Abstract
The mitogen-activated protein kinase Hog1 gene (Kmhog1) was isolated from Kluyveromyces marxianus strain NBRC 1777 by degenerate PCR and genome walking, and then disrupted to construct a mutant strain hog1∆. The mutant was now more sensitive to acetic acid and its growth was nearly completely inhibited by 0.5 M NaCl (97%) and 10 mM H2O2 (93%) as compared with the wild-type cells. However, neither strain grew at 47°C. Kmhog1 may thus be required for adaptation to acetic acid, osmotic, and oxidative stress but is not involved in thermotolerance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
With characters such as thermotolerance, a broad substrate spectrum, high growth rates, and less tendency to ferment when exposed to excess sugar, Kluyveromyces marxianus has been investigated for various biotechnological applications (Fonseca et al. 2008). Among these applications, large scale production of bioethanol at elevated temperature has received much attention (Boyle et al. 1997; Singh et al. 1998; Faga et al. 2010), as a 5°C increase in the fermentation temperature can greatly affect production costs (Abdel-Banat et al. 2010). However, to obtain a high productivity of ethanol fermentation, the ideal host would tolerate other multiple stresses including osmo-stress. Therefore, understanding the mechanism underlying the response of K. marxianus to environmental stresses may guide the design of rational strategies to increase ethanol production.
In Saccharomyces cerevisiae, as in all other eukaryotic cells, one important component of the signaling network is the high-osmolarity/glycerol (HOG), mitogen-activated protein kinase (MAPK) pathway. Upon hyperosmotic shock, the MAPK-HOG pathway is rapidly initiated through a phosphorylation cascade culminating in activation of the MAPK Hog1 by dual phosphorylation (Hohmann 2009). The activated Hog1 plays direct and indirect roles in a broad transcriptional response (O’Rourke and Herskowitz 2004) leading to increased production of intracellular glycerol as a counterbalancing osmolyte. In addition, Hog1 is also required for adaptation to other stress conditions, such as oxidative stress (Bilsland et al. 2004), arsenite (Thorsen et al. 2006), cold stress (Panadero et al. 2006; Hayashi and Maeda 2006) and acetic acid stress (Mollapour and Piper 2007). Consequently, Hog1 plays an important role in orchestrating the adaptation of yeast cells to several kinds of environmental stresses.
Little information is, however, available about Hog1 in K. marxianus. Therefore, we cloned the Hog1 gene from K. marxianus using degenerate PCR and genome walking. And the role of Hog1 in the cell response to acetic acid, osmotic stress, oxidative stress, and elevated temperature was investigated by creating a hog1∆ mutant through targeted gene inactivation.
Materials and methods
Strains and culture conditions
Kluyveromyces marxianus NBRC 1777 (Hong et al. 2007) was cultured at 37°C in 25 ml YPD medium (pH 6.0), containing 2% (w/v) glucose, 2% (w/v) peptone, and 1% (w/v) yeast extract, in 250 ml shake-flasks. Escherichia coli DH5α was used as a host cell for the propagation of plasmids.
Responses of K. marxianus to acetic acid, osmotic and oxidative stresses and temperature were tested in YPD medium as indicated in Figs. 2, 3, and 4. Experiments were carried out in triplicate and data presented are the average of three independent cultivations.
DNA manipulation
DNA manipulation in E. coli, as well as genomic DNA extraction from K. marxianus, was performed according to standard methods. For the amplification of the core sequence of K. marxianus Hog1 gene, 50 ng total DNA was used as template in 50 μl reaction mixture containing 5 μl 10× PCR buffer (10 mM Tris/HCl; pH: 9.0, 50 mM KCl, 1.5 mM MgCl2), 2 μM each of forward (Kmc-F) and reverse (Kmc-R) primers and 2.5 units of Taq DNA polymerase. PCR cycling profiles were 1 cycle at 94°C for 3 min, 30 cycles of 94°C for 1 min, 54°C for 1 min and 72°C for 1 min, followed by a final extension step at 72°C for 7 min. Restriction and modification enzymes were used following the recommendations of the manufacturer (Takara Biotechnology). DNA fragments were purified from agarose gels using the AcyPrep DNA gel extraction kit (Axygen Biosciences). Plasmid DNA was isolated using the AxyPrep plasmid miniprep kit (Axygen Biosciences).
Disruption of Kmhog1 gene in K. marxianus
Plasmid pHK-1 was constructed to disrupt Kmhog1 gene. The Kmhog1 gene, starting—800 nucleotides (nt) 5′ to the start codon and ending 1,538 nt 3′ to the start codon, was amplified from genomic DNA of K. marxianus using primers Km-F and Km-R (Table 1), and cloned into pMD19-T Easy (Takara Biotechnology) vector, yielding pMD19-Kh. The bacterial kanamycin gene (kan from Tn903) that confers resistance to geneticin (G418 sulfate) in K. marxianus was amplified from pPIC9K (Invitrogen) using primers Kan-F and Kan-R (Table 1). Then, the purified PCR product was digested with EcoRI/XbaI and inserted into the Kmhog1 coding region in pMD19-Kh through EcoRI/XbaI site, generating pHK-1.
Plasmid pHK-1 was linearized with SpeI, transformed into K. marxianus strains NBRC 1777 using the electroporation protocol according to the “Pichia Expression Kit” (Invitrogen). Candidate clones in which the Kmhog1 gene was disrupted were selected on YPD agar plates containing G418 (200 μg ml−1), and the insertion of kan gene into the chromosome of K. marxianus hog1∆ by double crossovers was confirmed by colony screening using PCR, and further verified by sequence analysis of the PCR product.
Analytical methods
Growth was monitored from the OD600 values that were then correlated to the cell dry weight. DNA sequence was analyzed and alignment was performed in BLAST web (http://www.ncbi.nlm.nih.gov/Blast/).
Results and discussion
Cloning of the K. marxianus hog1 gene
For isolation of full-length hog1 gene in K marxianus, the core sequence was first amplified. Through alignment of ten published hog1 genes from yeast species including K. lactis and S. cerevisiae, the conserved regions were identified for designing degenerate primers Kmc-F and Kmc-R (Table 1). A 0.9 kb PCR product was amplified using the primers Kmc-F and Kmc-R from the genomic DNA of K marxianus NBRC 1777. The PCR product was then cloned and sequenced. Alignment of the amino acid sequence of the PCR product with K. lactis and S. cerevisiae homologues showed more than 90% homology, suggesting that the PCR product contained the core sequence of the K marxianus hog1 gene. Then, the core sequence was used to design 6 gene-specific primers (Km5GW1, 2, 3 and Km3GW1, 2, 3) to obtain its upstream and downstream sequences using a Genome Walking Kit (Takara Biotechnology) according to the manufacturer’s instruction. After amplification using primer pairs Km5GW3/AP1 and Km3GW3/AP2, two DNA fragments, 1.1 and 0.6 kb, respectively, were obtained. The PCR products were purified and cloned into pMD19-T vector and then sequenced to finally assemble the full-length K marxianus hog1 gene Kmhog1. The nucleotide sequence was submitted to GenBank and assigned accession number EU625288.
The Kmhog1 gene encodes a putative protein of 465 amino acids with a predicted molecular weight of 52.8 kDa and a theoretical isoelectric point (pI) of 5.21. Homology studies on the deduced amino acid sequence revealed a high identity to MAP kinases of K. lactis (93%), Candida albicans (77%), S. cerevisiae (81%), and Zygosaccharomyces rouxii (91%). The sequence encompassing amino acids 28–36 of the encoded polypeptide perfectly matches the conserved ATP-binding signature of protein kinases. And a TGY motif, characteristic of hyperosmolarity-activated MAP kinases (Cano and Mahadevan 1995), is found at amino acids 173–175.
Construction of the Kmhogl-negative mutant
Plasmid pHK-1 was linearized with SpeI, transformed into K. marxianus strains NBRC 1777 to obtain its hog1 deletion derivative, K. marxianus hog1∆ (Fig. 1a). The positive transformants were selected for G418 resistance, and the integration of the kan gene was confirmed by PCR. As shown in Fig. 1b, a larger PCR product (3.4 kb) containing the inserted kan gene from hog1∆ (lane 2) using the primer pair Km-F/Km-R demonstrated the insertion in the hog1 gene. And the disruption of the hog1 gene was verified by the presence of the PCR products in hog1∆ with the primer pairs Kan-F/Kan-R (lane 4) and Km-F/Kan-R (lane 6), in contrast to no amplification from the wild-type strain (lane 3, 5). Then, the insertion of kan gene was confirmed using primers Kan-F and Kan-R. A 1.4-kb fragment was amplified from chromosomal DNA of hog1∆ (lane 8), while no amplification could be observed from the original strain NBRC 1777 (lane 7). Finally, the amplified PCR product was sequenced to further confirm the correct insertion of the kan gene in chromosome (data not shown).
Schematic presentation of targeted Kmhog1 inactivation via homologous recombination and confirmation of Kmhog1 gene disruption by PCR. a In plasmid pHK-1, the bacterial kanamycin gene (kan, dark) was inserted between 495 nt and 813 nt 3′ to the start codon of Kmhog1; Kmhog1 coding sequence (gray) in wild-type K. marxianus NBRC1777 was disrupted by gene replacement, resulting in the insertion of kan gene and thus G418 resistance of mutant strain K. marxianus hog1∆. b Confirmation of targeted gene disruption in mutant strains by PCR analysis. Lane 1 wild-type strain NBRC1777: PCR product size is 2.3 kb with the primer pair Km-F/Km-R, Lane 2 mutant strain hog1∆: 3.4 kb PCR product obtained with the primer pair Km-F/Km-R, Lane 3 NBRC1777: no amplification with the primer pair Kan-F/Km-R, Lane 4 hog1∆: 2.1 kb PCR product obtained with the primer pair Kan-F/Km-R, Lane 5 NBRC1777: no amplification with the primer pair Km-F/Kan-R, Lane 6 hog1∆: 2.7 kb PCR product obtained with the primer pair Km-F/Kan-R, Lane 7 NBRC1777: no amplification with the primer pair Kan-F/Kan-R, Lane 8 hog1∆: 1.4 kb PCR product obtained with the primer pair Kan-F/Kan-R, M Marker 3 (Dongsheng Biotech)
Effect of Kmhogl gene disruption on stress response of K. marxianus
To investigate the putative involvement of Kmhog1 in the integration and transduction of various environmental stress signals, we analyzed the acetic acid tolerance, osmotolerance, oxidative stress response, and sensitivity to high temperature of the K. marxianus hog1∆.
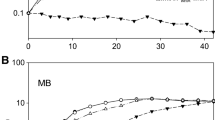
Acetic acid, as a byproduct of biomass hydrolysis and cell metabolite, is a major inhibitor in ethanol fermentation and Hog1 is required for yeast adaptation to acetic acid stress (Mollapour and Piper 2007). The role of Hog1 in acetate-stress resistance of K. marxianus was investigated by measuring growth kinetics of wild-type and hog1∆ cells exposed to increasing concentrations of acetate from 0.1 to 0.4% (v/v) (Fig. 2). In comparison to the wild-type strain, the acetic acid was more inhibitory to hog1∆ cells, lower acetate levels could result in comparable growth inhibition in hog1∆ cells, showing Hog1 is required for cell resistance to acetic acid stress in K. marxianus. When exposed to 0.5 M NaCl, about 97% growth inhibition could be observed in hog1∆ cells, as compared to the wild-type strain (Fig. 3). Similarly, we observed that the hog1∆ mutant showed sensitivity to oxidative stress, as indicated by its marked growth inhibition (93%) in 10 mM H2O2 compared to the growth of wild-type cells (Fig. 3). K. marxianus NBRC1777 can grow at 50°C (Hong et al. 2007) but, as shown in Fig. 4, neither the wild-type nor mutant cells grew well at 47°C.
Conclusions
To explore the involvement of Kmhogl in the cell response to various stress conditions, we inactivated Kmhogl gene by targeted gene disruption in K. marxianus strains NBRC 1777. Acetic acid, NaCl and H2O2 stressors strongly blocked the growth of the hog1∆ mutant, whereas elevated temperature showed similar growth inhibition to both wild-type and hog1∆ strains. Kmhogl is thus a HOG-type MAPK gene with multi-stress signaling functions, and a deeper understanding of the molecular mechanisms of Hog1-mediated cell adaptation to multiple stresses may increase the ability to delineate the genetic engineering of K. marxianus strains and process strategies aiming at increasing ethanol fermentation performance.
References
Abdel-Banat BMA, Hoshida H, Ano A, Nonklang S, Akada R (2010) High-temperature fermentation: how can processes for ethanol production at high temperatures become superior to the traditional process using mesophilic yeast? Appl Microbiol Biotechnol 85:861–867
Bilsland E, Molin C, Swaminathan S, Ramne A, Sunnerhagen P (2004) Rck1 and Rck2 MAPKAP kinases and the HOG pathway are required for oxidative stress resistance. Mol Microbiol 53:1743–1756
Boyle M, Barron N, McHale AP (1997) Simultaneous saccharification and fermentation of straw to ethanol using the thermotolerant yeast strain Kluyveromyces marxianus imb3. Biotechnol Lett 19:49–51
Cano E, Mahadevan LC (1995) Parallel signal processing among mammalian MAPKs. Trends Biochem Sci 20:117–122
Faga BA, Wilkins MR, Banat IM (2010) Ethanol production through simultaneous saccharification and fermentation of switchgrass using Saccharomyces cerevisiae D5A and thermotolerant Kluyveromyces marxianus IMB strains. Bioresour Technol 101:2273–2279
Fonseca GG, Heinzle E, Wittmann C, Gombert AK (2008) The yeast Kluyveromyces marxianus and its biotechnological potential. Appl Microbiol Biotechnol 79:339–354
Hayashi M, Maeda T (2006) Activation of the HOG pathway upon cold stress in Saccharomyces cerevisiae. J Biochem 139:797–803
Hohmann S (2009) Control of high osmolarity signalling in the yeast Saccharomyces cerevisiae. FEBS Lett 583:4025–4029
Hong J, Wang Y, Kumagai H, Tamaki H (2007) Construction of thermotolerant yeast expressing thermostable cellulase genes. J Biotechnol 130:114–123
Mollapour M, Piper PW (2007) Hog1 mitogen-activated protein kinase phosphorylation targets the yeast Fps1 aquaglyceroporin for endocytosis, thereby rendering cells resistant to acetic acid. Mol Cell Biol 27:6446–6456
O’Rourke SM, Herskowitz I (2004) Unique and redundant roles for HOG MAPK pathway components as revealed by whole-genome expression analysis. Mol Biol Cell 15:532–542
Panadero J, Pallotti C, Rodríguez-Vargas S, Randez-Gil F, Prieto JA (2006) A downshift in temperature activates the high osmolarity glycerol (HOG) pathway, which determines freeze tolerance in Saccharomyces cerevisiae. J Biol Chem 281:4638–4645
Singh D, Banat IM, Nigam P, Marchant R (1998) Industrial scale ethanol production using the thermotolerant yeast Kluyveromyces marxianus IMB3 in an Indian distillery. Biotechnol Lett 20:753–755
Thorsen M, Di Y, Tangemo C, Morillas M, Ahmadpour D, Van der Does C, Wagner A, Johansson E, Boman J, Posas F, Wysocki R, Tamas MJ (2006) The MAPK Hog1p modulates Fps1p-dependent arsenite uptake and tolerance in yeast. Mol Biol Cell 17:4400–4410
Acknowledgments
We are grateful to National Natural Science Foundation of China (20976050) and National Basic Research Program (973 Program 2007CB714306) for their financial supports to this research.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Qian, J., Qin, X., Yin, Q. et al. Cloning and characterization of Kluyveromyces marxianus Hog1 gene. Biotechnol Lett 33, 571–575 (2011). https://doi.org/10.1007/s10529-010-0458-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-010-0458-7