNA sequencing, and plasmids of positive clones were transformed into competent E. coli BL21(DE3) cells for expression.
Enzyme transformation
Recombinant strains E. coli BL21(DE3)/pET28a-ilvA, E. coli BL21(DE3)/pET28a-leudh and E. coli BL21(DE3)/pET28a-fdh were individually cultivated in lysogeny broth (LB) with 50 μg kanamycin mL−1 and grown to OD600 = 0.6–0.8. Expression of l-TD, l-LeuDH and FDH was then induced by adding 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), followed by incubation at 28 °C for 10 h. After harvesting, recombinant cells were washed and suspended in 0.1 M Tris/HCl buffer (pH 7.5), then disrupted by sonication. The lysate was removed by centrifugation for 20 min at 12,000×g at 4 °C. The supernatant was obtained as an enzyme solution and directly used for activity measurement.
The activity of l-TD was measured by determining the generation of α-ketobutyric acid (Yin et al. 2012). The activity of l-LeuDH was measured by determining the consumption of NADH in reductive amination (Ansorge and Kula 1999). The activity of FDH was measured from the yield of NADH catalysed by FDH (Berríos-Rivera et al. 2002).
The biotransformation of l-threonine to l-ABA was carried out in a shake flask. The original system contained 1 M l-threonine, 1 M ammonium formate, 6400 U wild-type l-TD L−1, 6400 U l-LeuDH L−1, 3200 U FDH L−1 and various NAD+ concentration in the range of 0.04–0.08 g L−1. The pH was maintained at 7.5 with 10% (v/v) formic acid. Samples were taken during the transformation. The amounts of l-threonine, α-ketobutyric acid and l-ABA in the system were measured by HPLC using a Rspak DE-613 column (6 μm, 4.6 × 150 mm, Shodex). The mobile phase (pH 2.8) was 5% (v/v) methanol with 1.4 mM perchloric acid. The column was eluted at 30 °C at 0.5 mL min−1. The eluent was analysed with G1314 UV–vis detector at 210 nm.
In the revised system, the cell lysate was incubated at 50° C for 60 min before being added into the reaction mixture. The revised system contained 1 M l-threonine, 1 M ammonium formate, 6400 U G323D/F510L/T344A L−1, 6400 U l-LeuDH L−1, 3200 U FDH L−1 and 0.04 g NAD+ L−1.
Results and discussion
Modification of l-TD
Out of generation screening, several mutants, namely A14T, T344A and R449C, showed higher thermostabilities without considerable loss in activity (Table 1). Site-saturated mutagenesis was performed on these three residues, but no better mutant was found.
The thermostable proteins within the same superfamily would share residues besides the definitive domain for this family, and this kind of similarity could be explained for their extra thermostability (Qiao et al. 2015). Through BLAST, four thermostable threonine deaminases were selected to perform MSAs with l-TD from E. coli K12 (Fig. 2). The residues Val31, Ala114, Phe157, Ala193, Glu240, Val281, Gly323, Ser443 and Phe510 were selected for simulated mutagenesis on DS 3.0. Among the residues above, Ala114, Glu240, Gly323, Ser443 and Phe510 showed big mutation energy change. Thus, these residues were subjected to site-saturated mutagenesis. After screening by using the directed evolution platform, mutants G323D and F510L were identified with greater thermostability (Table 1).
Amino acid sequences alignment of l-TD from E. coli K12 and four thermally stable enzymes. The four thermally stable threonine deaminases from Deinococcus-Thermus (Accession ID: WP_058978819.1), Chloroflexus sp. MS-G (Accession ID: WP_031459137.1), Hydrogenophilales bacterium (Accession ID: OGU19679.1) and Pseudomonas aeruginosa (Accession ID: NP_250017.1) were selected to perform MSAs with l-TD from E. coli K12 (Accession ID: CQR83192.1). The symbols under the residues indicate conservation of the residues. Asterisk indicates position that has a single and fully conserved residue. Colon and period indicates that one of the following “strong” groups is fully conserved and four-fifths of residues are identical. Residues surrounded by red rectangles were subjected to simulated mutagenesis
As the thermostability of mutant G323D was the best among the single point mutations, mutants A14T, T344A, R449C and F510L were combined with G323D separately using site-directed mutagenesis. Compared with the mutant G323D, mutant G323D/F510L showed the best improvements in T 1550 (temperature at which the activity of the enzyme decreased by 50% in 15 min) and t1/2,42°C (half-life at 42 °C) among the combined double-site mutants. Next, T344A, R449C and T344A/R449C were each combined with G323D/F510L. The T 1550 and t1/2,42°C values of the “best” mutant G323D/F510L/T344A were 0.5 °C higher and 9 min longer, respectively, than those of G323D/F510L. In addition, the t1/2,50°C of the purified enzyme (G323D/F510L/T344A) was also determined to be 61.6 min (Fig. 3). Compared to the kcat/Km of wild-type l-TD (9.7 × 104 M−1 s−1), G323D/F510L/T344A showed a 50% decrease, mainly due to the Km of G323D/F510L/T344A being increased. According to the molecular modeling study, the enzyme–substrate complex of G323D/F510L/T344A was more unstable than that of wild-type l-TD (Supplementary Fig. 1), which could account for the increase of Km. However, Km had a great influence on the reaction rate only if the substrate concentration was far less than Km. Hence, the effect of the decrease in kcat/Km could be ignored in terms of the whole reaction process, while the initial substrate concentration was much higher than Km.
Thermal inactivation of mutant G323D/F510L/T344A and wild-type l-TD. The wild-type l-TD (filled square) and mutant G323D/F510L/T344A (filled circle) were incubated at 50 °C and measured every 10 min at 30 °C under optimum pH (0.1 M Tris/HCl, pH 7.5). The original activity before pre-incubation was taken to be 100% (108 ± 1.8 U mg−1 and 99.55 ± 2.1 U mg−1 for wild-type and G323D/F510L/T344A, respectively). The data are presented mean ± SD from triplicate experiments
Analysing the non-covalent bonds surrounding the regions of each mutated site suggests that mutants A14T, G323D, R449C and F510L individually introduced additional hydrogen bonds in the region of mutant residues individually, which made the region more compact and stable (Fig. 4). For mutant T344A, the alanine residue could have altered hydrophobic interactions on the l-TD surface, which could pull the region away from the surface and make it more stable (Fig. 4e). Previous studies have demonstrated that the hydrophobic interactions and hydrogen bonds are essential for the improved thermal stability of modified enzymes (Pezeshgi Modarres et al. 2016). The additional hydrogen bond formation and hydrophobic interaction changes surrounding mutant residues could explain the significantly improved thermostability of mutations.
Computational analysis of the wild-type l-TD and mutations. Distribution of mutant amino acid residues a on the structure of l-TD (PDB: 1TDJ). Changes in the electronic interaction of mutants A14T (c), G323D (d), T344A (e), F510L (f) and R449 (b) in locally enlarged figures. Green dashes denote hydrogen bonds, and yellow arrows denote hydrophobic interactions
Effect of indigenous enzymes on NADH
The thermostability of mutant l-TD (G323D/F510L/T344A), l-LeuDH and FDH (Fig. 5) suggested that 50 °C was appropriate as the pretreatment temperature. The impact of pretreatment duration of mesophilic expression hosts on NADH decomposition showed that the NADH concentration in the crude enzyme decreased rapidly, with NADH totally consumed after 2 h (Fig. 6). Prolonged preheating duration rendered NADH relatively stable, with almost no NADH consumed after preheating for 60 min. Therefore, the irreversible degradation of NADH by the indigenous enzymes from expression hosts was efficiently eliminated by preheating the crude enzyme at 50 °C for 60 min.
Thermal inactivation of G323D/F510L/T344A, l-LeuDH and FDH. The supernatant G323D/F510L/T344A (filled square), l-LeuDH (filled circle) and FDH (filled triangle) were incubated at various temperatures (40, 45, 50, 55, 60 and 65 °C) for 15 min, and the residual activity was measured under standard assay conditions. The original activity before incubation was taken as 100% (107 ± 2.1, 69 ± 1.8 and 3.5 ± 0.8 U mL−1 for G323D/F510L/T344A, l-LeuDH and FDH, respectively). The data are presented mean ± SD from three independent experiments
Impact of indigenous enzymes from expression host on NADH. One gram of NADH l−1 was incubated at 35 °C in 0.1 M Tris/HCl buffer (pH 7.5) with (filled circle, filled triangle, open square, open circle, open triangle and open pentagon) and without (filled square) the crude enzyme of the expression host E. coli BL21(DE3). The crude enzyme was incubated at 50 °C for 0 (filled circle), 15 (filled triangle), 30 (open square), 45 (open circle), 60 (open triangle) and 75 min (open pentagon) before mixing with NADH. Samples were taken at 1-h intervals, and NADH concentration was measured from the absorbance at 340 nm. Values represent the mean of three experiments, and error bars represent the standard deviation
Evaluation of original and revised systems for l-ABA production
The thermostability of G323D/F510L/T344A has been significant improved, therefore it could tolerate preheat treatment. The revised system contained the preheated G323D/F510L/T344A, l-LeuDH and FDH was performed to synthesis l-ABA, with the original system consisting of wild-type l-TD, l-LeuDH and FDH as a control. After 10 h, the original system converted 81% of the l-threonine substrate and produced 0.635 M l-ABA (Fig. 7a) and the total turnover number (TTN, defined as the number of moles of product formed with a consumption of a complete reaction) of NAD+/NADH was 10531 (Supplementary Table 3). Moreover, l-ABA formation only reached 0.771 M with a TTN of 6393, despite the NAD+ dosage being drastically increased to 0.08 g L−1 (Supplementary Table 3). In contrast, l-threonine was transformed completely by the revised system, and 0.993 M l-ABA was obtained within 12 h (Fig. 7b) with a TTN of 16,469, which was a 56% increase compared to that of the original system (10,531). Comparing the bioconversion of the original and revised systems, the biosynthetic rate of l-ABA in the original system was decreased in the late stage of reaction, and the intermediate α-ketobutyric acid was accumulated gradually until the deamination reaction reached equilibrium. However, the biosynthetic rate of l-ABA in the revised system was relatively constant, and the intermediate α-ketobutyric acid was rarely accumulated. These results suggested that the revised system was advantageous for the biosynthesis of l-ABA. Our study showed that preheating the recombinant mesophiles having a heterologous thermophilic enzyme was an efficient solution to diminish the loss of NAD(H) and reduce the dosage of coenzyme. Similar process could be applied to other biosynthesis systems involving NAD(H), since these systems might also suffer from excessive coenzyme consumption.
Time course reactions to produce l-ABA by the original and revised one-pot systems. Time profiles of l-ABA (filled triangle), α-ketobutyric acid (filled circle) and l-threonine (filled square). a The original one-pot system contained 6400 U wild-type l-TD l−1, 6400 U l-LeuDH l−1, 3200 U FDH l−1 cell extract, 1 M l-threonine, 1 M ammonium formate and 0.04 g NAD+ l−1. b The revised one-pot system contained 6400 U G323D/F510L/T344A L−1, 6400 U l-LeuDH L−1, 3200 U FDH L−1 cell extract, 1 M l-threonine, 1 M ammonium formate and 0.04 g NAD+ L−1
Conclusion
To summarise, a thermostable mutant l-TD (G323D/F510L/T344A) was constructed by directed evolution and rational design without considerable loss in activity, and a one-pot system was constituted by the mutant G323D/F510L/T344A, thermophilic l-LeuDH and FDH. Using preheat treatment, this system quantitatively converted l-threonine to l-ABA with a significantly lower NAD+ demand. This study has greatly reduced the amount of NAD+ required for this biosynthesis by developing an alternative system that regenerated NAD+ efficiently.
References
An Z, Gu X, Liu Y, Zhu Q (2017) Bioproduction of L-2-aminobutyric acid by a newly-isolated strain of Aspergillus tamarii ZJUT ZQ013. Catal Lett 147:837–844
Ansorge MB, Kula MR (1999) Production of recombinant l-leucine dehydrogenase from Bacillus cereus in pilot scale using the runaway replication system E. coli [pIET98]. Biotechnol Bioeng 65:557–562
Berríos-Rivera SJ, Bennett GN, San KY (2002) Metabolic engineering of Escherichia coli: increase of NADH availability by overexpressing an NAD+-dependent formate dehydrogenase. Metab Eng 4:217–229
Eisenstein E (1991) Cloning, expression, purification, and characterization of biosynthetic threonine deaminase from Escherichia coli. J Biol Chem 266:5801–5807
Galkin A, Kulakova L, Yoshimura T, Soda K, Esaki N (1997) Synthesis of optically active amino acids from α-keto acids with Escherichia coli cells expressing heterologous genes. Appl Environ Microb 63:4651–4656
Gallagher D, Gilliland G, Xiao G, Zondlo J, Fisher K, Chinchilla D, Eisenstein E (1998) Structure and control of pyridoxal phosphate dependent allosteric threonine deaminase. Structure 6:465–475
Hatfield GW, Umbarger HE (1970) Threonine deaminase from Bacillus subtilis. J Biol Chem 245:1742–1747
Heuser F, Schroer K, Lütz S, Bringer-Meyer S, Sahm H (2007) Enhancement of the NAD(P)(H) pool in Escherichia coli for biotransformation. Eng Life Sci 7:343–353
Krutsakorn B, Honda K, Ye X, Imagawa T, Bei X, Okano K, Ohtake H (2013) In vitro production of n-butanol from glucose. Metab Eng 20:84–91
Li Z, Tao R, Wang Y, Jiang Y, Lin X, Yang Y, Zheng H, Jiang W, Yang S (2011) Removal of l-alanine from the production of l-2-aminobutyric acid by introduction of alanine racemase and d-amino acid oxidase. Appl Microbiol Biotechnol 90:903–910
Park ES, Dong JY, Shin JS (2013) ω-Transaminase-catalyzed asymmetric synthesis of unnatural amino acids using isopropylamine as an amino donor. Org Biomol Chem 11:6929–6933
Pezeshgi Modarres H, Mofrad MR, Sanati-Nezhad A (2016) Protein thermostability engineering. RSC Adv 6:115252–115270
Qiao P, Wu M, Zhu L, Zhang Y, Yang L, Fei H, Lin J (2015) Enhancing the thermal tolerance of a cis-epoxysuccinate hydrolase via combining directed evolution with various semi-rational redesign methods. J Mol Catal B Enzym 121:96–103
Sin JS, Kim BG (2009) Transaminase-catalyzed asymmetric synthesis of l-2-aminobutyric acid from achiral reactants. Biotechnol Lett 31:1595–1599
Tao R, Jiang Y, Zhu F, Yang S (2014) A one-pot system for production of L-2-aminobutyric acid from l-threonine by l-threonine deaminase and a NADH-regeneration system based on l-leucine dehydrogenase and formate dehydrogenase. Biotechnol Lett 36:835–841
Yin L, Hu X, Xu D, Ning J, Chen J, Wang X (2012) Co-expression of feed-back-resistant threonine dehydratase and acetohydroxy acid synthase increase l-isoleucine production in Corynebacterium glutamicum. Metab Eng 14:542–550
Acknowledgements
This work was funded by the National Hi-Tech Research and Development Program of China (2014AA022105).
Supporting information
Supplementary Table 1—Bacterial strains and plasmids used in this study.
Supplementary Table 2—Primers used in this study.
Supplementary Table 3—Biotransformation of l-ABA by the original and revised systems.
Supplementary Fig. 1—Electronic interaction between substrate l-threonine and (a) wild-type l-TD or (b) G323D/F510L/T344A.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, Y., Li, GS., Qiao, P. et al. Increased productivity of l-2-aminobutyric acid and total turnover number of NAD+/NADH in a one-pot system through enhanced thermostability of l-threonine deaminase. Biotechnol Lett 40, 1551–1559 (2018). https://doi.org/10.1007/s10529-018-2607-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-018-2607-3