Abstract
Most high-affinity phosphate transporter genes (OsPTs) in rice were highly induced in roots when phosphate was depleted. OsPT1, however, was highly expressed in primary roots and leaves regardless of external phosphate concentrations. This finding was confirmed histochemically using transgenic rice plants that express the GUS reporter gene under the control of the OsPT1 promoter, which exhibited high GUS activity even in the phosphate sufficient condition. Furthermore, transgenic rice plants overexpressing the OsPT1 gene accumulated almost twice as much phosphate in the shoots as did wild-type plants. As a result, transgenic plants had more tillers than did wild-type plants, which is a typical physiological indicator for phosphate status in rice.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Phosphate (Pi) is a constituent of key molecules such as ATP, nucleic acids, and phospholipids, which play crucial roles in energy transfer, metabolic regulation, and protein activation. Plants have evolved a number of different adaptive strategies to maximize Pi acquisition under Pi-limiting conditions. Biochemical and metabolic adaptations, including the secretion of organic acids, RNases, and acid phosphatases from roots, enhance the availability of Pi in the rhizosphere (Poirier and Bucher 2002). Developmental responses, such as increases in the root/shoot ratio, root hair proliferation and increased length, and lateral root number, increase the surface area for absorption, thereby increasing uptake efficiency (Poirier and Bucher 2002).
Many high-affinity Pi transporter genes have been isolated from Arabidopsis, potato, Medicago, tomato, maize and barley. Most of these genes are expressed predominantly in roots and are induced by Pi depletion, indicating that they are involved in the acquisition of Pi through the roots when low external Pi concentrations exist in the soil. However, the recent completion of plant genome projects has allowed for the detailed description of members of this family in Arabidopsis (Misson et al. 2004) and in rice (Paszkowski et al. 2002). Fusions of the promoters of AtPT family members to the GUS or GFP reporter genes have shown that these transporters are active in a variety of developmental stages and in various organs, including shoot tissues (Misson et al. 2004). These results suggest that the high-affinity Pi transporters of Arabidopsis have distinct roles in the internal translocation of Pi throughout the plant, in addition to the primary uptake of Pi from the soil.
In this study, we investigated the expression patterns of nine high-affinity Pi transporters from Oryza sativa in response to Pi depletion. Of this set of genes, the OsPT1 gene clearly showed a different response to Pi deficiency, including a different expression pattern. Using transgenic rice plants that overexpressed the OsPT1 gene, we demonstrated that OsPT1 is a bona-fide Pi transporter that is essential for the accumulation of high concentrations of Pi in the shoot.
Materials and methods
Plant materials and growth conditions
Plant materials and hydroponics growth conditions were prepared as described by Hur et al. (2007). A field experiment was performed in the Field Experiment Station of the Yeongnam Agricultural Research Institute (Milyang, South Korea) from June to October 2006. Twenty-day-old wild-type and transgenic seedlings (overexpressing OsPT1) were transplanted at 15- to 20-cm spacing into three plots. Two different fertilization conditions were applied to the soil of each plot before transplanting: normal fertilization (N:P:K = 4.5:4.5:4.0 kg/10a) or fertilization without Pi (N:K = 4.5:4.0 kg/10a). Three plants per plot were randomly harvested to investigate the contents of various nutrient elements at three growth stages (tillering, heading and harvesting).
Cloning of the OsPT1 gene from rice, vector construction, and generation of transgenic rice plants
Using an oligonucleotide primer set (For: TGTCTAGACATGGCGGGAGGGCAGCTC, Rev: GCTCTAGAATTACTTCGGGTAGGCCGCC), the coding region of the OsPT1 gene was amplified from rice genomic DNA by PCR. The PCR product was sequenced and cloned into pCAMBIA1300-35S for expression under the control of the CaMV 35S promoter. The OsPT1 promoter region (including 1.2 kb upstream from the OsPT1 start codon) was amplified from rice genomic DNA by PCR, using an oligonucleotide primer set that added EcoRI and NcoI sites (For: TGGGGATCCGGCCACCATTAGCAAGTG, Rev: CCCGCCATGGCTTCCCAACTCTTTGAG). After digestion of the PCR product with EcoRI and NcoI, the promoter region was fused with the initiating ATG of the β-glucuronidase (GUS) reporter gene in the binary vector pCAMBIA1381Z. The expression constructs were introduced into A. tumefaciens (EHA105) by tri-parental mating. Rice plants were transformed with A. tumefaciens as described by Hiei et al. (1994). Homozygote transgenic lines overexpressing OsPT1 gene were selected by hygromycin resistance (50 mg/l) and used for field tests.
RNA extraction and northern blot analyses
Total RNA was isolated from rice plants using TRIzol reagent (Invitrogen). Total RNA (10 μg) was separated by electrophoresis on a 1.2% (w/v) denaturing formaldehyde agarose gel, blotted onto nylon transfer membranes (Amersham), and crosslinked with UV light. The membranes were hybridized at 65°C for 12 h with a [32P]-dATP-labeled probe (Amersham) in a solution containing 7% (w/v) SDS, 1.5× SSPE, 100 g PEG (8,000 MW)/l, 250 mg/l heparin, and 10 ml herring sperm DNA/l (10 mg/ml). After hybridization, the membrane was washed twice with 2× SSC and 0.1% SDS for 30 min each at 65°C, and then twice with 0.1× SSC and 0.1% SDS for 10 min each at room temperature.
Histochemical analysis of GUS expression
Histochemical analysis of GUS activity was performed essentially as described by Jefferson et al. (1987). Whole rice transgenic plants were grown hydroponically in nutrient solutions supplemented with 1 mM Pi (Pi+) or 35 μM Pi (Pi−) for 2 days, then incubated at 37°C for 12 h in an X-gluc staining solution. The resulting GUS expression patterns were assessed using a dissecting microscope.
Measurement of Pi concentrations and other nutrient elements
Shoots of wild-type and transgenic plants grown under two different fertilization conditions were sampled at three different growth stages, dried at 70°C for 2 days, and ground with a Willey mill (Tomas Scientific, USA); biomass was determined as dry weight. For determination of total nitrogen contents, the plant samples (0.5 g) were digested with 5 ml of concentrated HClO4–H2SO4 and processed by the Kjeldahl protocol (Auto Kjeldahl, FOSS 2300, USA). Pi concentrations were measured with the phosphomolybdenum blue reaction of the Lancaster protocol. Potassium contents were measured by inductively-coupled plasma spectroscopy (ICP, Perkin Elmer Optimer 3300).
Results and discussion
Expression patterns of the OsPT genes in rice plants
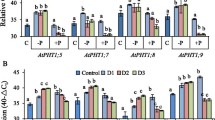
To examine the tissue-specific expression of the OsPT genes in Pi-starved rice plants, we performed northern blot analyses with total RNA isolated from the roots and shoots of rice plants grown in hydroponics with 1 mM (P+) or 35 μM (P−) Pi. The expression of OsPT2, OsPT4, OsPT6, OsPT7, OsPT8, and OsPT12 was upregulated predominantly in roots exposed for two days to Pi-deficient conditions (Fig. 1). These results were consistent with the expression patterns of other plant high-affinity phosphate transporter genes, and strongly suggested that these OsPT genes function as major high-affinity Pi transporters involved in the uptake of Pi from the rhizosphere under Pi-limited conditions.
The expression patterns of OsPT genes in rice plants in response to Pi starvation. Total RNA isolated from roots (R) and shoots (S) of rice plants grown hydroponically in nutrient solutions supplemented with 1 mM Pi (P+) or 35 μM Pi (P−) were hybridized with probes specific for the OsPT genes. Equal loading of RNA was demonstrated by ethidium bromide staining of the ribosomal RNAs
In contrast, the OsPT1 gene was expressed constitutively in the shoot regardless of Pi concentration, although OsPT1 expression in roots was slightly elevated by Pi deficiency (Fig. 1). The distinct expression pattern of OsPT1 was confirmed with callus suspension cells derived from rice seeds that were incubated in the presence of different concentrations of Pi (μM). After two days of incubation, the callus suspension cells were harvested, total RNA was extracted, and the transcriptional levels of OsPT1 and OsPT6 genes were investigated. A strong inverse relationship between the Pi concentration and the expression of the OsPT6 gene was observed. However, OsPT1 was not significantly regulated by Pi concentration (Supplementary Fig. 1). Similarly, LePT1 (Liu et al. 1998) and StPT1 (Leggewie et al. 1997), which are tomato and potato high-affinity Pi transporters, respectively, are constitutively expressed not only in roots but also in other plant organs, including leaves, stems, tubers, and flowers. The distinct spatial and temporal expression patterns of these Pi transporter genes imply that they may function beyond the acquisition of Pi from the soil.
Histochemical analysis of the OsPT1 promoter in rice
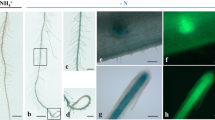
To determine the expression pattern and organ specificity of OsPT1, an OsPT1 genomic fragment containing a 1.5 kb promoter with a start codon was fused in frame with the β-glucuronidase (GUS) reporter gene; the resulting construct was used to make transgenic rice plants. A histochemical analysis of GUS expression in T2 transgenic plants indicated that OsPT1 was ubiquitously expressed in leaves, stems, and roots of plants grown hydroponically even in the phosphate sufficient condition. In contrast, a control plant containing OsPT4 promoter-GUS fusion showed a weak GUS activity in the leaf (Fig. 2). These results were consistent with the northern blot analysis. Specifically, stronger GUS expression was observed in primary roots as compared to lateral roots and root hairs of the transgenic plants containing OsPT1 promoter-GUS fusion (Fig. 2c), where Pi uptake takes place in response to Pi deficiency. This observation suggests a possible role for OsPT1 in the acquisition and mobilization of Pi at the basal level, irrespective of the Pi concentration in the rhizosphere.
Tissue specificity of the OsPT1 promoter as determined by GUS staining in transgenic rice plants. Transgenic rice plants transformed with the OsPT1 or OsPT4 promoter-GUS fusion were grown hydroponically in nutrient solutions supplemented with 1 mM Pi (P+) for 3 weeks and stained for GUS activity. The OsPT4 promoter-GUS fusion transgenic line exhibited GUS activity marginally in the junction of leaf and stem (a). In contrast, strong GUS activity was detected ubiquitously in the leaf (a), stem (b), and roots (c) of rice transgenic plants containing the OsPT1 promoter-GUS fusion
Overexpression of the OsPT1 gene in rice enhances Pi acquisition
To test the possibility that OsPT1 can contribute to Pi acquisition in the shoots independent of Pi concentrations, we generated transgenic rice plants that overexpressed the OsPT1 gene under the control of the CaMV 35S promoter (OsPT1-OX). Expression levels of the OsPT1 gene in the transgenic lines were confirmed by northern blot analysis, and independent OsPT1-OX lines (#1 and #2) were selected for further study (Fig. 3a). Interestingly, as shown in Fig. 3b, c, the OsPT1-OX plants had better root growth and more tillers than wild-type plants; these phenotypes were strongly correlated with Pi content and considered to be indicators of Pi status in plants (Yi et al. 2005) Therefore, it is likely that OsPT1-OX plants accumulated more Pi than wild-type plants.
Effect of the overexpression of OsPT1 on plant growth. (a) Northern blot analysis of OsPT1 for wild-type and independent transgenic lines transformed with OsPT1 under the control of the CaMV 35S promoter. Ethidium bromide staining of ribosomal RNA was shown at the bottom and used as a control for equal loading. Growth phenotypes of wild-type and transgenic rice plants grown in pots containing acid clay soil (135 mg Pi/kg soil) with normal fertilization (N:P:K = 4.5:4.5:4.0 kg/10a) for 5 weeks (b) or 12 weeks (c)
Using the wild-type and OsPT1-OX line #2 homozygote plants grown under different fertilization conditions in the field, we measured the Pi content in the shoots of plants randomly taken during three growth stages (tillering, heading, and harvesting). At each growth stage, the OsPT1-OX plants accumulated almost twice as much Pi as did wild-type plants, independent of fertilization conditions (Fig. 4). Fertilization with Pi (P; N:P:K = 4.5:4.5:4.0 kg/10a) slightly increased Pi content in wild-type plants, but had no detectable effect on the OsPT1-OX plants, indicating that the existing Pi content in the soil (135 mg Pi/kg soil) was high enough for the OsPT1-OX plants to take up sufficient Pi (Fig. 4b). These results suggested that OsPT1 is a ubiquitous transporter that functions in Pi translocation within the whole plant rather than in direct Pi uptake from the soil, consistent with an increased number of tillers in the transgenic lines. Furthermore, the transgenic plants had generated 20% more panicles than wild-type plants at the harvest stage (Table 1).
Effect of the overexpression of OsPT1 on Pi content in shoots. Three plants per plot were randomly taken for determination of Pi content at three growth stages (tillering, heading and harvesting). (a) Pi content of plants fertilized without Pi (N:K = 4.5:4.0 kg/10a), (b) Pi content of plants fertilized with Pi (N:P:K = 4.5:4.5:4.0 kg/10a)
However, the height of the transgenic plants during harvesting time was 30% shorter than that of wild-type plants, independent of fertilization conditions (Table 1). It is likely that the increased acquisition of Pi by overexpression of OsPT1, without a concomitant increase in other nutrients such as nitrogen (N) and potassium (K; Supplementary Table 1) could induce a deficiency of the other nutrients by a dilution effect. Because the critical deficiency contents of N and K increase as the P content increases, the OsPT1-OX plants may be under a condition of N and P deficiency, leading to shorter height as compared to wild-type. Optimal ratios between nutrients in plants are often as important as absolute contents (Marschner 1995).
Previous studies have indicated that increased expression of high-affinity Pi transporter genes (Pht1) enhances Pi acquisition from Pi-limited environments, as shown by increased Pi uptake rates in BY2 suspension cells and in rice suspension cells overexpressing ARAth;Pht1;1 (AtPT1) (Mitsukawa et al. 1997) or HORvu;Pht1;1 (HvPT1) (Rae et al. 2003), respectively. However, the successful use of high-affinity Pi transporter genes to enhance Pi acquisition in plants has not previously been reported (Rae et al. 2004). In this study, we provided molecular evidence demonstrating that the distinct class of the OsPT1 gene was able to enhance the efficiency of Pi acquisition by the plant. We expect this finding to contribute to the development of transgenic plants able to adapt to Pi-deficient conditions by manipulating OsPT1, in combination with other nutrient transporter genes. New transgenic plants might lead to improved crop yields and productivity, with a lower input of P fertilizer, thereby protecting the environment.
References
Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6:271–282
Hur YJ, Lee HG, Jeon EJ, Lee YY, Nam MH, Yi G, Eun MY, Nam J, Lee JH, Kim DH (2007) A phosphate starvation-induced acid phosphatase from Oryza sativa: phosphate regulation and transgenic expression. Biotechnol Lett 29:829–835
Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6:3901–3907
Leggewie G, Willmitzer L, Riesmeier JW (1997) Two cDNAs from potato are able to complement a phosphate uptake-deficient yeast mutant: identification of phosphate transporters from higher plants. Plant Cell 9:381–392
Liu H, Trieu AT, Blaylock LA, Harrison MJ (1998) Cloning and characterization of two phosphate transporters from Medicago truncatula roots: regulation in response to phosphate and to colonization by arbuscular mycorrhizal (AM) fungi. Mol Plant Microbe Interact 11:14–22
Marschner H (1995) Mineral nutrition of higher plants, 2nd edn. Academic Press, London
Misson J, Thibaud MC, Bechtold N, Raghothama K, Nussaume L (2004) Transcriptional regulation and functional properties of Arabidopsis Pht1;4, a high-affinity transporter contributing greatly to phosphate uptake in phosphate deprived plants. Plant Mol Biol 55:727–741
Mitsukawa N, Okumura S, Shirano Y, Sato S, Kato T, Harashima S, Shibata D (1997) Overexpression of an Arabidopsis thaliana high-affinity phosphate transporter gene in tobacco cultured cells enhances cell growth under phosphate-limited conditions, Proc Natl Acad Sci USA 94:7098–7102
Paszkowski U, Kroken S, Roux C, Briggs SP (2002) Rice phosphate transporters include an evolutionarily divergent gene specifically activated in arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci USA 99:13324–13329
Poirier Y, Bucher M (2002) Phosphate transport and homeostasis in Arabidopsis. In: Somerville CR, Meyerowitz EM (eds) The Arabidopsis book, American Society of Plant Biologists, Rockville. www.aspb/publications/arabidopsis/
Rae AL, Jarmey JM, Mudge SR, Smith FW (2004) Over-expression of a high-affinity phosphate transporter in transgenic barley plants does not enhance phosphate uptake rates. Funct Plant Biol 31:141–148
Yi K, Wu Z, Zhou J, Du L, Guo L, Wu Y, Wu P (2005) OsPTF1, a novel transcription factor involved in tolerance to phosphate starvation in rice. Plant Physiol 138:2087–2096
Acknowledgements
This work was supported by grants to J. Nam from the Korea Research Foundation of the Korean Government (KRF-2003-015-C00640).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Seo, HM., Jung, Y., Song, S. et al. Increased expression of OsPT1, a high-affinity phosphate transporter, enhances phosphate acquisition in rice. Biotechnol Lett 30, 1833–1838 (2008). https://doi.org/10.1007/s10529-008-9757-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-008-9757-7