Abstract
Escherichia coli strain CAR001 that produces β-carotene was genetically engineered to produce lycopene by deleting genes encoding zeaxanthin glucosyltransferase (crtX) and lycopene β-cyclase (crtY) from the crtEXYIB operon. The resulting strain, LYC001, produced 10.5 mg lycopene/l (6.5 mg/g dry cell weight, DCW). Modulating expression of genes encoding α-ketoglutarate dehydrogenase, succinate dehydrogenase and transaldolase B within central metabolic modules increased NADPH and ATP supplies, leading to a 76 % increase of lycopene yield. Ribosome binding site libraries were further used to modulate expression of genes encoding 1-deoxy-d-xylulose-5-phosphate synthase (dxs) and isopentenyl diphosphate isomerase (idi) and the crt gene operon, which improved the lycopene yield by 32 %. The optimal strain LYC010 produced 3.52 g lycopene/l (50.6 mg/g DCW) in fed-batch fermentation.
Similar content being viewed by others

Avoid common mistakes on your manuscript.
Introduction
Lycopene is a carotenoid that is used in the pharmaceutical, nutraceutical, cosmetic, and food industries (Nagao 2009). It plays an important role in human health as a biological antioxidant that prevents oxidation of low-density lipoprotein and cholesterol (Arab and Steck 2000; Mordente et al. 2011). Further, evidence acquired from in vitro and in vivo studies suggests that lycopene prevents cancers of the skin, breast, lung, and liver (Khan et al. 2008; Seren et al. 2008).
Lycopene is produced by direct extraction from tomatoes, chemical synthesis, and microbial fermentation. Because the lycopene content of tomatoes is low (approx. 0.02 g/kg) and many other carotenoids are present (Clinton 1998), the extraction method is relatively expensive. Chemical synthesis is complex and involves using hazardous materials. Therefore, the interest in producing lycopene by microbial fermentation has increased and Escherichia coli is widely used for this purpose (Farmer and Liao 2001; Alper et al. 2005a, b, 2006; Kang et al. 2005; Yoon et al. 2006, 2007; Jin and Stephanopoulos 2007; Alper and Stephanopoulos 2008; Choi et al. 2010; Kim et al. 2011).
Several strategies have improved lycopene production by E. coli. First, overexpression of genes required for isoprenoid synthesis that encode components of the 2-C-methyl-d-erythritol-4-phosphate (MEP) pathway, such as 1-deoxy-d-xylulose-5-phosphate synthase (dxs) and isopentenyl diphosphate isomerase (idi), increase supplies of the isoprenoid precursors, isopentenyl pyrophosphate (IPP) and dimethylallyl pyrophosphate (DMAPP) (Jin and Stephanopoulos 2007; Choi et al. 2010). Second, systematic and combinatorial methods identified gene knockouts or amplification targets for improving lycopene production (Farmer and Liao 2001; Alper et al. 2005a, b; Kang et al. 2005; Jin and Stephanopoulos 2007; Alper and Stephanopoulos 2008; Choi et al. 2010). Third, a heterologous mevalonate pathway was introduced into E. coli to increase the IPP supply (Yoon et al. 2006, 2007). Fourth, fermentation processes were optimized to improve lycopene production, such as adding auxiliary carbon sources (Kim et al. 2011) and maintaining high O2 levels and pH values (Yoon et al. 2006).
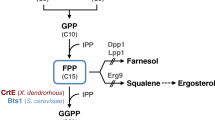
The genetically-stable E. coli strain CAR001 that produces β-carotene was constructed by integrating into its chromosome the genes encoding components of the β-carotene pathway of Pantoea agglomerans (crtEXYIB) and modulating the expression of dxs, idi, and crt genes with multiple regulatory parts (Zhao et al. 2013). In the present study, the genes encoding zeaxanthin glucosyltransferase (crtX) and lycopene β-cyclase (crtY) (Fig. 1) derived from the crtEXYIB operon of strain CAR001 were deleted to program a switch from β-carotene to lycopene synthesis. Further, the expression of certain genes of the pentose phosphate and tricarboxylic acid (TCA) pathways that increase NADPH and ATP supplies (Zhao et al. 2013) were modulated to increase lycopene production. Finally, ribosome binding site (RBS) libraries were used to modulate the expression of dxs, idi, and crt genes in a further effort to improve lycopene production.
Materials and methods
Strains and culture conditions
The bacterial strains and plasmids used here are listed in Table 1. During strain construction, cultures were grown aerobically at 30, 37, or 39 °C in LB broth (per liter: 10 g tryptone, 5 g yeast extract, and 5 g NaCl). For lycopene production, single colonies were picked from a plate and inoculated into 15 × 100 mm tubes containing 4 ml LB and grown overnight at 37 °C with shaking at 250 rpm. This seed culture was used to inoculate a 100 ml flask containing 10 ml LB (initial OD600 = 0.05) that was incubated at 37 °C with shaking at 250 rpm. After 24 h, cells were collected to measure lycopene production.
Deletion of crtX and crtY genes from strain CAR001
A two-step homologous recombination method (Zhang et al. 2007; Jantama et al. 2008) was used to delete crtX and crtY genes from strain CAR001 (Zhao et al. 2013) for lycopene production. In the first recombination, the cat-sacB cassette was amplified from plasmid pXZ-CS (Tan et al. 2013) using primer set CrtE-taa-cat-f/CrtI-atg-cat-r to replace the crtXY genes in CAR001. In the second recombination, a DNA fragment was amplified from CAR001 using primer set CrtEI-RBS-f/CrtI-484-r. Primer CrtEI-RBS-f included 50 nucleotides identical to those at the 3′-end of crtE followed by an artificial RBS sequence (GGAGGATTACTAT). Primer CrtI-484-r included 484 nucleotides identical to those at the 5′-end of crtI. This amplicon was used to replace the cat-sacB cassette by selection for resistance to sucrose. Colonies were verified using PCR amplification with primer set ldhA-up/CrtI-484-r, and the colony with the correct sequence was designated LYC001 (Table 1). Primers used in this study are listed in Supplementary Table 1.
Modulating expression of sucAB, sdhABCD and talB genes
A two-step recombination method (Shi et al. 2013) was used for markerless modulation of the expression of sucAB, sdhABCD and talB genes with regulatory part M1-46 as described previously (Zhao et al. 2013).
Modulating expression of dxs, idi, and crt genes with RBS library
The native RBS of the crt operon of strain LYC005 was replaced by a RBS library through a two-step homologous recombination method. First, the cat-sacB cassette was amplified from pXZ-CS (Tan et al. 2013) using primer set ldhA-up-cat/crtE-sacB-down and inserted upstream of the ATG translation initiation codon of crt gene. A DNA fragment containing the RBS library was amplified from genomic DNA of recombinant E. coli M1-93 (Lu et al. 2012) using primer set ldhA-up-P/crtE-RBSL-down and used for the second recombination to replace the cat-sacB cassette. E. coli M1-93 was selected from a previously constructed regulatory parts library, which had a regulatory part inserted in front of lacZ gene of E. coli ATCC 8739, and its strength was five times that of the induced E. coli lacZ promoter (Lu et al. 2012). The sequence of the RBS library is CAGGAGRNNNNNN. The upstream region of crtE (from −50 to 0 relative to the translation start site of crtE) was used as the upstream homologous arm, and the partial coding region of crtE gene (from +1 to +50 relative to the translation start site of crtE) was used as the downstream arm. After the second recombination, colonies sensitive to chloramphenicol were picked for PCR verification using primer set p-up/crtE-340-down. Fifteen colonies with the correct sequences were selected from each library. The same procedures were used to modulate expression of dxs and idi genes.
Analysis of lycopene production
Cells were harvested by centrifugation at 4,000×g for 10 min, suspended in acetone (1 ml), incubated at 55 °C for 15 min in the dark, centrifuged at 14,000×g for 10 min and the acetone supernatant containing lycopene was transferred to a new tube. Lycopene production was analyzed using HPLC with a variable wavelength detector set to 480 nm and a Symmetry C18 column (250 mm × 4.6 mm, 5 μm, Waters). Methanol/acetonitrile/dichloromethane (21:21:8, by vol) was used as the mobile phase at 0.8 ml/min at 30 °C (Yoon et al. 2009). The results represent the mean ± standard deviation (SD) of three independent experiments. Dry cell weight (DCW) was calculated as follows: 1 OD600 = 0.379 g DCW/l.
Fed-batch fermentation
Strains LYC001, LYC005 and LYC010 were used to produce lycopene through fed-batch fermentation. The medium used for seed preparation and fermentation was described previously (Zhao et al. 2013). The seed culture was prepared by inoculating several colonies into a 250 ml flask containing 50 ml culture medium and incubating at 37 °C with shaking at 250 rpm for 14 h. The seed was then transferred to a fermentor (7 l) (Infors Biotechnology Co. Ltd) containing medium (3 l) with an initial OD600 = 0.4. Fermentation was carried out at 30 °C with an air flow of 3 l/min. Dissolved O2 was maintained at 30 % by adjusting the agitation speed from 300 to 1,200 rpm. The pH was maintained at 7 by automatic addition of 5 M NH4OH. A fed solution containing (per liter) 500 g glycerol, 15 g peptone, 30 g yeast extract, and 30 g MgSO4·7H2O, was added to the fermentor at an average rate of 20 ml/h.
Results and discussion
Deletion of crtXY genes from strain CAR001 for lycopene production
Strain CAR001 was previously engineered to produce β-carotene (Zhao et al. 2013). The gene operon required for β-carotene synthesis by P. agglomerans (crtEXYIB) was integrated into the E. coli chromosome at the ldhA site, and its expression was controlled by regulatory part M1-93 (Lu et al. 2012). Further, dxs and idi genes were modulated by regulatory parts M1-37 and M1-46, respectively, to increase IPP and DMAPP supplies. Strain CAR001 produces 25.7 mg β-carotene/l, equivalent to 18.4 mg/g DCW (Zhao et al. 2013).
IPP and DMAPP are converted to β-carotene in strain CAR001 by farnesyl diphosphate synthase (IspA), geranylgeranyl diphosphate synthase (CrtE), phytoene synthase (CrtB), phytoene desaturase (CrtI), and lycopene β-cyclase (CrtY) (Fig. 1). To switch β-carotene to lycopene production, the crtY gene should be deleted. Further, zeaxanthin glucosyltransferase (CrtX) is not required for lycopene production. Strain LYC001 was created by deleting crtX and crtY genes from strain CAR001. This strain produced 10.49 mg lycopene/l, equivalent to 6.52 mg/g DCW (Table 2).
Engineering central metabolic modules to improve lycopene production
ATP and NADPH are two cofactors for terpenoid synthesis. The synthesis of β-carotene is enhanced by modulating the expression of genes encoding α-ketoglutarate dehydrogenase (sucAB), succinate dehydrogenase (sdhABCD) and transaldolase B (talB) that are components of central metabolic modules (Zhao et al. 2013). Modulating sucAB and sdhABCD genes could enhance carbon flux towards TCA cycle and increase NADPH and ATP supply. Modulating talB gene could enhance carbon flux towards pentose phosphate module and increase NADPH supply. Therefore, the expression of these genes was modulated in LYC001 to investigate whether lycopene synthesis was improved.
Strain LYC002 was created by introducing regulatory part M1-46 into strain LYC001 to modulate the expression of sucAB gene. Lycopene production increased 46 % to 15.4 mg/l and yield increased 64 % to 10.7 mg/g DCW (Table 2). Strain LYC003 was created by introducing regulatory part M1-46 into strain LYC002 to modulate the expression of talB gene, which increased lycopene from 10.7 mg/g DCW to 11 mg/g DCW (Table 2). Strain LYC005 was created by inserting regulatory part M1-46 into LYC003 to modulate the expression of the sdhABCD operon. This strain produced 18.9 mg lycopene/l, equivalent to 11.5 mg/g DCW (Table 2). The lycopene yield increased 76 % after modulating the expression of sucAB, sdhABCD and talB genes (Table 2). These results indicate that engineering central metabolic modules shows promise as a general strategy to improve the efficiency of the terpenoid synthetic pathways, which require ATP and NADPH as key cofactors.
Modulating expression of dxs, idi, and crtE genes of strain LYC005 with RBS library
During the construction of strain CAR001 for β-carotene production, three regulatory parts were selected for modulating the expression of dxs and idi genes, and two regulatory parts were selected for modulating the expression of crt operon (Zhao et al. 2013). Modulating dxs, idi, and crt genes with regulatory parts M1-37, M1-46 and M1-93, respectively, was identified to be the best combination for β-carotene production. However, the strengths of these regulatory parts may not be optimal for lycopene production. It was suggested that modulating expression of these important genes with regulatory parts libraries would have more opportunities to obtain higher lycopene production.
For this purpose, a RBS library (CAGGAGRNNNNNN) was inserted to the upstream of the ATG translation initiation codon to modulate the expression of dxs, idi, and crt genes. Inserting sequences from this RBS library upstream of E. coli ATCC 8739 lacZ altered lacZ expression levels (after induction of the lacZ promoter) by factors ranging from 0.17 to 8.6 (Chen et al. 2013). Using this approach, these RBS library sequences were inserted into strain LYC005 to modulate expression of dxs, idi, or crt gene. Fifteen colonies were randomly selected from each library (Fig. 2). For dxs gene, lycopene yields ranged by factors of 0.73–1.13 compared with that of LYC005 (Fig. 2a), and the best strain, dxs8, produced 18.3 mg lycopene/l, equivalent to 13.1 mg/g DCW. For idi, lycopene yields ranged by factors of 0.67–1.13 compared with that of LYC005 (Fig. 2b), and the best strain, idi9, produced 18.3 mg lycopene/l, equivalent to 13 m/g DCW. For the crt operon, lycopene yields ranged by factors of 0.88–1.14 compared with that of LYC005 (Fig. 2c), and the best strain, crtE9 (designated LYC008), produced 20.9 mg lycopene/l, equivalent to 13.2 mg/g DCW (Table 2). Modulating dxs, idi and crt genes with RBS libraries led to 13, 13 and 14 % improvement of lycopene yield compared to parent strain, suggesting that this gene modulation strategy would have more opportunities to obtain optimal strength for production of target compound.
Because the lycopene yields of strain LYC008 were highest, it was subjected to further manipulation of dxs and idi genes using the RBS library. After modulating dxs gene in strain LYC008, the lycopene yields of the resulting strains ranged by factors of 0.77–1.15 compared with that of LYC005 (Fig. 3a). The best strain, crtE9-dxs12 (designated LYC009), produced 24.3 mg lycopene/l, equivalent to 13.3 mg/g DCW (Table 2). After modulating idi gene in strain LYC009, the lycopene yields of the resulting strains ranged by factors of 1.05–1.32 times compared with that of LYC005 (Fig. 3b). The best strain, crtE9-dxs12-idi7 (designated LYC010), produced 26.3 mg lycopene/l, equivalent to 15.2 mg/g DCW (Table 2). Combined manipulation of these three genes with RBS libraries resulted in 32 % improvement of the lycopene yield. The RBS sequences of the dxs, idi, and crt genes of strains LYC006, LYC007, LYC008, LYC009, and LYC010 are listed in Supplementary Table 2.
Fed-batch fermentation of strains LYC001, LYC005 and LYC010 for lycopene production
To increase lycopene production, fed-batch fermentation of strains LYC001, LYC005 and LYC010 was performed in a 7 l fermentor at pH 7.0. Strain LYC001 produced 0.47 g lycopene/l and the yield was 11.1 mg/g (Fig. 4a). Strain LYC005 produced 0.95 g lycopene/l and the yield was 21.3 mg/g (Fig. 4b). Strain LYC010 produced 3.52 g lycopene/l and the yield was 50.6 mg/g (Fig. 4c). Lycopene yields of strains LYC001, LYC005 and LYC010 under fed-batch condition were 79, 85 and 233 % higher than those under shake flask conditions, respectively. This might be due to deficient nutrient and oxygen supply under shake flask conditions. It was also suggested that shake flask conditions limited the improvement of lycopene yield by modulating dxs, idi and crt genes with RBS library. To the best of our knowledge, lycopene concentration and yield of strain LYC010 are the highest for engineered E. coli strains.
References
Alper H, Stephanopoulos G (2008) Uncovering the gene knockout landscape for improved lycopene production in E. coli. Appl Microbiol Biotechnol 78:801–810
Alper H, Fischer C, Nevoigt E, Stephanopoulos G (2005a) Tuning genetic control through promoter engineering. Proc Natl Acad Sci USA 102:12678–12683
Alper H, Jin YS, Moxley JF, Stephanopoulos G (2005b) Identifying gene targets for the metabolic engineering of lycopene biosynthesis in Escherichia coli. Metab Eng 7:155–164
Alper H, Miyaoku K, Stephanopoulos G (2006) Characterization of lycopene-overproducing E. coli strains in high cell density fermentations. Appl Microbiol Biotechnol 72:968–974
Arab L, Steck S (2000) Lycopene and cardiovascular disease. Am J Clin Nutr 71:1691S–1695S
Chen J, Zhu X, Tan Z, Xu H, Tang J, Xiao D, Zhang X (2013) Activating C4-dicarboxylate transporters DcuB and DcuC for improving succinate production. Appl Microbiol Biotechnol 98:2197
Choi HS, Lee SY, Kim TY, Woo HM (2010) In silico identification of gene amplification targets for improvement of lycopene production. Appl Environ Microbiol 76:3097–3105
Clinton SK (1998) Lycopene: chemistry, biology and implications for human health and disease. Nutr Rev 56:35–51
Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 97:6640–6645
Farmer WR, Liao JC (2001) Precursor balancing for metabolic engineering of lycopene production in Escherichia coli. Biotechnol Prog 17:57–61
Jantama K, Zhang X, Moore JC, Shanmugam KT, Svoronos SA, Ingram LO (2008) Eliminating side products and increasing succinate yields in engineered strains of Escherichia coli C. Biotechnol Bioeng 101:881–893
Jin YS, Stephanopoulos G (2007) Multi-dimensional gene target search for improving lycopene biosynthesis in Escherichia coli. Metab Eng 9:337–347
Kang MJ, Lee YM, Yoon SH, Kim JH, Ock SW, Jung KH, Shin YC, Keasling JD, Kim SW (2005) Identification of genes affecting lycopene accumulation in Escherichia coli using a shot-gun method. Biotechnol Bioeng 91:636–642
Khan N, Afaq F, Mukhtar H (2008) Cancer chemoprevention through dietary antioxidants: progress and promise. Antioxid Redox Signal 10:475–510
Kim YS, Lee JH, Kim NH, Yeom SJ, Kim SW, Oh DK (2011) Increase of lycopene production by supplementing auxiliary carbon sources in metabolically engineered Escherichia coli. Appl Microbiol Biotechnol 90:489–497
Lu J, Tang J, Liu Y, Zhu X, Zhang T, Zhang X (2012) Combinatorial modulation of galP and glk gene expression for improved alternative glucose utilization. Appl Microbiol Biotechnol 93:2455–2462
Mordente A, Guantario B, Meucci E, Silvestrini A, Lombardi E, Martorana GE, Giardina B, Bohm V (2011) Lycopene and cardiovascular diseases: an update. Curr Med Chem 18:1146–1163
Nagao A (2009) Absorption and function of dietary carotenoids. Forum Nutr 61:55–63
Seren S, Lieberman R, Bayraktar UD, Heath E, Sahin K, Andic F, Kucuk O (2008) Lycopene in cancer prevention and treatment. Am J Ther 15:66–81
Shi A, Zhu X, Lu J, Zhang X, Ma Y (2013) Activating transhydrogenase and NAD kinase in combination for improving isobutanol production. Metab Eng 16:1–10
Tan Z, Zhu X, Chen J, Li Q, Zhang X (2013) Activating phosphoenolpyruvate carboxylase and phosphoenolpyruvate carboxykinase in combination for improving succinate production. Appl Environ Microbiol 79(16):4838–4844
Yoon SH, Lee YM, Kim JE, Lee SH, Lee JH, Kim JY, Jung KH, Shin YC, Keasling JD, Kim SW (2006) Enhanced lycopene production in Escherichia coli engineered to synthesize isopentenyl diphosphate and dimethylallyl diphosphate from mevalonate. Biotechnol Bioeng 94:1025–1032
Yoon SH, Kim JE, Lee SH, Park HM, Choi MS, Kim JY, Shin YC, Keasling JD, Kim SW (2007) Engineering the lycopene synthetic pathway in E. coli by comparison of the carotenoid genes of Pantoea agglomerans and Pantoea ananatis. Appl Microbiol Biotechnol 74:131–139
Yoon SH, Lee SH, Das A, Ryu HK, Jang HJ, Kim JY, Oh DK, Keasling JD, Kim SW (2009) Combinatorial expression of bacterial whole mevalonate pathway for the production of beta-carotene in E. coli. J Biotechnol 140:218–226
Zhang X, Jantama K, Moore JC, Shanmugam KT, Ingram LO (2007) Production of l-alanine by metabolically engineered Escherichia coli. Appl Environ Microbiol 77:355–366
Zhao J, Li Q, Sun T, Zhu X, Xu H, Tang J, Zhang X, Ma Y (2013) Engineering central metabolic modules of Escherichia coli for improving beta-carotene production. Metab Eng 17:42–50
Acknowledgments
This research was supported by grants from Tianjin Key Technology R&D program of Tianjin Municipal Science and Technology Commission (12ZCZDSY14700), National Basic Research Program of China (2011CBA00800) and National High Technology Research and Development Program of China (2012AA02A704). Xueli Zhang was supported by the Hundred Talent Program of the Chinese Academy of Sciences.
Conflict of interest
This work has been included in a patent application by Tianjin Institute of Industrial Biotechnology.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Tao Sun and Liangtian Miao have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sun, T., Miao, L., Li, Q. et al. Production of lycopene by metabolically-engineered Escherichia coli . Biotechnol Lett 36, 1515–1522 (2014). https://doi.org/10.1007/s10529-014-1543-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-014-1543-0