Abstract
To develop an economical industrial medium, untreated cane molasses (UCM) was tested as a carbon source for fermentation culturing of Escherichia coli. To test the industrial application of this medium, we chose a strain co-expressing a carbonyl reductase (PsCR) and a glucose dehydrogenase (BmGDH). Although corn steep liquor (CSL) could be used as an inexpensive nitrogen source to replace peptone, yeast extract could not be replaced in E. coli media. In a volume of 40 ml per 1-l flask, a cell concentration of optical density (OD600) 15.1 and enzyme activities of 6.51 U/ml PsCR and 3.32 U/ml BmGDH were obtained in an optimized medium containing 25.66 g/l yeast extract, 3.88 g/l UCM, and 7.1% (v/v) CSL. When 3.88 g/l UCM was added to the medium at 6 h in a fed-batch process, the E. coli concentration increased to OD600 of 24, and expression of both PsCR and BmGDH were twofold higher than that of a batch process. Recombinant cells from batch or fed-batch cultures were assayed for recombinant enzyme activity by testing the reduction of ethyl 4-chloro-3-oxobutanoate to ethyl (S)-4-chloro-3-hydroxybutanoate (CHBE). Compared to cells from batch cultures, fed-batch cultured cells showed higher recombinant enzyme expression, producing 560 mM CHBE in the organic phase with a molar yield of 92% and an optical purity of the (S)-isomer of >99% enantiomeric excess.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Escherichia coli is the most widely used prokaryotic system for the production of heterologous proteins because it is easily genetically manipulated, and inexpensive to culture, and can produce substantial quantities of protein in a single day (Studier 2005). Culture medium is a key factor in maximizing heterologous protein expression in E. coli. To reduce medium costs, corn steep liquor (CSL), a by-product of wet milling corn for starch, is extensively used in industrial fermentation as a component of microorganism culture medium. It provides a rich but inexpensive source of nutrients, vitamins, and minerals (Ji et al. 2009). Molasses, an agro-industrial by-product that contains abundant sugar, amino acids, organic acids, inorganic compounds, and vitamins, is often used as a carbon source in E. coli fermentation. For example, pretreated beet molasses gave high cell yields and high recombinant protein production when used in E. coli fermentation (Çalik and Levent 2009a). Although fermentation of E. coli using pretreated molasses as a carbon source has been described (Agarwal et al. 2006, 2007; Çalik and Levent 2009b), few studies have addressed the use of untreated molasses in E. coli medium. Engineered E. coli might be assumed to be unable to metabolize untreated molasses, possibly because they cannot hydrolyze sucrose, a major molasses component.
Ethyl (S)-4-chloro-3-hydroxybutanoate [(S)-CHBE] is a precursor to enantiopure intermediates in the production of chiral drugs, including cholesterol-lowering hydroxymethylglutaryl-CoA reductase inhibitors such as Pfizer’s Lipitor (atorvastatincalcium; Asako et al. 2009). This is the largest selling prescription medicine worldwide, with annual sales of over $10 billion (Lee and Park 2009). We previously described a novel carbonyl reductase (PsCR) from Pichia stipitis that produces (S)-CHBE at >99% enantiomeric excess (Ye et al. 2009a). Analysis of the amino acid sequence of PsCR revealed <60% identity to similar known reductases (Ye et al. 2009b). An efficient and balanced NADPH regeneration system was constructed for the production of (S)-CHBE (Ye et al., under review). To develop an economical and industrial medium for our strain, we used untreated cane molasses (UCM) as a carbon source and CSL as a nitrogen source for E. coli fermentation. We also developed a fed-batch process using medium with UCM that could achieve high cell concentrations and high expression of recombinant enzymes. Finally, asymmetric reduction of ethyl 4-chloro-3-oxobutanoate (COBE) to (S)-CHBE using recombinant E. coli in a water/butyl acetate system was examined.
Materials and methods
Cells, medium, and cultivation
E. coli Rosetta (DE3)pLysS co-expressing PsCR from P. stipitis (CBS 6054) and BmGDH (DSM 2894, E. coli Rosetta pBP) with a balanced and efficient NADPH regeneration system was used (Ye et al. 2009, under review). The strain was maintained on Luria–Bertani (LB) agar slants with yeast extract 5 g/l, tryptone 10.0 g/l, NaCl 10.0 g/l, and agar 15 g/l. Cells from newly prepared slants were inoculated into 1-l flasks containing MC medium which was, per liter, 1.5 g Na2HPO4⋅12H2O, 1.0 g NaH2PO4⋅2H2O, 1.0 g NaCl, and 2 g lactose. Ampicillin (100 µg/ml) and chloramphenicol (34 µg/ml) were added for plasmid maintenance.
UCM components
UCM was obtained from the Guilin sugar refinery (Guangxi, PRC). The composition, by weight, was 45% sucrose, 10% converted sugars (5% glucose and 5% fructose), 3.3% other carbohydrates, 1.5% crude protein, 0.5% crude fat, 5.1% ash, 2.6% salt, 6.9% metal ions, and 25% water.
Utilization of different carbon and nitrogen sources by recombinant E. coli
LB medium was supplemented with either 10 g/l of either glucose, fructose, sucrose, lactose, a sugar mixture, or with the equivalent of 10 g/l of sugar from UCM. Different mixtures of nitrogen sources were added to MC medium containing 5 g/l UCM. Fermentation medium was initially pH 7.0. For fermentation experiments, 2% seed culture was added to 60 ml medium in a 1-l flask at 37°C for 16 h at 220 rpm.
Cultivation in flasks
Different culture volumes (40–100 ml/l) were tested for recombinant E. coli Rosetta pBP fermentation. Cells were grown in MC medium containing molasses (5 g/l), CSL (5%, v/v), and yeast extract (5 g/l). For different yeast extract concentrations, cells were grown in 40 ml in a 1-l flask in MC medium with molasses (5 g/l), CSL (5%, v/v), and yeast extract (5–25 g/l). For different molasses concentrations, cells were grown as above in MC containing CSL (5%, v/v), yeast extract (20 g/l), and molasses from 2.5 to 20 g/l. For different CSL concentrations, cells were grown as above in MC medium containing yeast extract (5 g/l), molasses (5 g/l), and CSL from 1–10% (v/v).
Central composite design
Central composite design was employed to optimize three media components, yeast extract, UCM, and CSL, with the goal of enhancing the expression of the recombinant enzymes. The three independent factors were studied at five different levels (−1.68, −1, 0, +1, and +1.68) for a set of 20 experiments (Table 1). The factors were coded according to the following equation:
where x i was the coded factor, X i was the real factor, X 0 was the value of X i at the center point, and ΔX was the step change value.
The behavior of the system was explained by the following second-order polynomial equation:
where Y was the predicted response, β 0 was the intercept, x i and x j were the coded independent factors, β i was the linear coefficient, β ii was the quadratic coefficient, and β ij was the interaction coefficient.
Statistical analysis and experimental validation of the optimized conditions
The statistical software package STATISTICA 6.0 (StatSoft Inc., Tulsa, OK, USA) was used for the experimental design and regression analysis data. Analysis of variance was used to evaluate the model. The quality of the polynomial model equation was judged statistically using the coefficient of determination R 2, and its statistical significance was determined by an F test. The significance of the regression coefficients was determined with a t test. To validate the optimization of the medium composition, three tests were carried out under the optimized conditions and the results analyzed statistically.
Batch process and fed-batch-process cultivation with UCM as a carbon source
Batch process and fed-batch process with UCM as a carbon source were investigated. Cells were grown in 40 ml in a 1-l flask in MC medium containing yeast extract (25.66 g/l), molasses (3.88 g/l), and CSL (7.1%, v/v). For fed-batch process, 3.88 g/l UCM was added at 6 h. Recombinant cells were cultured for 20 h at 37°C.
Biocatalysis of COBE to (S)-CHBE in a two-phase system
Biocatalysis of COBE to (S)-CHBE was carried out by E. coli Rosetta pBP from batch or fed-batch cultures. Recombinant cells from 500 ml of culture were harvested by centrifugation (15,000×g, 20 min, 4°C) and washed twice with 100 mM potassium phosphate buffer (pH 6.0). A 100-ml reaction of 100 mM potassium phosphate buffer (pH 6.0), COBE (609 mM, 100 g/l), NADP (0.1 mM), glucose (1.0 M, 180 g/l), Triton X-100 (1%, v/v), and the recombinant E. coli cells was mixed with an equal volume of butyl acetate in a 250-ml reaction vessel. The reaction was stirred at 30°C, and pH was held at 6.0 with 10 M sodium hydroxide.
Enzyme assay, COBE, and CHBE analysis and other analytical methods
PsCR and GDH activities were determined spectrophotometrically as described previously (Ye et al. 2009b; Kizaki et al. 2001). The concentrations of COBE and CHBE were determined by gas chromatography and the optical purity of (S)-CHBE determined by HPLC (Ye et al. 2009a). SDS-PAGE was performed on 10% slab gels that were stained with Coomassie brilliant blue R-250 for protein. The OD of cells was measured at 600 nm (spectrophotometer Lambda 25 PerkinElmer, USA). Sugar concentrations were measured with an Agilent 1200 HPLC (Agilent, USA) with a reflective index detector using a Shodex SP0810 column (Showa Denko Co., Ltd., Tokyo, Japan). The mobile phase was water at a flow rate of 1 ml/min at 80°C.
Results
Use of different carbon sources by recombinant E. coli Rosetta pBP
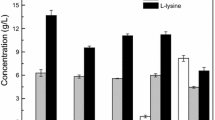
To study the feasibility of using UCM as a low cost substrate in E. coli Rosetta pBP fermentation, several different sugars were tested as initial carbon sources (Fig. 1). An E. coli cell density of OD600 of 13.0 and residual sugar concentration of 2 g/l were obtained using glucose, indicating the efficiency of glucose as a carbon source. Cell density was approximately OD600 of 11.5 using sucrose, so the cells could use sucrose as a carbon source. Little difference was observed when adding fructose or sucrose to the E. coli fermentation medium. Electronic supplementary materials, Fig. S1 shows different carbon sources with UCM. Since the inexpensive UCM consists primarily of sucrose, glucose, and fructose, a mixture of these three sugars at approximately the ratio they occur in molasses was used for fermentation. Figure 1 shows that this mixture was fermented efficiently by recombinant E. coli. In addition, the highest concentration (OD600 of 14.3) of E. coli and the lowest residual sugar (<1 g/l) were observed using UCM as a carbon source at a sugar concentration equivalent to the controls.
Effect of different carbon sources on recombinant E. coli Rosetta pBP fermentation. Cells were grown in LB medium containing 10 g sugar (glucose, fructose, sucrose, lactose, or sugar mixture) or an equivalent amount of sugar from UCM. Mixtures contained glucose, fructose, and sucrose at approximately the ratio found in molasses. For details, see “Materials and methods.” Results are means ± SEM of triplicate samples
Use of different nitrogen sources by recombinant E. coli Rosetta pBP
Because of its low price and nutritional abundance, CSL (5%, v/v) was used as a nitrogen source in MC medium containing 5 g/l UCM. Figure 2 shows the use of different nitrogen sources by recombinant E. coli, and the results indicate little difference between adding yeast extract alone and adding peptone and yeast extract to medium, with final OD600 measurements of approximately 10. PsCR activity was 3.1 U/ml and BmGDH was 1.5 U/ml, but the concentration of cells and the enzyme activities decreased dramatically in the medium containing CSL and peptone. Furthermore, almost no recombinant PsCR and BmGDH activities were observed when only CSL was used as a nitrogen source.
Effect of different nitrogen sources. For the control, no extra nitrogen source was added. Peptone and yeast extract were 20 g/l, and the concentration of CSL was 5% (v/v). For details, see “Materials and methods.” Results are means ± SEM of triplicate samples
Cultivation in flasks
Fermentation of recombinant E. coli in different liquid volumes was carried out, and the highest cell concentration (OD600 of 12.3) and maximal enzyme activities (4.1 U/ml PsCR and 1.6 U/ml BmGDH) were obtained with 40 ml in a 1-l flask (Fig. 3a). With increasing volume, cell density slowly decreased, with enzyme activities decreasing sharply above the optimum volume, with almost 75% loss of activity in 100 ml medium in a 1-l flask.
Effect of culture volume and composition on growth. a Effect of culture volume on recombinant E. coli Rosetta pBP fermentation. b Effect of yeast extract concentration. c Effect of molasses concentration. d Effect of CSL concentration. For details, see “Materials and methods.” Results are means ± SEM of triplicate samples
Since yeast extract was essential for growth and expression of recombinant E. coli, different concentrations of yeast extract were tested for the effects on cell growth. As shown in Fig. 3b, increasing yeast extract concentrations from 5 to 25 g/l markedly increased the cell density and enzyme activities. At 25 g/l yeast extract, PsCR activity was 4.7 U/ml and BmGDH was 2.4 U/ml, almost double the results for 5 g/l yeast extract. However, compared to 25 g/l yeast extract, addition of 30 g/l yeast extract had no effect on cell density or enzyme expression.
Fermentation of recombinant cells with different amounts of UCM was assayed. As shown in Fig. 3c, increase of initial cane molasses concentrations from 2.5 to 25 g/l increased cell density. Addition of 5 g/l initial cane molasses gave maximum enzyme activities, with PsCR 5 U/ml and BmGDH 2.5 U/ml. When the concentration of UCM increased from 5 to 25 g/l, the activities of the two enzymes decreased markedly. When the UCM concentration reached 25 g/l, the highest density of E. coli was obtained (OD600 of 17.4), while almost no recombinant enzyme activity was observed.
At a molasses concentration of 5 g/l, different concentrations of CSL were investigated. Figure 3d shows that PsCR (5.9 U/ml) and BmGDH (2.9 U/ml) exhibited maximal activities at 7.5 ml/l CSL and cell density of OD600 = 14.4. As CSL concentration decreased, cell concentration decreased notably. The activities of the two enzymes at 7.5 ml/l CSL were more than triple the activity yielded by 1 ml/l CSL, but cell density and enzyme expression showed little difference between 7.5 and 10 ml/l CSL.
Central composite design and experimental validation of the optimal condition
Central composite design was used to determine the optimal levels of yeast extract, UCM, and CSL and the effect of their interactions on PsCR expression. The experimental design and results are in Table 1. The results were fitted with the second-order polynomial:
where Y is the predicted response and X 1, X 2, and X3 are coded values of yeast extract, UCM, and CSL concentrations, respectively.
Table 2 shows that among the independent variables, X 21 , X 2, X 22 , and X 23 had significant effects (P < 0.05) and negative coefficients, suggesting that decreasing their concentration increased PsCR production (Eq. 3). The interactions (X 1 X 2, X 2 X 3, and X 1 X 3), however, had no significant effect on PsCR expression (P > 0.05, Table 2). In this model, R 2 was 0.9516, implying that the sample variation of 95.16% for PsCR yield could be attributed to the independent variables. The value of Adj R 2 was 0.8791, suggesting the significance of the model, since normally, a regression model with an R 2 value higher than 0.9 is considered to have a very high correlation (Ying et al 2009). The model predicted that the optimal values for the test factors of the coded units were X 1 = 0.23, X 2 = −0.56, and X 3 = 0.32 or 25.66 g/l yeast extract, 3.88 g/l UCM, and 7.1% CSL (v/v). The maximum predicted value of PsCR activity was 6.44 U/ml.
To confirm the ability of the model to predict the maximum PsCR expression, three experiments in shaking flasks in the optimized medium composition were performed. The mean value for PsCR activity was 6.51 U/ml, which was in excellent agreement with the predicted value (6.44 U/ml). The correlation between these two results verifies the validity of the response model and the existence of an optimal point. BmGDH showed an activity of 3.32 U/ml in the optimized medium at cell density of OD600 = 15.1.
Fed-batch with UCM as a carbon source
Figure 3c shows that at molasses concentrations >5 g/l, the cell density of E. coli Rosetta pBP was increased markedly, but the activities of the two recombinant enzymes decreased dramatically. Therefore, a fed-batch process with molasses was developed. Figure 4 shows a comparison of batch process and fed-batch process cultures using UCM as a carbon source. SDS-PAGE analysis of recombinant PsCR and BmGDH expression over time is shown in Fig. 5. At 6 h, sugar was almost depleted, and 3.88 g/l UCM was added to the medium. From 6 to 11 h, cells in fed-batch medium grew faster than cells in batch medium, although the expression of recombinant enzymes in the batch medium culture was higher than in the fed-batch process. At 11 h, the supplemental sugar was almost depleted in the fed-batch medium (data not shown). Enzyme expression increased significantly and was higher than in the non-fed-batch medium from 11 to 20 h, while at 16 h, PsCR and BmGDH activities showed maximum values of 13.5 and 6.4 U/ml, which were twofold higher than in the batch culture. At 20 h, the activities of the two enzymes decreased, with almost 40% loss of original activity in both the batch and fed-batch process. A maximum cell density of OD600 of 24 was obtained for the fed-batch process at 14 h and for the batch process at 11 h.
Comparison of batch process and fed-batch process with UCM as a carbon source. For details, see “Materials and methods.” Results are means ± SEM of triplicate samples
COBE reduction with recombinant E. coli Rosetta pBP in a two-phase system reaction
Water/butyl acetate two-phase reactions were carried out using recombinant E. coli Rosetta pBP with an initial COBE concentration of 609 mM. At 0.5 h, 120 mM CHBE was obtained from E. coli cells from fed-batch fermentation, while 60 mM CHBE was achieved by E. coli cells from batch fermentation. The maximum reaction rate was observed for both E. coli culture from 0 to 2 h (Fig. 6). Recombinant E. coli cells from the batch fermentation gave a 502-mM product with a molar yield of 82%. In contrast, cells from fed-batch fermentation produced 560 mM CHBE in the organic phase with a molar yield of 92% and an optical purity of the (S)-isomer of >99% enantiomeric excess. In this system, the total turnover number, defined as moles (S)-CHBE formed per mole NADP+, was 5,600 mol/mol.
Reduction of ethyl 4-chloro-3-oxobutanoate (COBE) to ethyl (S)-4-chloro-3-hydroxybutanoate (CHBE) using recombinant batch culture or fed-batch culture cells in a water/butyl acetate system. For details, see “Materials and methods.” Results are means ± SEM of triplicate samples
Discussion
Several strains can ferment molasses, including Aspergillus niger, Zymomonas mobilis, Saccharopolyspora erythraea, Actinobacillus succinogenes, and E. coli (Ikram-ul et al. 2004; Cazetta et al. 2005; Xiao et al. 2007; El-Enshasy et al. 2008; Liu et al. 2008). However, most bacteria, including engineered E. coli strains, cannot use untreated molasses as a carbon source. This work focused on the fermentation of engineered E. coli strain using UCM as a carbon source. Calik and co-workers systematically studied the fermentation of E. coli BL21 (DE3) using pretreated beet molasses and concluded that untreated molasses contains too much sucrose to be metabolized by E. coli BL21 (DE3), unless the sucrose is hydrolyzed to glucose and fructose (Çalik and Levent 2009a). However, several nutrient substances are degraded by these chemical processes (Liu et al. 2008). In this study, a recombinant E. coli Rosetta strain grew well on sucrose, a major ingredient of UCM (Fig. 1). A sucrose hydrolase gene that would allow utilization of sucrose as the sole carbon source may be present in the genome of this E. coli Rosetta strain (Jahreis et al. 2002). Direct catabolism of untreated molasses would be beneficial because pretreatment steps for molasses including ultrafiltration or sucrose hydrolysis would not be required. Thus, E. coli Rosetta can use untreated molasses as a carbon source. Furthermore, molasses has obvious advantages as a carbon source over other sugar mixtures for generating E. coli density and minimizing residual sugar (Fig. 1). It contains abundant sugar, amino acids, organic acids, inorganic compounds, and vitamins, while metal ions such as calcium, sodium, iron, magnesium, or copper and suspended colloids in UCM are apparently not toxic or inhibitory for E. coli Rosetta cells. This was different from several reports that untreated molasses contained undesirable substances that may negatively affect growth (Xiao et al. 2007). To the best of our knowledge, this is the first investigation of fermentation of engineered E. coli using untreated molasses, and the results are favorable for its use for industrial production.
CSL is an inexpensive by-product of cornstarch production (Lee et al. 2003a; Agarwal et al. 2006). Yeast extract and peptone are expensive when used for scale production, so to develop a cheap and industrial medium, CSL was added to the medium instead of yeast extract or peptone. CSL was demonstrated to be an inexpensive nitrogen source that could replace peptone (Fig. 2). Very poor growth of our E. coli strain, with reduced expression of PsCR and BmGDH, was observed in medium containing CSL as the sole nitrogen source because of deficiency of essential micronutrients. However, when CSL was supplemented with yeast extract, dense growth and enhanced expression of the recombinant enzymes was observed, consistent with several reports (Lee et al. 2003b). Previous studies also found that yeast extract is an essential component in E. coli growth medium (Agarwal et al. 2006).
Oxygen is an important factor for growth and protein expression of E. coli. We found an optimal liquid volume for E. coli Rosetta pBP fermentation of 40 ml per 1-l flask. With increasing liquid volume, cells density decreased slowly and enzyme activities decreased sharply. If the culture becomes sufficiently dense that oxygen consumption exceeds the rate of aeration, oxygen limitation triggers complex regulatory responses that adjust the metabolic capacities of the cell to oxygen, carbon, and energy availability (Studier 2005). Therefore, the higher the aeration rate, the higher is the culture density, from 40 to 100 ml per 1-l flask.
Central composite design and analysis were used to determine the optimal levels of three factors (yeast extract, UCM, and CSL), all of which significantly influenced PsCR expression. The close agreement between the experimental results and the theoretical values predicted by the model equation (Table 1) indicated that the regression model was useful for analyzing the trends in the responses. The R 2 value reflected a very good fit between the observed and predicted responses, indicating that the regression model could be used to analyze trends of the responses. Regression coefficient significance was determined using a t test, and results are in Table 2. P values were used to check the significance of each coefficient to determine the pattern of mutual interactions between the best factors. The smaller the P value, the bigger is the significance of the corresponding coefficient (Li et al. 2007). The optimal culture condition for heterologous protein production was MC medium containing 25.66 g/l yeast extract, 3.88 g/l UCM, and 7.1% (v/v) CSL. Using these optimized conditions, the activity of PsCR was significantly enhanced from 4.01 to 6.51 U/ml, which was 1.8-fold higher than the original medium. Since the activity of BmGDH was less than half the activity of PsCR (Ye et al. 2009, under review), accordingly, the activity of BmGDH increased from 1.6 to 3.32 U/ml, or 2.0-fold higher than with the original medium.
Another factor with a profound growth influence is the availability of molasses. We found that 3.88 g/l untreated molasses consisting of 2.2 g/l sugar was the optimal initial concentration for E. coli expression of recombinant enzymes. This differed from another E. coli BL21 (DE3) strain as reported by Çalik and Levent (2009a). This study found that the highest cell concentration and recombinant benzaldehyde lyase activity of E. coli were obtained in medium containing 30 g/l pretreated beet molasses consisting of 7.5 g/l glucose and 7.5 g/l fructose. Figure 3a shows that a high concentration of molasses was beneficial for cell growth, but PsCR and BmGDH activities decreased substantially at high molasses concentrations because of the inhibitory effect of the glucose in molasses. Glucose can support growth to high density, but excessive glucose prevents induction by lactose of the lac operon and T7 expression system (Peti and Page 2007). Hence, to achieve high cell concentration and maximize heterologous protein expression in E. coli, a fed-batch process was developed. In this strategy, when the sugar was consumed at 6 h, 3.88 g/l UCM was added. Glucose caused catabolite repression and inducer exclusion, preventing the uptake of lactose by lactose permease, the product of the lac Y gene which is thought to be the only means of lactose uptake in wild-type cells (Gombert and Kilikian 1998). Hence, compared with the batch process, the PsCR and BmGDH activities were low when glucose in the medium blocked induction by lactose from 6 to 11 h in the fed-batch fermentation, while growth was rapid. When the glucose was depleted at 11 h, a high concentration of cells was achieved in the fed-batch culture, while lactose could be taken up by the small amount of lacY present in uninduced cells. Conversion to allolactose, the natural lac inducer, occurred through β-galactosidase, the product of the lacZ gene (Huber et al. 1976; Neubauer et al. 1992). Therefore, high-level expression was seen for both PsCR and BmGDH, which were twofold higher than in the batch process. These observations are consistent with previous studies showing that glucose prevents lactose from inducing the lac operon (Shiloach et al. 1996; Pflug et al. 2007; Çalik and Levent 2009b). Furthermore, the decrease in recombinant enzyme activity at 20 h may result from degradation by proteases in E. coli (Makrides 1996).
Since COBE and CHBE were both inhibitory for enzymes and cells in an aqueous monophase system, biosynthesis of (S)-CHBE using recombinant E. coli was examined using a water/organic solvent diphasic system. Butyl acetate was used as an organic phase for efficient partitioning of COBE and CHBE (Shimizu et al. 1990). In this reaction system, NADPH was regenerated by coupling glucose oxidation with BmGDH. The resulting gluconate was neutralized with NaOH to maintain an initial pH of 6.0 at which purified PsCR exhibited maximum activity (Ye et al. 2009b). Cells from fed-batch cultures reduced COBE to (S)-CHBE more rapidly than cells from batch cultures (Fig. 6). However, as CHBE increased to 300 mM, the velocity of the reactions decreased in both systems. Cells from the fed-batch culture produced 560 mM CHBE in the organic phase with a molar yield of 92% and an optical purity of the (S)-isomer of >99% enantiomeric excess. Compared to an aqueous monophase system used in our previous study, a higher concentration of product and TTN of CHBE forming NADP+ was obtained with the water/butyl acetate system.
References
Agarwal L, Isar J, Meghwanshi GK, Saxena RK (2006) A cost effective fermentative production of succinic acid from cane molasses and corn steep liquor by Escherichia coli. J Appl Microbiol 100:1348–1354
Agarwal L, Isar J, Dutt K, Saxena RK (2007) Statistical optimization for succinic acid production from E. coli in a cost-effective medium. Appl Biochem Biotechnol 142:158–167
Asako H, Shimizu M, Itoh N (2009) Biocatalytic production of (S)-4-bromo-3-hydroxybutyrate and structurally related chemicals and their applications. Appl Microbiol Biotechnol 84:397–405
Çalik P, Levent H (2009a) Effects of pretreated beet molasses on benzaldehyde lyase production by recombinant Escherichia coli BL21 (DE3) pLySs. J Appl Microbiol 107:1536–1541
Çalik P, Levent H (2009b) Effects of pulse feeding of beet molasses on recombinant benzaldehyde lyase production by Escherichia coli BL21 (DE3). Appl Microbiol Biotechnol 85:65–73
Cazetta ML, Celligoi MAPC, Buzato JB, Scarmino IS, da Silva RSF (2005) Optimization study for sorbitol production by Zymomonas mobilis in sugar cane molasses. Process Biochem 40:747–751
El-Enshasy HA, Mohamed NA, Farid MA, El-Diwany AI (2008) Improvement of erythromycin production by Saccharopolyspora erythraea in molasses based medium through cultivation medium optimization. Bioresour Technol 99:4263–4268
Gombert AK, Kilikian BV (1998) Recombinant gene expression in Escherichia coli cultivation using lactose as inducer. J Biotechnol 60:47–54
Huber RE, Kurz G, Wallenfels K (1976) A quantitation of the factors which affect the hydrolase and transgalactosylase activities of β-galactosidase (E. coli) on lactose. Biochem 15:1994–2001
Ikram-ul H, Ali S, Qadeer MA, Iqbal J (2004) Citric acid production by selected mutants of Aspergillus niger from cane molasses. Bioresour Technol 93:125–130
Jahreis K, Bentler L, Bockmann J, Hans S, Meyer A, Siepelmeyer J, Lengeler JW (2002) Adaptation of sucrose metabolism in the Escherichia coli wild-type strain EC3132. J Bacteriol 184:5307–5316
Ji XJ, Huang H, Du J, Zhu JG, Ren LJ, Li S, Nie ZK (2009) Development of an industrial medium for economical 2,3-butanediol production through co-fermentation of glucose and xylose by Klebsiella oxytoca. Bioresour Technol 100:5214–5218
Kizaki N, Yasohara Y, Hasegawa J, Wada M, Kataoka M, Shimizu S (2001) Synthesis of optically pure ethyl (S)-4-chloro-3-hydroxybutanoate by Escherichia coli transformant cells coexpressing the carbonyl reductase and glucose dehydrogenase genes. Appl Microbiol Biotechnol 55:590–595
Lee SH, Park OJ (2009) Uses and production of chiral 3-hydroxy-γ-butyrolactones and structurally related chemicals. Appl Microbiol Biotechnol 84:817–828
Lee PC, Lee SY, Hong SH, Chang HN (2003a) Batch and continuous cultures of Mannheimia succiniciproducens MBEL55E for the production of succinic acid from whey and corn steep liquor. Bioprocess Biosyst Eng 26:63–67
Lee PC, Lee SY, Hong SH, Chang HN, Park SC (2003b) Biological conversion of wood hydrolysate to succinic acid by Anaerobiospirillum succiniciproducens. Biotechnol Lett 25:111–114
Li JS, Ma CQ, Ma YH, Li Y, Zhou W, Xu P (2007) Medium optimization by combination of response surface methodology and desirability function: an application in glutamine production. Appl Microbiol Biotechnol 74:563–571
Liu YP, Zheng P, Sun ZH, Ni Y, Dong JJ, Zhu LL (2008) Economical succinic acid production from cane molasses by Actinobacillus succinogenes. Bioresour Technol 99:1736–1742
Makrides SC (1996) Strategies for achieving high-level expression of genes in Escherichia coli. Microbiol Rev 60:512–538
Neubauer P, Hofmann K, Holst O, Mattiasson B, Kruschke P (1992) Maximizing the expression of a recombinant gene in Escherichia coli by manipulation of induction time using lactose as inducer. Appl Microbiol Biotechnol 36:739–744
Peti W, Page R (2007) Strategies to maximize heterologous protein expression in Escherichia coli with minimal cost. Protein Expr Purif 51:1–10
Pflug S, Richter SM, Urlacher VB (2007) Development of a fed-batch process for the production of the cytochrome P450 monooxygenase CYP102A1 from Bacillus megaterium in E. coli. J Biotechnol 129:481–488
Shiloach J, Kaufman J, Guillard AS, Fass R (1996) Effect of glucose supply strategy on acetate acumulation, gowth, and rcombinant potein poduction by Escherichia coli BL21 (λDE3) and Escherichia coli JM 109. Biotechnol Bioeng 49:421–428
Shimizu S, Kataoka M, Katoh M, Morikawa T, Miyoshi T, Yamada H (1990) Stereoselective reduction of ethyl 4-chloro-3-oxobutanoate by a microbial aldehyde reductase in an organic solvent–water diphasic system. Appl Environ Microbiol 56:2374–2377
Studier FW (2005) Protein production by auto-induction in high-density shaking cultures. Protein Expr Purif 41:207–234
Xiao ZJ, Liu PH, Qin JY, Xu P (2007) Statistical optimization of medium components for enhanced acetoin production from molasses and soybean meal hydrolysate. Appl Microbiol Biotechnol 74:61–68
Ye Q, Yan M, Xu L, Cao H, Li ZJ, Chen Y, Li SY, Ying HJ (2009a) A novel carbonyl reductase from Pichia stipitis for the production of (S)-4-chloro-3-hydroxybutanoate ethyl. Biotechnol Lett 31:537–542
Ye Q, Yan M, Yao Z, Xu L, Cao H, Li ZJ, Chen Y, Li SY, Bai JX, Xiong J, Ying HJ, Ouyang PK (2009b) A new member of the short-chain dehydrogenases/reductases superfamily: purification, characterization and substrate specificity of a recombinant carbonyl reductase from Pichia stipitis. Bioresour Technol 100:6022–6027
Ying H, Chen X, Cao H, Xiong J, Hong Y, Bai J, Li Z (2009) Enhanced uridine diphosphate N-acetylglucosamine production using whole-cell catalysis. Appl Microbiol Biotechnol 84:677–683
Acknowledgments
This work was supported by the Major Basic Reserach Program of China (grant no. 2009CB724700), National Key Technology RD Program (grant no. 2008BAI63B07). Qi YE was supported by the Innovation Fund for Doctoral Dissertation of Nanjing University of Technology. We would like to express appreciation to the referees for suggestions on improving our paper. We also thank Prof. Hong Xu and Dr. Sha Li for revising the paper.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 72 kb)
Rights and permissions
About this article
Cite this article
Ye, Q., Li, X., Yan, M. et al. High-level production of heterologous proteins using untreated cane molasses and corn steep liquor in Escherichia coli medium. Appl Microbiol Biotechnol 87, 517–525 (2010). https://doi.org/10.1007/s00253-010-2536-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-010-2536-0