Abstract
Using National Longitudinal Survey of Youth (NLSY) fertility variables, we introduce and illustrate a new genetically-informative design. First, we develop a kinship linking algorithm, using the NLSY79 and the NLSY-Children data to link mothers to daughters and aunts to nieces. Then we construct mother–daughter correlations to compare to aunt–niece correlations, an MDAN design, within the context of the quantitative genetic model. The results of our empirical illustration, which uses DF Analysis and generalized estimation equations (GEE) to estimate biometrical parameters from NLSY79 sister–sister pairs and their children in the NLSY-Children dataset, provide both face validity and concurrent validity in support of the efficacy of the design. We describe extensions of the MDAN design. Compared to the typical within-generational design used in most behavior genetic research, the cross-generational feature of this design has certain advantages and interesting features. In particular, we note that the equal environment assumption of the traditional biometrical model shifts in the context of a cross-generational design. These shifts raise questions and provide motivation for future research using the MDAN and other cross-generational designs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Behavior genetics (BG) is certainly among the most design-oriented of disciplines. The prototype BG research design involves comparing correlations among monozygotic (MZ) to those among dizygotic (DZ) twins. A variant of the twin study was the original biometrical design, proposed by Galton in 1876 (Plomin et al. 1990, p. 28 provides discussion). Other standard designs include the adoption design, in which adoptive sibling correlations are compared to biological sibling correlations (or parent–child correlations are compared across adoptive and biological parents) and pedigree analysis, based on kinship relationships across an extended family structure. Such designs use natural manipulations across different levels of kinship that vary on levels of both genetic and environmental factors.
Of course the use of the term “design” within this context refers to a quasi-experimental design, or a design-of-nature (Shadish et al. 2002). Research manipulations and random assignment to groups are ethically, pragmatically, and biologically impossible, and so BG researchers rely on clever applications of quasi-experimental design principles to reduce threats to internal and external validity (we note that some biological assignment processes can be assumed random, which provides useful leverage in eliminating threats to validity). Part of the BG disciplinary agenda has been to specify design weaknesses and other threats to internal validity, and to address those. It is, apparently, not an accident that Sir Ronald Fisher, the father of modern research design (Fisher 1935), was also the father of modern quantitative genetics (Fisher 1930).
In addition to twin and adoption designs, other innovative and unusual design structures have been used to support biometrical modeling. The MZ-twins-raised-apart design (Bouchard et al. 1990) is a simple design yet difficult to use, because of the natural limitation on the numbers of such MZ twin pairs. However, the payoff is substantial, because such MZ twin correlations directly estimate h 2 or heritability (under assumptions of the quantitative genetic model). Designs that compare same-sex and opposite-sex kinship pairs can be used to test for sex-linked genetic etiology (Neale and Cardon 1992; Van Hulle et al. 2007). Another new and innovative design is the virtual twin design, using correlations of approximately same-age adoptive siblings raised in the same family. They share 100% of their environment, including age effects, but 0% of their trait-relevant genes (Segal et al. 2006); their kinship correlation directly estimates c 2 or shared family environmental variance (under assumptions of the model).
Family designs include naturally-occurring kinship relationships among children within a household, including adoptive siblings, cousins, half-siblings, full-siblings, and twins. Ideally, such designs are approximately representative of the occurrence of those kinship categories in the population, resulting in substantial increases in external and ecological validity compared to the more specialized designs. The National Longitudinal Survey of Youth 1979 survey (NLSY79) and the children of the NLSY79 mothers (NLSYC) have supported a number of studies based on these types of family design structures (e.g., Rodgers et al. 1999; Rodgers et al. 1994b; van den Oord and Rowe 2000; D’Onofrio et al. 2007). The flexible and useful design features of the NLSY will be used in the current research as well.
Another powerful design structure is the Children-of-Twins (COT) design (see Nance and Corey 1976; Heath et al. 1985 for early references). Here correlations from the offspring of twins are compared to one another (e.g., D’Onofrio et al. 2003; Silberg and Eaves 2004), which controls for many threats to validity caused by family background differences. In many COT analyses, the children of MZ and DZ twins discordant for a trait are compared to one another (Harden et al. 2007). COT designs have been used with a number of different phenotypes (Gottesman and Bertelsen 1989; Magnus et al. 1985; Mendle et al. 2006). D’Onofrio et al. delineated the logic underlying various outcome patterns and extended the design in several important ways relevant to the presentation in this paper. First, they used quantitative instead of binary measures from the parents’ generation. Second, they expanded the COT design to include cross-generational as well as within-generation comparisons.
To be biometrically informative, a BG design usually requires multiple kinship categories; the correlations from those different categories are compared to one another, informing and allowing parameter estimation in the context of the standard biometrical model (Falconer 1979; Jinks and Fulker 1970). Or, in unusual circumstances (e.g., the MZ-twins-raised-apart design, the virtual twin design), a single kinship category is biometrically informative under the standard model assumptions. But across the different design structures reviewed above, a common feature is that the correlations and kinship structures are defined within-generation (except for pedigree analysis and comparisons of adoptive/biological children to their parents). This feature is primarily related to data availability. Cross-generational kinship structures are highly informative of biometrical processes, if data to support such structures can be obtained.
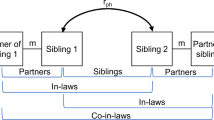
In this paper, we propose and illustrate a new BG design that has never been used, as far as we are aware. The design takes advantage of cross-generational kinship structure, and compares two different types of kinship categories, mothers–daughters and aunt–nieces. We call this a Mother–Daughter–Aunt–Niece (MDAN) design, and we illustrate and apply the design using data from the NLSY79 and NLSYC databases. The MDAN design is a special case of a broader conceptual framework of cross-generational designs that could be used to estimate biometrical models, given data availability. A Father–Son–Uncle–Nephew (FSUN) design would be the male counterpart of the MDAN design. Or, more broadly, both of these are special cases of a Children-of-Siblings (COS) design, which also includes the COT design as a special case. Past applications of the COT design have used cross-generational kinship structure to create kinship links, but the correlations/covariances that are used to estimate the model are within-generation measures from the children generation. In the COS design, the correlations may be either within- or cross-generational correlations, or both (also see D’Onofrio et al. 2003).
In the first section of this paper, we describe the MDAN design, and illustrate it using female respondents from the NLSY79 and NLSYC data. In the next section, we analyze female fertility precursors based on the MDAN design. Following, we discuss validity issues, including how some of the standard assumptions shift (especially the equal environments assumption) within the context of an MDAN design. In addition, we discuss MDAN design extensions, which motivate a research program with broad implications.
The MDAN design, applied to the NLSY data
The data
Before new or unusual design structures are useful within BG, there must be data available to estimate biometrical parameters within the context of the design. The NLSY79 and NLSYC data are based on a complex survey design that allows innovative within- and cross-generational structures to be created using standard data management procedures.
On December 31, 1978, a random sample of 3,000 U.S. households was drawn, conditional on the household containing one or more adolescents in the age range of 14–21 years. Households containing poor white and minority youth were oversampled, and a military sample was obtained. The original NLSY79 survey had 12,686 respondents between ages 14–22 when the interviews occurred in 1979. The respondents were followed yearly from 1979 until 1986, and then bi-annually; the 2004 data were available at the time of this writing, and the 2006 NLSY79 data have been collected, but not yet released.
Starting in 1986, a new survey was begun of all biological children born to females in the NLSY79 sample, the NLSYC sample (Chase-Lansdale et al. 1991). Since then, on a biannual basis, these children have been administered various assessments, including measures of the home environment, cognitive development, and risk taking. Measures of timing of puberty and age at first intercourse will be used in the current research, because of instrumentation links across the two generations. By 2004 the NLSY79 females were 39–47 years old; thus, virtually all childbearing had been completed (over 95%, according to Center for Human Resource Research 2006). In earlier rounds, however, an important and substantial source of bias has existed, caused by the children in the NLSYC survey being born to disproportionately young mothers. Obviously, this type of bias still exists when measures from adolescents are used (e.g., age at menarche), because even though virtually all of the NLSY79 female childbearing is complete, many of their children have not reached adolescence.
One of the powerful features of the NLSY79/NLSYC data is the availability of measures in which NLSY79 mothers and NLSYC children were assessed using (approximately) the same instruments. Our measures of fertility precursors (age at menarche; age at first intercourse) are almost identical across the two surveys. Although instrumentation differences are an important threat to validity in most cross-generational research, this threat can be minimized for certain measures in the NLSY79/NLSYC cross-generational fertility research design.
The NLSY79 and NLSYC kinship links
Because of the household sampling feature of the design, there are thousands of sibling pairs, of various types, within the NLSY79 files. Further, because all biological children of NLSY79 mothers comprise the NLSYC data, there are also thousands of sibling pairs available in the NLSYC files. However, the NLSY survey questions have never explicitly distinguished categories of siblings. There are questions, though, that implicitly identify these kinship links, including ones accounting for the NLSY79 respondents’ yearly living status in relation to their biological mother and father. Using these types of questions, NLSY79 kinship links have been constructed (Rodgers 1996; Rodgers et al. 1999) that categorized 90% of the kinship pairs in one version and 65% in another. A number of past publications have been based on these kinship pairs; the smaller 65% sample has usually shown validity advantages.
NLSYC sibling pairs are twins, full, or half-siblings, because they share a biological mother. At least three different research teams have used linking algorithms to define NLSYC kinship pair links (see Baydar and Greek 2001; Rodgers et al. 1994a; van den Oord and Rowe 1997). Those earlier links, however, were constructed when NLSY79 female fertility was incomplete, and the NLSYC sample was much smaller. Recently, we updated this effort, and defined objective kinship links among the 11,438 biological children in the 2002 NLSYC database, resulting in identified levels of genetic relatedness for 12,532 kinship pairs, almost 100% of the available links (Rodgers et al. 2005). Both NLSY79 and NLSYC kinship linking files are publicly available from the first author.
These two sets of kinship links are required to define and demonstrate the MDAN design with the NLSY. We use those two sets of within-generational links to create cross-generational links for mothers–daughters and aunt–nieces. In our analyses within this paper, we will only use the cross-generational links. Research is currently in progress to capitalize on elaborate kinship structure available within the NLSY data files to use in the MDAN (and broader COS) designs.
The measures of fertility precursors
Recent work in the demography and biometrical literature has studied fertility and fertility precursors. Research has been devoted to whether there is reliable genetic variance underlying fertility-related variables (see Kohler et al. 2006; Rodgers et al. 2000; Rodgers and Kohler 2003), and the relationship of such findings to Fisher’s Fundamental Theorem of Natural Selection (the FTNS; see Fisher 1930 for specification; Rodgers et al. 2001a, for discussion). The NLSY has played an important role in this research program (e.g., Doughty and Rodgers 2000; Rodgers et al. 2007; Rodgers et al. 1999; Bard and Rodgers 2006). Several NLSY variables have been investigated using within-generational analyses, including age at menarche and age at first intercourse.
There are several advantages to using variables from the NLSY that have been the basis of previous research. First, the previous results can be used to evaluate concurrent validity. Second, the psychometric properties of these variables are well-know from previous research. Third, current results can be interpreted in the context of previous findings, and so the results have more status than simply to validate the new design proposed within this paper.
The NLSY79 females reported age at menarche in 1984, when they were 19–26 years old. The question was re-asked in 1985 for those not answering in 1984. Age at first intercourse was assessed for all NLSY79 respondents at least once in 1984, 1985, and 1986. For most females in 1984 and 1985, the question was asked in both years, permitting computation of a test–retest reliability measure, r = .84 (N = 4,801). These two phenotypes are self-reported. If there are multiple indicators that differ we take the mean of those as our dependent variable values.
The NLSYC females were first asked their age at menarche in 1986, and then all non-reporters (missing in 1986, or who aged into the appropriate assessment during this period) were asked the question between 1992 until 2004. 604 respondents were asked age at menarche twice. The correlation between these responses—an estimate of test-retest reliability—is r = .65. This is a somewhat low reliability for this type of response, although 75% of the sample gave responses at the two different time points that were at or within 1 year of one another. For all who did not respond in a previous survey year, age at first intercourse was asked during survey administration every other year from 1986 until 2004.
Previous research has established the reliability and validity of retrospective indicators of age at menarche and first intercourse. Wording for both measures was virtually identical across the two generations. Further, the age structure was similar (though not identical) across the two generations. For example, the NLSY79 females were 19–26 when they first reported retrospective age at menarche, whereas the NLSYC females had their first opportunity at age 15 (though the majority did not first respond until age 16 or later). Age at menarche has been established in previous research as a highly memorable event that most women can reliably and validly report many years after its occurrence (Brooks-Gunn et al. 1987; Damon et al. 1969; Moffitt et al. 1992). Reliability/validity studies of age at first intercourse can be found in Rodgers et al. (1982, 1992), Siegel et al. (1998), and Upchurch et al. (2002).
Constructing the MDAN links
There are several options that must be defined in creating MDAN kinship links. We used a form of the design that provides strong control over unobserved heterogeneity caused by family background. Mothers and aunts are linked by design; substantial control of background heterogeneity is achieved by using only sister–sister pairs from the mother/aunt generation. Thus, all mother–daughter pairs have (at least) one matching aunt–niece pair in which the mothers and aunts grew up in the same household. This process references a third generation, the original households of the mothers/aunts. In terms of control of bias due to endogeneity and selection, this is the type of environmental control provided by the COT design as well (see D’Onofrio et al. 2003).
Within sister–sister pairs, one or the other sister was required to have a female child (a daughter) old enough to have reported age at menarche. There were 932 NLSY79 sister–sister pairs (at least one of whom had an eligible daughter), from 755 separate families. We limited our sister–sister database to the 552 sister–sister pairs who were identifiably twins, full siblings, or ambiguous siblings. (Note that using half-siblings or cousin pairs would result in a slightly different design, e.g., a mother–daughter–aunt–half–niece design.) When these NLSY79 mothers who had age-at-menarche data were linked to the NLSYC daughters who had age-at-menarche data, the database contained 498 mother–daughter pairs. When these same sister–sister links were used to link NLSY79 sisters to their sisters’ daughters (their nieces), that database contained 524 aunt–niece pairs. We present in Table 1 a flow chart of how these sample sizes were obtained from the original samples. All of the links had a match on the age-at-menarche question, by construction; most also had a match on age-at-first-intercourse question. We call all of our respondent pairs mother–daughter or aunt–niece pairs, but there slight variants of these categories among these kinship pairs. Mother–daughters share 50% of their genes, whereas typical aunt–nieces share on average 25% of their genes. If the aunts came from sister–sister pairs who were twins of unknown zygosity (R = .75), we assigned an aunt–niece genetic coefficient of R = .375. If the aunts came from sister–sister pairs of ambiguous relatedness (either full or half-siblings, R = .375), the matching aunt–niece pair was assigned R = .1875. More complex linking approaches in relation to these kinship pairs is ongoing, and will be publicly available in the future. The MDAN cross-generational kinship links used in this study may be obtained by request from the first author. Other broader MDAN links will be similarly available in the future.
Biometrical analyses
There are several methods to estimate biometrical parameters from genetically informative samples. Each uses the information in kinship links to define expected correlations/covariances within a model, which is estimated by optimizing the fit between those parameter estimates and empirical observations using an objective loss function (e.g., maximum likelihood estimation (MLE) or least squares estimation). The typical parameters-of-interest that are estimated from a biometrical model are different sources of variance, the proportion of shared environmental variance (c 2), and the proportion of nonshared environmental variance (which is usually confounded with measurement error, the combination of which is designated e2). Models that focus on these three sources of variance are often called ACE models (A = Additive genetic variance, C = Common environmental variance, E = unique variance/measurement error; Neale and Cardon 1992). If the shared environmental component is dropped, the model becomes an AE model, and if the genetic variance is dropped, it becomes a CE model. Originally, correlations were compared to estimate these parameters (see Plomin 1990, for formulas). More powerful analytic estimation routines were developed based on structural equations modeling (SEM; e.g., Loehlin 1989). Mx was developed (Neale and Cardon 1992) as a SEM package dedicated to BG analysis.
DF Analysis (DeFries and Fulker 1985) is a simple yet powerful approach to estimating biometrical parameters, based on multiple regression and least squares estimation. Application of DF Analysis is described by Rodgers et al. (2001a, b). Several variants have been proposed (e.g., Cherny et al. 1992; Kohler and Rodgers 1999; Purcell and Sham 2003; Rodgers et al. 1994a; Rodgers and Kohler 2005). Waller (1994) adapted DF Analysis to estimate models of genetic dominance, LaBuda and DeFries (1990) accounted for group differences, and Purcell and Koenen (2005) improved DF Analysis methods to model measured nonshared environmental differences.
In the current paper, we use DF Analysis to estimate parameters in the context of the MDAN design. These parameters emerge from the basic DF Analysis model:
where KIN1 is the fertility score of the first member of the kinship pair, KIN2 is the score of the second member, R is the genetic coefficient (e.g., R = .50 for mothers–daughters, R = .25 for aunt–nieces), the b’s are least squares regression weights, and e is the residual. DeFries and Fulker (1985) and Rodgers and McGue (1994) showed that b3 estimates h 2, and b1 estimates c 2. We use the reparameterization from Rodgers and Kohler (2005) within PROC GENMOD to estimate the biometrical parameters.
In an “unselected” DF Analysis, the kinship pairs must be double-entered, so that there is an observation for each individual (equivalent to using intra-class correlations; see Rodgers et al. 2001c, for double entry rationale, and Kohler and Rodgers 2001, for statistical treatment). In the MDAN design, there is logical separation within kinship pair based on generation. We estimated these models using single-entered data in which mother/aunt scores were treated as probands within the KIN2 variables in Eq. 1.
Testing statistical hypotheses within the context of this design has some problematic features. For example, there is error dependency across observations caused by the clustering of extended families and the within-family repetition of offspring who act as both daughters and nieces. In response, we adjusted our standard errors and statistical tests by the sandwich estimators (Huber 1967; White 1980), popularized by the methods of generalized estimating equations (GEE; e.g., Liang and Zeger 1986; Dunlop 1994). Biometrical treatment of this situation is presented in Kohler and Rodgers (1999), although only double-entry DF Analysis is treated there. The DF Analysis models were run in PROC GENMOD in SAS, adjusting for the clustering at the family level (and ignoring additional within-individual covariance structure across repeated offspring outcomes). In fitting these models, we chose to specify an independent repeated error correlation matrix in the absence of a known structure for the unique and complex structuring of these dyadic observations (see Hardin and Hilbe 2003, for details and elaboration). In Appendix A, we include the SAS code used to estimate these models.
Of course there are limitations to the use of DF Analysis as well. MLE modeling (using Mx or Mplus, for example) is considerably broader and more flexible in supporting multivariate analysis. DF Analysis was designed for univariate analysis, and has slight potential for multivariate modeling (e.g., Rodgers et al. 2001c). But complex biometrical models, including competing pathways and choleski (time-oriented) models, are not compatible with the DF Analysis model. However, for evaluating the validity of the MDAN design, and for demonstration purposes, DF Analysis is conceptually simple and also has the advantage of matching the analysis method used in most of the comparison research used to evaluate concurrent validity. We note that more complex analytic routines have been used often with the NLSY data (e.g., see Rodgers et al. 2007).
Results
Descriptive analyses
In Table 2, we present means and standard deviations for each of the two fertility variables—age at menarche and age at first intercourse—for both generations. Mean age at menarche is almost identical across the two generations, and age at first intercourse is somewhat lower in the daughter/niece generation.
Analysis of fertility precursors
Table 3 shows kinship correlations for mother–daughter and aunt–niece pairs. The true mother–daughter (R = .50; N = 498) and aunt–niece (R = .25; N = 443) pairs carry most of the weight in this analysis; aunt–niece pairs obtained from mothers who were twins (R = .375, N = 11) and aunt–niece pairs from ambiguous sibling mothers (R = .1875, N = 70) have smaller sample sizes, and less reliable correlations. Overall, there were 1022 kinship pairs involving 2044 individuals (not quite half of whom were overlapping through the mother/aunt sister–sister pairs by design). In Table 3, we present kinship correlations for the two large mother–daughter and aunt–niece categories (though all four categories are used in the biometrical analysis). Table 3 shows univariate kinship correlations, and also bivariate kinship correlations between age-at-menarche (AgeMen) and age-at-first-intercourse (AFI). The (bolded) univariate correlation patterns are suggestive of genetic variance, especially for age-at-menarche, because the mother–daughter correlations are notably higher than the aunt–niece correlations. The bivariate correlations are not suggestive of genetic variance (because mother–daughter correlations are no higher than aunt–niece). There is some slight suggestion of overlapping shared environmental variance, because half of the biviariate correlations (those including AFI from the second generation) are of moderate size.
Table 4 presents results of a standard univariate ACE model fit using DF Analysis. In the age-at-first-intercourse analysis, h 2 is moderate and nonsignificant, and c 2 is small and nonsignificant (using a GEE approach to test hypotheses). In the age-at-first-menarche analysis, the h 2 is moderate-to-substantial and significant, and the c 2 is slightly negative. Each model was re-estimated by dropping the shared environmental component of variance. In these AE models, h 2 = .37 for age-at-first-intercourse, and h 2 = .39 for age-at-menarche (both significant).
Comparison to previous fertility results
Previous research has estimated h 2 and c 2 in each of the two generations separately. Rodgers et al. (1999) presented a biometrical analysis of age-at-first-intercourse, using the NLSY79 data, and Doughty and Rodgers (2000) did the same type of analysis using the NLSY79 age-at-menarche variable. Bard and Rodgers (2006) presented biometrical models of the same two variables using the updated NLSYC kinship links.
In Table 5, we present results from these four different sources. There is no necessary constraint imposed on the cross-generational analysis by the within-generational findings. In fact, as we will discuss, there are assumptions that are different in the cross-generational analysis than in the separate within-generational analyses. If these changing assumptions affect the outcomes, we would expect different results in the MDAN design than in the within-generational family designs. The comparisons in Table 5 might, in fact, be interpreted at least partially as a test of these assumptions. A comparison of patterns in Table 5 shows, however, that the h 2 and c 2 values are similar across the four different studies.
Discussion
The primary goal of the current paper is to present and illustrate a new behavior genetic design. In addition, useful substantive results have emerged and some broader implications of behavior genetic modeling have been suggested. We now summarize the validation and demonstration analysis, evaluate the substantive findings in relation to fertility research, and discuss assumptions and expansion of the MDAN design within a cross-generational perspective.
Validation of the MDAN design
The primary goal, to define and illustrate the MDAN design, has been achieved. As discussed, there are many different ways to implement such a design. We have presented one, based on a straightforward approach to linking mothers and daughters, and aunts–nieces, in which these two categories themselves are linked because the mothers and aunts are sisters. This design structure, which is similar to the one on which the children-of-twins design is based, controls for threats to validity caused by disparate family background. Of course there is a sample size cost.
This demonstration illustrates several useful features of the MDAN design. First, in both the kinship correlations and within the DF Analysis, the biometrical results were plausible and sensible, which helps establish the face validity of this design. Second, MDAN analysis results match closely ones obtained in previous traditional within-generational analyses, which provides evidence for concurrent validity of this design. Third, the mother–daughter kinship links, and the aunt–niece kinship links, once defined, can be used in future research, just as the NLSY79 and NLSYC links have been used by different research teams besides the developers (e.g., McCartan 2007). These links could also be of value outside the context of the MDAN design; for example, a researcher studying the aunt–niece relationship could use these links. Researchers can obtain a file of these cross-generational links (as well as the NLSY79 and NLSYC kinship links) by contacting the first author.
We now suggest several other adaptations. The first proposed expansion of the MDAN design is to fit models simultaneously using both within-generational and cross-generational kinship links. We have begun development of these links and expect to present results in future papers. The second expansion involves using mothers who may not have a sister match (and who therefore do not have NLSYC nieces); these links are, nevertheless, informative of the mother–daughter relationship. The third expansion is to use a broader set of kinship links. To keep the design/data management simple, we eliminated cousin and half-sibling pairs in the mothers’ generation, and the corresponding aunt–niece pairs as well. The full use of this broad set of potential kinship links begins to approach the complexity of a pedigree analysis (but with tighter bounds on age ranges and generational specification).
A fourth expansion of MDAN designs would be to include statistical features to model variance. For example, we treated the kinship units using sandwich estimators and GEE estimation to adjust for family-level clustering. However, these and other types of clustering (e.g., clustering of first-level sampling units, or sampling design effects) can bias model estimates. Whether such adjustments to achieve external validity are ultimately useful is partly an empirical question, because of the restrictions that are in place as a result of the requirements of the design. Most other biometrical analyses have a similar or greater restriction. For example, most twin studies are highly select in a similar fashion. But these types of clustering concerns can be naturally handled using a mixed model/multi-level analysis, rather than constructing sibling pairs (Guo and Wang 2002; McArdle and Prescott 2005; van den Oord 2001), which allows broader control over the correlated portions of the design.
Finally, as noted, the MDAN design is a special case of a Children of Siblings (COS) design. All of the features of the MDAN analysis illustrated in the Results section can be implemented more broadly with cross-generational links that also include fathers, sons, uncles, and nephews (using within- and cross-gendered links to mothers, aunts, daughters, and nieces). The practical limitation to implementing a COS design is the availability of data. At this time, few data sources other than the NLSY would support cross-generational analyses, especially with (partially) equivalent instrumentation. The NLSY, because it only collects information on children of NLSY79 mothers, would be restricted to MDAN designs, or to expanded versions including nephews within the second generation (mother–son/daughter and aunt–niece/nephew designs). Unfortunately, little information exists about the fathers of NLSYC children.
Substantive fertility findings
We identified moderate heritability underlying age at first intercourse, and moderate/substantial heritability underlying age at menarche. Neither phenotype had any shared environmental variance; the remaining variance was attributed to nonshared environmental variance/measurement error. The finding of meaningful and interpretable genetic variance underlying fitness-related phenotypes has become a common finding in recent years (for empirical support, see Bricker et al. 2006; Kirk et al. 2000; Rodgers et al. 2001c; Trumbetta et al. 2007). Despite the apparent implication of Fisher’s fundamental theory (the FTNS) that fitness traits should not have significant heritability, explanations for these findings have begun to emerge that make sense evolutionarily (Hughes and Burleson 2000), sociologically (Udry 1996; Rodgers et al. 2007), and genetically (Rodgers et al. 2001a).
Broader implications of cross-generational designs
A secondary goal with potentially interesting implications involves the role of the standard biometrical assumptions within the MDAN design. Cross-generational designs are, in a sense, “closer” to the basic units of biometrical analysis than within-generational designs, because they meet more of the assumptions underlying biometrical analyses. Full siblings share a biological mother and father, and therefore share 50% of their genes identical by descent (i.b.d.)—on the average. But mothers and daughters share exactly 50% of their genes i.b.d., which reduces the sampling variability associated with the R coefficient. Typically, this sampling variability is assumed away in biometrical modeling, so that models can be identified, but assuming it away is not necessary in considering the mother–daughter links. Another way to conceptualize this is to note that the transmission of genetic information is naturally a cross-generational process; the mother–daughter correlation is exemplary of this transmission. Evans et al. (2002) discuss biological and genetic bases for this transmission process.
The equal environments assumption is a standard assumption of biometrical modeling. It originally emerged from considering the twin design. Plomin (1990) defined the assumption in relation to the twin design: “if identical twins are treated more similarly for non-genetic reasons than are fraternal twins, greater behavioral resemblance of identical twins could be due to environment rather than heredity” (p. 49). But the assumption applies to all ACE modeling, no matter what kinship pairs are used. The problem specified by the assumption occurs if inferred genetic similarity is, rather, caused by environmental similarity. Considerable research has been done investigating the nature, effect, and legitimacy of the equal environments assumption.
In a cross-generational design, the assumption takes on a different nature. The assumption would be violated if mother–daughter correlations are inflated compared to aunt–niece correlations because of environmental factors that are treated by the model as genetic. One example of this would occur if grandparents provided more support—financial or otherwise—to a daughter who is a mother than to a daughter who is not a mother (i.e., an aunt within our design). If this support translated into mother–child similarity that was inflated compared to the aunt–child similarity, then this would be an example of a violation of the equal environments assumption.
In other words, in a cross-generational design, an interesting implication is that the equal environments assumption moves back a generation, or at least includes the previous generation prior to the mother/aunts generation for consideration. Further, if within- and cross-generational results converge (as they did for our analysis of fertility precursors) the patterns may be fairly robust to the location of the equal environments assumption. This implication is one of the most potentially interesting substantive finding that emerges from the MDAN modeling. Past research has focused on the nature of the transmission of cultural values across generations (e.g., Heath et al. 1985; Truett et al. 1994). Loehlin (1989) discussed the differences between “indirect correlations,” those between individuals raised in the same family environment, and “direct correlations” from individuals not raised in the same family. The NLSY and MDAN design provide a rich structure of genetic relationships that can be used to empirically evaluate questions related to the EEA and other biometrical assumptions.
We conclude by noting that a new, flexible, and theoretically interesting design comparing and modeling mother–daughter and aunt–niece correlations provides a number of challenging and fascinating research questions within the behavior genetic perspective. A cross-generational research agenda appears to be a fruitful ways to expand biometrical modeling in new directions.
References
Bard DE, Rodgers JL (2006) Use of discrete-time survival analysis for modeling multivariate ACE models of fertility precursors from the children of the NLSY. Poster presented at the June, 2006 meetings of the Behavior Genetic Association, Storrs, CN
Baydar N, Greek A (2001) Analysis of data from related individuals. Working paper, Battell Centers for Public Health Research and Evaluation, Seattle WA
Bouchard TJ, Lykken DT, McGue M, Segal NL, Tellegen A (1990) Sources of human psychological differences: the Minnesota Study of Twins Reared Apart. Science 250:223–228. doi:10.1126/science.2218526
Bricker JB, Stallings MC, Corley RP, Wadsworth SJ, Bryan A, Timberlake DS, Hewitt JK, Caspi A, Hofer SM, Rhea SA, DeFries JC (2006) Genetic and environmental influences on age at sexual initiation in the Colorado adoption project. Behav Genet 36:820–832. doi:10.1007/s10519-006-9079-2
Brooks-Gunn J, Warren MP, Rosso J, Gargiulo J (1987) Validity of self-report measures of girls’ pubertal status. Child Dev 58:829–841. doi:10.2307/1130220
Center for Human Resource Research (2006) NLSY79 child & young adult data users guide. Center for Human Resource Research, Columbus
Chase-Lansdale PL, Mott FL, Brooks-Gunn J, Phillips DA (1991) Children of the National Longitudinal Survey of Youth: a unique research opportunity. Dev Psychol 27:918–931. doi:10.1037/0012-1649.27.6.918
Cherny SS, DeFries JC, Fulker DW (1992) Multiple regression of twin data: a model-fitting approach. Behav Genet 22:489–497. doi:10.1007/BF01066617
Damon A, Damon ST, Reed RB, Valadian I (1969) Age at menarche of mothers and daughters with a note on accuracy of recall. Hum Biol 38:255–273
DeFries JC, Fulker DW (1985) Multiple regression analysis of twin data. Behav Genet 15:467–473. doi:10.1007/BF01066239
D’Onofrio BM, Turkheimer EN, Eaves LJ, Corey LA, Berg K, Solaas MH, Emery RE (2003). J Child Psychol Psychiatry Allied Discipl 44:1130–1144. doi:10.1111/1469-7610.00196
D’Onofrio BM, Van Hulle CA, Waldman ID, Rodgers JL, Rathouz PJ, Lahey BB (2007) Causal inferences regarding prenatal alcohol exposure and childhood externalizing problems. Arch Gen Psychiatry 64:1296–1304. doi:10.1001/archpsyc.64.11.1296
Doughty D, Rodgers JL (2000) Behavior genetic modeling of menarche in U.S. females. In: Rodgers JL, Rowe D, Miller WB (eds) Genetic influences on fertility and sexuality. Kluwer Academic Press, Boston
Dunlop DD (1994) Regression for longitudinal data: a bridge from least squares regression. Am Stat 48:299–303. doi:10.2307/2684838
Evans DM, Gillespie NA, Martin NG (2002) Biometrical genetics. Biol Psychol 61:33–51. doi:10.1016/S0301-0511(02)00051-0
Falconer DS (1979) Introduction to quantitative genetics. Longman, New York
Fisher RF (1930) The genetical theory of natural selection. Clarendon Press, Oxford
Fisher RF (1935) The design of experiments. Olyver and Boyd, Edinburgh
Gottesman II, Bertelsen A (1989) Confirming unexpressed genotypes for schizophrenia. Arch Gen Psychiatry 46:867–872
Guo G, Wang JM (2002) The mixed or multilevel models for behavior genetic analysis. Behav Genet 32:37–49. doi:10.1023/A:1014455812027
Harden KP, Turkheimer E, Emery RE, D’Onofrio BM, Slutske WS, Heath AC, Martin NG (2007) Marital conflict and conduct problems in children of twins. Child Dev 78:1–18. doi:10.1111/j.1467-8624.2007.00982.x
Hardin JW, Hilbe JM (2003) Generalized estimating equations. Chapman & Hall/CRC, Boca Raton
Heath AC, Kendler AS, Eaves LJ, Markell D (1985) The resolution of cultural and biological inheritance: Informativeness of different relationships. Behav Genet 15:439–465. doi:10.1007/BF01066238
Huber PJ (1967) The behavior of maximum likelihood estimates under non-standard conditions. In Proceedings of the fifth Berkeley symposium on mathematical statistics and probability, vol 1, pp 221–233
Hughes KA, Burleson MH (2000) Evolutionary causes of genetic variation in fertility and other fitness components. In: Rodgers JL, Rowe DC, Miller WB (eds) Genetic influences on human fertility and sexuality. Kluwer, Boston, pp 7–34
Jinks JL, Fulker DW (1970) A comparison of the biometrical-genetical, MAVA and classical approaches to the analysis of human behavior. Psychol Bull 73:311–349. doi:10.1037/h0029135
Kirk KM, Blomberg SP, Duffy DL, Heath AC, Owens IPF, Martin NG (2000) Natural selection and quantitative genetics of life history traits in western women: a twin study. Evol Int J Org Evol 55:423–435
Kohler H-P, Rodgers JL (1999) DF analysis of binary, ordered, and censored variables using probit and tobit approaches. Behav Genet 29:221–232. doi:10.1023/A:1021686005855
Kohler H-P, Rodgers JL (2001) DF-analysis of heritability with double-entry twin data: Asymptotic standard errors and efficient estimation. Behav Genet 31:179–192. doi:10.1023/A:1010253411274
Kohler H-P, Rodgers JL, Miller WB, Skytthe A, Christensen K (2006) Bio-social determinants of fertility. Int J Androl 29:46–53. doi:10.1111/j.1365-2605.2005.00606.x
LaBuda MC, DeFries JC (1990) Genetic etiology of reading disability: Evidence from a twin study. In: Pavlidis GT (ed) Persepctives on dyslexia, vol 1. Wiley, New York, pp 47–76
Liang K-Y, Zeger SL (1986) Longitudinal data analysis using generalized linear models. Biometrika 73:13–22. doi:10.1093/biomet/73.1.13
Loehlin JC (1989) Partitioning environmental and genetic contributions to behavioral development. Am Psychol 44:1285–1292. doi:10.1037/0003-066X.44.10.1285
Magnus P, Berg K, Bjerkedal T (1985) No significant difference in birth weight for offspring of weight discordant monozygotic female twins. Early Hum Dev 12:55–59. doi:10.1016/0378-3782(85)90137-9
McArdle JJ, Prescott CA (2005) Mixed-effects variance components models for biometrical family analyses. Behav Genet 35:631–652. doi:10.1007/s10519-005-2868-1
McCartan LM (2007) Inevitable, influential, or unnecessary? Exploring the utility of genetic explanation for delinquent behavior. J Crim Just 35:219–233. doi:10.1016/j.jcrimjus.2007.01.008
Mendle J, Turkheimer E, D’Onofrio BM, Lynch SK, Emergy RE, Slutske WS, Martin NG (2006) Family structure and age at menarche: a children-of-twins approach. Dev Psychol 42:533–542. doi:10.1037/0012-1649.42.3.533
Moffitt TE, Caspi A, Belsky J, Silva PA (1992) Childhood experience and the onset of menarche: a test of a sociobiological model. Child Dev 63:47–58. doi:10.2307/1130900
Nance WE, Corey LA (1976) Genetic models for the analysis of data from the families of identical twins. Genetics 83:811–826
Neale MC, Cardon LR (1992) Methodology for genetic studies of twins and families. Kluwer Academic Press, Dordrecht
Plomin R (1990) Nature and nurture: an introduction to human behavior genetics. Brooks/Cole, Pacific Grove
Plomin R, DeFries JC, McClearn GE (1990) Behavior genetics: a primer. Freeman and Company, New York
Purcell S, Koenen KC (2005) Environmental mediation and the twin design. Behav Genet 35:491–498. doi:10.1007/s10519-004-1484-9
Purcell S, Sham PC (2003) A model-fitting implementation of the DeFries–Fulker model for selected twin data. Behav Genet 33:271–278. doi:10.1023/A:1023494408079
Rodgers JL (1996) NLSY Youth linking algorithm. Internal Document
Rodgers JL, Kohler H-P (eds) (2003) The biodemography of human reproduction and fertility. Kluwer Academic Publishers, Boston
Rodgers JL, Kohler H-P (2005) Reformulating and simplifying the DF Analysis model. Behav Genet 35:211–217. doi:10.1007/s10519-004-1020-y
Rodgers JL, McGue M (1994) A simple algebraic demonstration of the validity of DeFries–Fulker analysis in unselected samples with multiple kinship levels. Behav Genet 24:259–263. doi:10.1007/BF01067192
Rodgers JL, Billy JO, Udry JR (1982) The rescission of behaviors: inconsistent responses in adolescent sexuality data. Soc Sci Res 11:280–296. doi:10.1016/0049-089X(82)90012-6
Rodgers JL, Harris D, Vickers KB (1992) Seasonality of onset of adolescent coitus. Soc Biol 39:1–14
Rodgers JL, Rowe DC, Li C (1994a) Beyond nature versus nurture: DF analysis of nonshared influences on problem behaviors. Dev Psychol 30:374–384. doi:10.1037/0012-1649.30.3.374
Rodgers JL, Rowe DC, May K (1994b) DF analysis of NLSY IQ/achievement data: nonshared environmental influences. Intelligence 19:157–177. doi:10.1016/0160-2896(94)90011-6
Rodgers JL, Rowe DC, Buster M (1999) Nature, nurture, and first sexual intercourse in the USA: fitting behavioral genetic models to NLSY kinship data. J Biosocial Sci 31:29–41.
Rodgers JL, Rowe DC, Miller WB (eds) (2000) Genetic influences on human fertility and sexuality: theoretical and empirical contributions from the biological and behavioral sciences. Kluwer Academic Publishers, Boston
Rodgers JL, Hughes K, Kohler H-P, Christensen K, Doughty D, Rowe DC, Miller WB (2001a) Genetic influence helps explain variation in human fertility: evidence from recent behavioral and molecular genetic studies. Curr Dir Psychol Sci 10:184–188. doi:10.1111/1467-8721.00145
Rodgers JL, Buster M, Rowe DC (2001b) Genetic and environmental influences on delinquency: DF analysis of NLSY kinship data. J Quant Criminol 17:145–168. doi:10.1023/A:1011097929954
Rodgers JL, Kohler H-P, Kyvik K, Christensen K (2001c) Behavior genetic modeling of human fertility: findings from a contemporary Danish twin study. Demography 38:29–42. doi:10.1353/dem.2001.0009
Rodgers JL, Johnson A, Bard DE (2005) NLSY-Children/Young Adult (1986–2000) kinship linking algorithm. Internal document
Rodgers JL, Bard DE, Miller WB (2007) Multivariate cholesky models of human female fertility patterns in the NLSY. Behav Genet 37:345–361. doi:10.1007/s10519-006-9137-9
Segal N, McGuire J, Havlena PG (2006) IQ similarity in virtual twins: Developmental trends. Paper presented at the June, 2006, meetings of the Behavior Genetic Association, Storrs, CN
Shadish WR, Cook TD, Campbell DT (2002) Experimental and quasi-experimental designs for generalized causal inference. Houghton-Mifflin, Boston
Siegel DM, Aten MJ, Roughmann KJ (1998) Self-reported honesty among middle and high school students responding to a sexual behavior questionnaire. J Adolesc Health 23:20–28. doi:10.1016/S1054-139X(97)00274-7
Silberg JL, Eaves LJ (2004) Analyzing the contribution of genes and parent-child interaction to childhood behavioral and emotional problems: a model for the children of twins. Psychol Med 34:347–356. doi:10.1017/S0033291703008948
Truett KR, Eaves LJ, Waiters EE, Heath AC, Hewitt JK, Meyer JM, Silberg J, Neale MC, Martin NG, Kendler KS (1994) A model system for analysis of family resemblance in extended kinships of twins. Behav Genet 24:35–49. doi:10.1007/BF01067927
Trumbetta SL, Markowitz EM, Gottesman II (2007) Marriage and genetic variation across the lifespan: not a steady relationship? Behav Genet 37:362–375. doi:10.1007/s10519-006-9132-1
Udry JR (1996) Biosocial models of low-fertility societies. In: Casterline JB, Lee RD, Foote KA (eds) Fertility in the United States: new patterns, new theories. The Population Council, New York
Upchurch DM, Lillard LA, Aneshensel CS, Li NF (2002) Inconsistencies in reporting the occurrence and timing of first intercourse among adolescents. J Sex Res 39:197–206
van den Oord EJCG (2001) Estimating effects of latent and measured genotypes in multilevel models. Stat Methods Med Res 10:393–407. doi:10.1191/096228001682157625
van den Oord EJCG, Rowe DC (1997) Continuity and change in children’s social maladjustment: A developmental behavior genetic study. Dev Psychol 33:319–332. doi:10.1037/0012-1649.33.2.319
van den Oord EJCG, Rowe DC (2000) Racial differences in birth health risk: a quantitative genetic approach. Demography 37:285–298. doi:10.2307/2648042
Van Hulle CA, Rodgers JL, D’Onofrio BM, Waldman ID, Lahey BB (2007) Sex differences in the causes of self-reported adolescent delinquency. J Abnorm Psychol 116:236–248. doi:10.1037/0021-843X.116.2.236
Waller NB (1994) A DeFries and Fulker regression model for genetic nonadditivity. Behav Genet 24:149–153. doi:10.1007/BF01067818
White H (1980) A heteroskedasticity-consistent covariance matrix estimator and a direct test for heteroskedasticity. Econometrica 48:817–838. doi:10.2307/1912934
Author information
Authors and Affiliations
Corresponding author
Additional information
Edited by Danielle Dick.
Appendix A: SAS Coding to estimate the DF Analysis model using GEE procedures to obtain sandwich standard errors
Appendix A: SAS Coding to estimate the DF Analysis model using GEE procedures to obtain sandwich standard errors


Rights and permissions
About this article
Cite this article
Rodgers, J.L., Bard, D.E., Johnson, A. et al. The Cross-Generational Mother–Daughter–Aunt–Niece Design: Establishing Validity of the MDAN Design with NLSY Fertility Variables. Behav Genet 38, 567–578 (2008). https://doi.org/10.1007/s10519-008-9225-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10519-008-9225-0