Abstract
A substrate-coupled biocatalytic process was developed based on the reactions catalyzed by an NADPH-dependent sorbose reductase (SOU1) from Candida albicans in which ethyl 4-chloro-3-oxobutanoate (COBE) was reduced to (S)-4-chloro-3-hydroxybutanoate [(S)-CHBE], while NADPH was regenerated by the same enzyme via oxidation of sugar alcohols. (S)-CHBE yields of 1,140, 1,150, and 780 mM were obtained from 1,220 mM COBE when sorbitol, mannitol, and xylitol were used as co-substrates, respectively. Optimization of COBE and sorbitol proportions resulted in a maximum yield of (S)-CHBE (2,340 mM) from 2,500 mM COBE, and the enantiomeric excess was 99.6 %. The substrate-coupled system driven by SOU1 maintained a stable pH and a robust intracellular NADPH circulation; thus, pH adjustment and addition of extra coenzymes were unnecessary.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The biosynthesis of ethyl (S)-4-chloro-3-hydroxybutanoate [(S)-CHBE], a chiral intermediate in the synthesis of pharmacologically active compounds, such as hydroxymethylglutaryl-CoA reductase inhibitors and 4-hydroxypyrrolidone (Karanewsky et al. 1990), has attracted significant attention. The asymmetric reduction of ethyl 4-chloro-3-oxobutanoate (COBE) by enantioselective oxidoreductases is a practical method for the production of (S)-CHBE (Ye et al. 2011). However, this method requires an efficient regeneration for the coenzyme NAD(P)H. To overcome this problem, enzyme-coupled systems were proposed and many coenzyme regeneration systems, including formate/formate dehydrogenase (FDH) (Yamamoto et al. 2004, 2005), glucose 6-phosphate (G6P)/glucose-6-phosphate dehydrogenase (G6PDH) (Yu et al. 2007), and glucose/glucose dehydrogenase (GDH) (Kizaki et al. 2001; Ye et al. 2010a), have been widely used to provide NAD(P)H coupling with oxidoreductase-catalyzed synthesis systems. The substrate-coupled method, using NAD(P)H-dependent alcohol dehydrogenase (ADH) (Wang et al. 2011; Yamamoto et al. 2002), was an alternative enzymatic approach for cofactor regeneration.
Substrate-coupled systems involve a single enzyme and are simpler than enzyme-coupled systems. However, only a limited number of such systems have been reported because the enzyme in substrate-coupled systems should exhibit broad substrate specificity and high enantioselectivity toward chiral compounds. Only ADH can be used as both a catalyst in chiral synthesis and a coenzyme regenerator by oxidation of alcohols, such as 2-propanol and ethanol (Wang et al. 2011; Yamamoto et al. 2002).
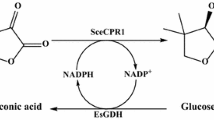
In a previous study, an NADPH-dependent sorbose reductase (SOU1, EC 1.1.1.289) from Candida albicans was reported that also had NADP+-dependent sorbitol dehydrogenase activity which converted sorbitol to sorbose by the concomitant reduction of NADP+ into NADPH (Greenberg et al. 2005). SOU1 sequence shares 74 % identity with another reported novel reductase from C. albicans which converts COBE to (S)-CHBE with high yield and enantioselectivity (An et al. 2012). The current study confirms that SOU1 can catalyze the asymmetric reduction of COBE to (S)-CHBE. SOU1 can also convert sorbitol, mannitol and xylitol into corresponding sugars. Thus, a substrate-coupled biocatalytic process around SOU1 can be developed for the biosynthesis of (S)-CHBE (Fig. 1).
Materials and methods
Chemicals
COBE, (R)-CHBE, and (S)-CHBE were obtained from Acros Organics. All other chemicals were of analytical grade and were commercially available.
Cloning and expression of the SOU1 gene
The SOU1 gene from C. albicans was amplified by PCR with designed primers of 5′-GGAATTCCATATGATGAGTGAAGAAATCATTTCA-3′ (NdeI site is underlined) and 5′-CCGGAATTCTTATGGACATGTATAACCCCCAT-3′ (EcoRI site is underlined). The PCR conditions were as follows: 35 cycles of 60 s at 94 °C, 30 s at 56 °C, and 60 s at 72 °C. The amplified product was ligated into the vector pET-22b at the NdeI and EcoRI sites, resulting in a recombinant plasmid designated as pET-22b-SOU1. The recombinant Escherichia coli Rosseta (DE3)/pET-22b-SOU1 was obtained by heat shock transformation.
The recombinant strains were grown at 37 °C for 3 h, with shaking at 200 rpm in TB medium plus 2 g glucose/l, 3 g lactose/l, 100 mg ampicillin/l, and 20 mg chloramphenicol/l. The temperature was then changed to 30 °C and expression was induced by lactose. After 12 h induction, the cells were harvested by centrifugation, washed with 100 mM potassium phosphate buffer (pH 6.2), and used for biotransformation.
Enzyme activity determination
1 ml culture broth of cells was harvested by centrifugation, suspended in 1 ml 100 mM potassium phosphate buffer (pH 6.2), and then disrupted by ultrasonication. The cell debris was removed by centrifugation; the supernatant was recovered as the cell-free extract which was used to determine enzymatic activity with different substrates. Cells harboring an empty pET-22b vector were treated by the same method and used as control. Enzyme activities were determined using either 10 mM COBE with 1 mM NADPH or 10 mM sugar alcohols with 1 mM NADP+, following changes in absorbance at 340 nm (Greenberg et al. 2005). The extinction coefficient of NADPH was 6.22 mM−1 cm−1. One unit of enzyme activity was defined as the disappearance or production of 1 μmol NADPH per minute. Protein concentration was measured using the Bradford method with BSA as standard. The repeatability was determined by three-time measurements.
Bioconversion of COBE to (S)-CHBE
The bioconversion of COBE to (S)-CHBE was performed in water/butyl acetate (1:1, v/v) biphasic system containing 100 mM potassium phosphate buffer (pH 6.2), 1,220 mM COBE, 1,300 mM co-substrate, Triton X-100 (1 ‰, v/v), and 0.2 g dry cell wt E. coli Rosseta (pET-22b-SOU1), which expressed SOU1, in 50 ml. The reaction was performed at 30 °C and 220 rpm for 20 h. The organic layer was isolated to determine the product concentration and optical purity.
COBE and CHBE analysis
COBE and CHBE concentrations were determined using gas chromatography and the optical purity of (S)-CHBE was determined by HPLC on a Chiralcel OB-H column as described previously (Ye et al. 2010b).
Results
SOU1 activity on different substrates
SOU1 exhibited an activity of 6.2 U mg−1 toward COBE. The activities of the enzyme on sorbitol, mannitol, and xylitol were 0.02, 0.03, and 0.05 U mg−1, respectively. The crude enzyme extract from control cells exhibited no activity on these substrates.
Bioconversion of COBE to (S)-CHBE
Sorbitol, mannitol, and xylitol were used as co-substrates, and the concentrations of (S)-CHBE produced were measured in the time intervals shown in Fig. 2. The bioconversion in the absence of co-substrate was carried out as control. The sorbitol/mannitol driven system produced more (S)-CHBE than the xylitol co-substrate system. After 20 h of bioconversion, 1,140, 1,150, and 780 mM (S)-CHBE were produced, resulting in yields of 93, 94, and 64 %, respectively, The enantiomeric excess was over 99 %. The system without adding co-substrates only produced a yield of 3 % because of low regeneration of NADPH.
Effects of the substrate concentration ratio on the (S)-CHBE yield
Because of differences in activity between COBE and the co-substrates, the substrate concentration ratio was adjusted to establish favorable conditions for both reactions. Since sorbitol is the natural substrate of SOU1 and is cheaper than mannitol or xylitol, it was chosen for further research.
The bioconversion protocol applied was identical to that described above, except that 2,500 mM COBE and different concentrations of sorbitol were used. The concentrations of sorbitol used were 100, 150, and 200 % of the COBE concentration. As shown in Fig. 3, as the sorbitol concentration increased, the yield of (S)-CHBE also increased. After 30 h of biotransformation, the maximum yield of (S)-CHBE (2,343 mM, 93.7 %) was achieved when COBE and sorbitol were added at a ratio of 1:2. The enantiomeric excess was 99.6 %.
Discussion
A substrate-coupled biocatalytic process for the asymmetric reduction of COBE to (S)-CHBE is proposed: it gives a high yield and optical purity by using SOU1 from C. albicans. In a previously described oxidoreductase-catalyzed COBE reduction system, glucose/GDH was used to regenerate NADPH but the accumulation of gluconate from GDH significantly decreased the pH and NaOH was then needed to control the pH of the reaction at 6.5 (An et al. 2012; Kizaki et al. 2001). In the substrate-coupled system driven by SOU1, the pH maintains 6.2 which is optimum for SOU1 over the entire reaction time without any adjustment. The substrate-coupled system driven by SOU1 maintains a stable pH and simplifies the reduction process for industrial application.
Although the enzyme exhibited low activity toward the co-substrates, the reactions performed by SOU1 in both directions were robustly circulated, and over 90 % of conversion rate was achieved using the system. The (S)-CHBE yield can be improved by optimizing the proportions of the substrate concentration. Increasing the co-substrate concentration improved the yield of the product. Under optimized conditions, 12 mM (S)-CHBE was produced per milligram of dry cell weight (DCW), which is similar to the production of a GDH system (13 mM/mg DCW) (Kizaki et al. 2001). However, GDH systems require additional NADP+ to promote bioconversion. In this process, no additional coenzyme was necessary. Since the proposed substrate-coupled system does not require extra coenzymes, it is more efficient in coenzyme regeneration and has potential use in industrial (S)-CHBE production.
References
An MD, Cai P, Yan M, Hao N, Wang SS, Liu H, Li Y, Xu L (2012) A novel reductase from Candida albicans for the production of ethyl (S)-4-chloro-3-hydroxybutanoate. Biosci Biotechnol Biochem 76(6):1210–1212. doi:10.1271/bbb.120048
Greenberg JR, Price NP, Oliver RP, Sherman F, Rustchenko E (2005) Candida albicans SOU1 encodes a sorbose reductase required for l-sorbose utilization. Yeast 22(12):957–969. doi:10.1002/yea.1282
Karanewsky DS, Badia MC, Ciosek CP Jr, Robl JA, Sofia MJ, Simpkins LM, DeLange B, Harrity TW, Biller SA, Gordon EM (1990) Phosphorus-containing inhibitors of HMG-CoA reductase. 1,4-[(2-arylethyl)hydroxyphosphinyl]-3-hydroxy-butanoic acids: a new class of cell-selective inhibitors of cholesterol biosynthesis. J Med Chem 33(11):2952–2956
Kizaki N, Yasohara Y, Hasegawa J, Wada M, Kataoka M, Shimizu S (2001) Synthesis of optically pure ethyl (S)-4-chloro-3-hydroxybutanoate by Escherichia coli transformant cells coexpressing the carbonyl reductase and glucose dehydrogenase genes. Appl Microbiol Biotechnol 55(5):590–595
Wang LJ, Li CX, Ni Y, Zhang J, Liu X, Xu JH (2011) Highly efficient synthesis of chiral alcohols with a novel NADH-dependent reductase from Streptomyces coelicolor. Bioresour Technol 102(14):7023–7028. doi:S0960-8524(11)00560-8
Yamamoto H, Matsuyama A, Kobayashi Y (2002) Synthesis of ethyl (R)-4-chloro-3-hydroxybutanoate with recombinant Escherichia coli cells expressing (S)-specific secondary alcohol dehydrogenase. Biosci Biotechnol Biochem 66(2):481–483
Yamamoto H, Mitsuhashi K, Kimoto N, Matsuyama A, Esaki N, Kobayashi Y (2004) A novel NADH-dependent carbonyl reductase from Kluyveromyces aestuarii and comparison of NADH-regeneration system for the synthesis of ethyl (S)-4-chloro-3-hydroxybutanoate. Biosci Biotechnol Biochem 68(3):638–649
Yamamoto H, Mitsuhashi K, Kimoto N, Kobayashi Y, Esaki N (2005) Robust NADH-regenerator: improved alpha-haloketone-resistant formate dehydrogenase. Appl Microbiol Biotechnol 67(1):33–39. doi:10.1007/s00253-004-1728-x
Ye Q, Cao H, Yan M, Cao F, Zhang Y, Li X, Xu L, Chen Y, Xiong J, Ouyang P, Ying H (2010a) Construction and co-expression of a polycistronic plasmid encoding carbonyl reductase and glucose dehydrogenase for production of ethyl (S)-4-chloro-3-hydroxybutanoate. Bioresour Technol 101(17):6761–6767. doi:S0960-8524(10)00576-6
Ye Q, Li X, Yan M, Cao H, Xu L, Zhang Y, Chen Y, Xiong J, Ouyang P, Ying H (2010b) High-level production of heterologous proteins using untreated cane molasses and corn steep liquor in Escherichia coli medium. Appl Microbiol Biotechnol 87(2):517–525. doi:10.1007/s00253-010-2536-0
Ye Q, Ouyang P, Ying H (2011) A review-biosynthesis of optically pure ethyl (S)-4-chloro-3-hydroxybutanoate ester: recent advances and future perspectives. Appl Microbiol Biotechnol 89(3):513–522. doi:10.1007/s00253-010-2942-3
Yu MA, Wei YM, Zhao L, Jiang L, Zhu XB, Qi W (2007) Bioconversion of ethyl 4-chloro-3-oxobutanoate by permeabilized fresh brewer’s yeast cells in the presence of allyl bromide. J Ind Microbiol Biotechnol 34(2):151–156. doi:10.1007/s10295-006-0179-z
Acknowledgments
This work was supported by the National Basic Research Program of China (No. 2011CBA00804) and the Innovation Fund for Doctor Degree Dissertation in Nanjing University of Technology (No. BSCX200809).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Cai, P., An, M., Xu, L. et al. Development of a substrate-coupled biocatalytic process driven by an NADPH-dependent sorbose reductase from Candida albicans for the asymmetric reduction of ethyl 4-chloro-3-oxobutanoate. Biotechnol Lett 34, 2223–2227 (2012). https://doi.org/10.1007/s10529-012-1029-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-012-1029-x