Abstract
The National Longitudinal Survey of Youth datasets (NLSY79; NLSY-Children/Young Adults; NLSY97) have extensive family pedigree information contained within them. These data sources are based on probability sampling, a longitudinal design, and a cross-generational and within-family data structure, with hundreds of phenotypes relevant to behavior genetic (BG) researchers, as well as to other developmental and family researchers. These datasets provide a unique and powerful source of information for BG researchers. But much of the information required for biometrical modeling has been hidden, and has required substantial programming effort to uncover—until recently. Our research team has spent over 20 years developing kinship links to genetically inform biometrical modeling. In the most recent release of kinship links from two of the NLSY datasets, the direct kinship indicators included in the 2006 surveys allowed successful and unambiguous linking of over 94 % of the potential pairs. In this paper, we provide details for research teams interested in using the NLSY data portfolio to conduct BG (and other family-oriented) research.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
We describe a set of high-quality kinship links available for the National Longitudinal Survey of Youth 1979 (NLSY79) and NLSY-Children/Young Adult (NLSY-C/YA) datasets that support high-quality biometrical research using these broad and powerful longitudinal databases. Our project site (http://liveoak.github.io/NlsyLinks/) contains multiple kinship link files, the linking algorithm source code, the NlsyLinks R package, and vignettes illustrating data management and biometric estimation procedures. In the future, we expect to develop similar kinship links for the third NLSY data, the NLSY97 (which is an 18 year approximate replication of the design and sampling strategy of the NLSY79).
The current paper follows the examples of other past publications that have provided detailed information on datasets relevant to specific types of researchers in the social and behavioral sciences. Chase-Landsdale et al. (1991) promoted the value of the NLSY-Children dataset for developmental and other behavioral science researchers. Harris et al. (2006) published a similar article describing the value of the Add Health dataset for researchers using twin samples. An article matching the aims of the current article even more closely focused on the Add Health sibling pairs (Harris et al. 2013).
The current article has a similar goal to these three earlier articles. It is broader in the sense that we have developed research databases using two of the NLSY datasets (with a third in development), and links between them, all of which are available (either upon request or online) to academic researchers. But this article is also quite focused because it is specifically targeted to researchers in behavior genetics (with application as well to those working within related methodological and family-studies arenas).
Few population-based family data sources based on probability sampling exist that are biometrically informed. Even rarer are such biometrically-informed data with many decades of longitudinal structure, including cross-generational links. The set of three databases comprising the National Longitudinal Survey of Youth (NLSY) provides a valuable resource for behavior genetic—and other demographic, aging, developmental, and family-based—researchers. The immediate purpose of this article is to document the history and development of the kinship links, and to illustrate the value of the NLSY datasets for behavior genetic (BG) research. The ultimate goal of the article is to stimulate BG and other research teams to incorporate within BG and family-based research studies the kinship information that is now easily available and accessible to NLSY researchers.
Background
The NLSY samples
The NLSY datasets are among the most popular and valuable of all social/behavioral science data sources. A part of the National Longitudinal Survey (NLS) portfolio, the NLSY contains three different data sources, two of which are linked cross-generationally (the NLSY79 and the NLSY-Children/Young Adult), and two of which are linked through intentional replication, including approximately matched sampling designs (the NLSY79 and NLSY97).
The NLSY79 originated as a household probability sample of 12,686 adolescents in the age range 14–21 on December 31, 1978, thus born between 1957 and 1964. The age range applies to the day the sample was drawn; interviews occurred several months later, so that there are a few 22-year-old respondents at first interview. The NLSY79 contained 6111 respondents from the original household probability sample, 5295 respondents who were part of the minority and poor white oversample, and 1280 respondents who were part of the military sample. The status of the NLSY79 as a probability sample is an extremely valuable feature of the NLSY portfolio. Because sampling weights are contained in each release of the dataset, the potential for high external validity in NLSY-research is an attraction for all investigators, but especially for behavior genetic researchers. We note that few datasets provide the kind of support for high levels of both internal and external validity that emerges from the NLSY.
The 12,686 respondents represented 8770 unique households, with 2862 households that contained two or more NLSY79 respondents; of these, 2448 were multiple-sibling households, with 414 households containing no siblings, but instead two or more young spouses, cousins, and other categories of relatedness. A total of 5263 NLSY79 respondents had siblings who were also NLSY79 respondents. Importantly, until 2006 there was no effort to separately identify full, half, step, and adoptive siblings; all of these categories were identified in the household roster as “sibling.” The NLSY79 designation of “sibling” did not originally discern the various levels of genetic relatedness among siblings. However, several variables were informative of this distinction, and previous NLSY research using kinship links has been based on inferred level of kinship using implicit indicators of relatedness.
The kinship links identify sibling and other kinship relationships among the multi-respondent households. These links are approximately representative of those that occurred among 14–21 year olds in households in the U.S. on the last day of 1978, and include twins, different types of siblings, cousins, uncle–nephews, step-siblings, and a number of other more unusual types of kinship pairs. Respondents in the NLSY79 were interviewed yearly from 1979 to 1986, at which point interviews were conducted on a biennial basis until 2014 and continuing. The 1643 respondents in the poor white oversample were dropped for budgetary reasons following the 1990 survey; 1079 respondents from the military sample were dropped following the 1986 survey (all but 201 randomly sampled military respondents who were maintained in the sample).
The NLSY-Children (NLSYC; we often refer to this dataset as “Generation 2,” or “Gen2”; some past researchers have referred to this dataset as the CNLSY data) consists of all biological children born to the 6283 females in the NLSY79 (which we often refer to as “Generation 1” or “Gen1”). Interviews with the NLSYC Gen2 children began in 1986, and have occurred biennially until 2014 and continuing. By the 2012 interviews the NLSY79 Gen1 females were 47–55, and childbearing can be considered complete from 2012 and continuing for this cohort of women. By 2010, 11,504 children had been identified as born to the NLSY79 females, 86 % of whom were interviewed in 2010; the NLSYC children ranged in age at the 2010 survey from 1 to 39. (Note that children born to females in the military sample after 1986, and to females in the poor white oversample after 1990, when these sub-samples were dropped, have no information in the NLSYC files.) Obviously, because of the design of the survey, there are many full- and half-siblings within the NLSYC sample. As well, by linking mothers to their sisters, half-sisters, and cousins, we can also identify cousins, half cousins, and several other cousin categories of relatedness within the NLSY-Children data. In fact, the cousin data within Gen2 has seldom been used as an exclusive data source in academic research (see van den Oord and Rowe 1999, and Goodnight et al. 2013, for exceptions), but has been used fairly often in combination with other kinship categories (including, in a few cases, half-cousins as well).
The cousin data within the NLSY are an interesting, underused, and underappreciated source of information. Fisher (1918) noted the value of cousin data in biometrical studies: “… the hypothesis of cumulative Mendelian factors seems to fit the facts very accurately. The only marked discrepancy from existing published work lies in the correlation for first cousins. … but until we have a record of complete cousinships measured accurately and without selection, it will not be possible to obtain satisfactory numerical evidence on this question” (p. 168). Within the current context, we note that our research team has observed several times that the cousin correlations obtained from the NLSY kinship links appear unusually high (or sometimes we have felt that the half-sibling correlations were surprisingly low, thus leading to the appearance of high cousin correlations). The NLSY data provide two different generations of cousin data, and the Gen2 cousins may be as close as can ever be obtained to a large cousin sample, emerging from probability sampling mechanisms and providing, in Fisher’s words, “a record of complete cousinships measured accurately and without selection.” Future NLSY researchers should pay particular attention to the NLSY cousins as the basis for a potentially fruitful research endeavor.
In 1994 and following, a separate survey has been administered to NLSYC respondents age 15 or older in a given survey year; this survey is referred to as the NLSY-Young Adult (NLSY-YA) survey. It contains different types of questions than the childhood surveys administered to the NLSYC respondents, including questions related to sexual development, dating, marriage, reproduction, and fertility; jobs and employment; delinquency, drinking, and smoking; and other adolescent and young adult behaviors. Though there are different names for the NLSYC and NLSY-YA surveys, it is important to emphasize that these are the same Gen2 respondents at different ages. In this paper, when the distinction between the two surveys does not matter, we refer to these respondents as NLSY-C/YA respondents.
The NLSY97 dataset was developed as an approximate replication of the NLSY79, for a cohort born almost two decades later. It consisted of a household probability sample that surveyed all adolescents between the ages of 12 and 17 on December 31, 1996 (therefore born in 1979–1984, and age 12–18 at time of first interview with an age range in 2012 of 27–33). The original sample size was 8984; almost 83 % were still being interviewed in 2012, which was round 15. Surveys have been annual, but occur biennially from 2012 on. The original sample included 6748 respondents from a cross-sectional household probability sample, and another 2236 in a minority oversample. Survey questions in NLSY97 are similar to those in the NLSY79, and in fact in many cases the original instrumentation is maintained.
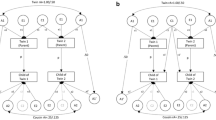
There are a number of features of these three datasets that provide cross-dataset information. Of particular note is that the link between the NLSY79 Gen1 mothers and the NLSY-C/YA Gen2 children creates a two-generational data source. (In addition, some information is available about the parents of the NLSY79 respondents, Gen0, including parents’ educational outcomes, birthdates, and place of birth; thus, limited three-generational information is available.) Further, the timing of the sampling of the NLSY97 causes the NLSY97 mean age to be fairly close to that of the NLSY-C/YA respondents (though there is obviously much more age variability in the NLSY-C/YA data). Figure 1 shows a schematic of the timing and pace of births, fertility, and NLSY data collection for these three surveys. All NLSY files are public access, and can be downloaded from http://www.bls.gov/nls/nlsy79.html.
The kinship links
History
In the early 1990’s three independent research teams recognized the potential value of the NLSY-Children data for behavior genetic research: Joe Rodgers and David RoweFootnote 1 in the U.S.; Nazli Baydar and colleagues in Turkey and the U.S.; and Edwin van den Oord in The Netherlands. Rodgers et al. (1994a) documented almost a thousand kinship pairs, which naturally increased as NLSY79 females (age 27–35 in the 1992 survey) continued childbearing. However, as described above, in the first 20 years of the NLSYC survey and the first 27 years of the NLSY79 survey, no questions were included to explicitly distinguish between sibling categories.
Nevertheless, several questions in the NLSYC data were informative of kinship relatedness, in particular questions asked of a NLSYC respondent’s NLSY79 biological mother about the biological father. The three teams independently developed algorithms using several survey questions to distinguish full and half siblings. Baydar and Greek (2001) published research using her links. Van den Oord produced a linking algorithm, but began collaborating with Rowe and Rodgers, and ultimately used their algorithm and kinship links for most of his NLSY research.Footnote 2
The first two research articles based on the kinship links were published in 1994 using biometrical modeling and the NLSYC dataset. Rodgers et al. (1994a) reported results for childhood problem behaviors for 822 Gen2 kinship pairs. Rodgers et al. (1994b) reported results for PIAT-Math, PIAT-Reading Recognition, and PIAT-Reading Comprehension using 855 Gen2 kinship pairs. Since 1994, over 30 research articles have been published using these NLSY-C/YA kinship links.
Success of the NLSYC linking algorithm motivated development of linking algorithms for the NLSY79 dataset. Gen1 provided a more difficult challenge, because of the many types of relatedness in the Gen1 households; successful linking required three different programming efforts between 1992 and 1996. The first two linking efforts were tested in validation exercises similar to the original NLSYC analysis (in which biometrical models were fit to phenotypes—height and weight—with known heritability), and rejected as a useful set of kinship links. The algorithm was re-written using overlapping and different survey questions; the third set of kinship links satisfied our validation goals. Of the targeted 3890 kinship pairs, 2470 (63 %) were successfully classified. The first NLSY79 kinship link publication was Rodgers et al. (1999), reporting heritabilities and shared environmental variance for age at first intercourse. Since 1999, over 20 papers have been published using NLSY79 kinship links.
Despite demonstrated concurrent validity with other research results, validation support, and a growing publication stream, a few behavior genetic researchers criticized the early kinship links (through reviewer comments, conference discussions, and direct e-mails). Some criticism was entirely justified, some arguably not. Critics tended to focus on the relatively low linking rates (63 % of eligible links were specified in the NLSY79 data) and on lack of direct ascertainment to distinguish full and half siblings. One set of critics included researchers who often used highly selected twin samples; we noted that for external validity, 63 % of a probability sample has certain slight advantages over volunteer twin samples. Other critics included researchers using survey-based zygosity determination, a method often of lower reliability/validity than the NLSY algorithms, which used maternal information about the biological fathers of their children. A third consistent criticism involved our use of a category of “ambiguous siblings,” NLSY79 and NLSYC kinship pairs who were definitely full or half siblings but who could not be further distinguished. In early kinship pair research, the ambiguous sibling categories had phenotypic kinship correlations midway between half- and full-sibling correlations. Given hundreds of ambiguous pairs in each dataset, the category was empirically useful, especially for subgroup analyses by race and gender.
We agreed with many early criticisms; some of these features indeed were threats to internal and external validity. Just as obviously, we rejected the view that threats to validity automatically disqualify data as scientifically valid. We prefer to use the tenets of quasi-experimental design methodology (e.g., Shadish et al. 2002) to carefully address threats to both internal and external validity. Further, we have consistently argued the irony of some criticisms, given that behavior genetics has built its foundation and reputation in part on the use of creative and opportunistic kinship links (e.g., adoptive siblings, MZ/DZ twins raised apart).
Starting in the mid-1990s, we spent considerable research effort attempting to get direct ascertainment questions into the NLSY surveys, but with little success. Economists at the Bureau of Labor Statistics (BLS, which has managed and financially supported the NLSY79 survey process) were concerned that asking about level of relatedness could potentially offend some respondent families, who might drop out. They recommended that we obtain independent funding for new direct ascertainment (one cost estimate was around $200,000 of direct cost funding in 1996 to add those questions to one survey). However, we could not obtain permission from cautious BLS administrators to move forward with a grant application, which made the cost a moot issue. To support this effort, we drafted direct ascertainment questions for each survey, pilot tested those questions, and set them aside. Eventually, these questions became useful.
By the early 2000’s, the NLSY-C/YA kinship links needed to be updated. Thousands of new children had been born to the NLSY79 females (though childbearing was not completed), and many in the original NLSYC kinship links had aged into the NLSY-YA, the separate survey for NLSYC respondents aged 15 and older. Between 2003 and 2005 we updated the NLSY-C/YA links, which were immediately put to use by a number of research teams. Important publications that emerged included Rodgers et al. (2008); D’Onofrio et al. (2008); Lahey et al. (2009); Mendle et al. (2009); Hart et al. (2010); Salsberry and Reagan (2010); Goodnight et al. (2012); Beaver et al. (2013); Cheung et al. (2014); Rodgers et al. (2015); and a number of other articles. This linking update was completed in 2005, and plain text (CSV) and SAS files containing the links were e-mailed to 50+ researchers who had expressed earlier interest, and then over the next several years to others upon request.
In 2006, an important new development emerged. Center for Human Resource Research (CHRR), the Ohio State affiliate institute that manages and distributes the NLS datasets, decided (with support from BLS) to proceed with direct ascertainment. Our original questions were included in the 2006 NLSY79 and the NLSY-YA surveys, and have continued to be included on the NLSY-YA for respondents who passed age 15 in 2008, 2010, 2012, 2014, and continuing. We implemented a three-year project from 2011 to 2014 to update both the NLSY79 and NLSYC kinship links. During this update, we used the new direct ascertainment questions; information from this type of ascertainment we refer to as “explicit indicators,” leading to “explicit links.” We also used the prior, indirect information that was used in earlier linking efforts. We refer to that type of ascertainment as “implicit indicators,” leading to “implicit links.” (Note that many kinship links are developed using both types of indicators.) Instead of SAS algorithms, we implemented object-oriented programming using C#. We also began a user services support office, which has assisted researchers with kinship linking projects since 2011, and continuing through at least 2017. We distributed updated NLSY-C/YA kinship links in May, 2012, sent by e-mail to approximately 100 NLSY researchers, and with online availability through SAS and CSV files. The NLSY79 kinship links were distributed in a beta version (using only new explicit indicators) in December, 2012. A completed update of all datasets was distributed in November, 2013, based on both explicit and implicit indicators.
The NLSY97 kinship links have not been developed (by fall, 2015). However, direct ascertainment was included in the original 1997 survey questions, reducing the time that will be required to produce quality links. We have a plan for defining NLSY97 links, implemented within the next few years, including integration of NLSY97 into the online kinship link system.
Modern linking mechanisms and validity studies
During the 20+ years since we first began developing NLSY kinship linking algorithms, we have implemented a number of logical and data management innovations that we describe in this section. During this period, programming methods have transitioned from SAS-oriented linking algorithms to object oriented programming using C#, and from storage on tapes (originally) and diskettes to online storage (along with online documentation and associated vignettes); there are many advantages to this transition for linking purposes. In these most recent versions, we produce linking files in CSV and SAS formats (which are readable by SAS, R, Excel, etc.). Furthermore, we have also developed the R package, NlsyLinks (http://cran.r-project.org/package=NlsyLinks), which bundles biometric estimation functions with NLSY data. Its examples and documentation can assist researchers new to BG and the NLSY to learn this type of modeling, and its basic functions can also save time and effort for experienced researchers/programmers. We also post our linking algorithm as an open-source code repository. Typical researchers will never read the C# code, but rather will deal with the product of the code (i.e., the kinship links themselves). Programming-oriented BG researchers or those with certain requirements may readily adapt the code for special purposes. (Earlier linking algorithms were considered proprietary; only links, not the algorithms, were distributed).
In addition to the linking information, we also post online a set of vignettes that serve two purposes. First, they perform basic data management necessary to use the kinship links, including doing extracts from the NLSY online files, producing double-entry data structure, and merging phenotype scores into the double-entry data structure. Second, they demonstrate and illustrate a number of basic biometrical methods, including fitting univariate ACE models, multivariate ACE models, and Cholesky models (see Neale and Cardon 1992).
The kinship links themselves have been created using several basic principles. First, we consider information from the 2006 explicit links to be primary. However, both old implicit links and implicit links developed from new indirect questions are used extensively. In many cases the implicit links solve problems that cannot be addressed through the explicit links (because of missing data or other logical weaknesses). Further, in the thousands of redundant links that can be assigned using both explicit and implicit information, there is high agreement between the two methods in how they assign kinship links. However, it is also clear that the combination of the two types of information—both explicit and implicit indicators of relatedness—provides higher-quality kinship links than those using only one or the other.
A second principle that we implement is to resolve inconsistencies within families when possible. Because there are many thousands of kinship links to manage, virtually any possible inconsistency that can occur has occurred. As a simple example, if the older two of three brothers indicated in their explicit links that they are full-siblings to their other two brothers, but the third and youngest brother indicated a half-sibling relationship to the older two, this is a logically impossible inconsistency. By using longitudinal structure, the implicit indicators that used information from maternal reports, and additional implicit information from the respondents themselves, we have been able to resolve most such inconsistencies.
In addition to those first two innovations, three additional principles are described here. The third, implemented in the latest revision cycle, is production of a master kinship linking file that includes all of the NLSY79 links, all of the NLSYC links, and also cross-generational links. Across the NLSY79 (Gen1) and NLSY-C/YA (Gen2) generations are links from mothers to daughters, aunts to nieces and nephews, and uncles to nieces and nephews (see Rodgers et al. 2008, for a paper based on cross-generational links).Footnote 3 It should be noted that, because only the children of NLSY79/Gen1 females are included in the NLSYC/Gen2 dataset, there are no father–child links. The uncle–niece/nephew links are available from the brothers of Gen1 females, whose children are in the Gen2 dataset. There are over 42,000 links within this master file, with variables indicating the type of link. Evaluating cross-dataset and cross-generational invariance properties within the NLSY data will become a new and important source of information for NLSY researchers; in some (perhaps many) cases, NLSYC and NLSY79 files can be combined. Fourth, we have produced both kinship pair structure, and also multi-level within-family data structure (consistent with methods developed by Guo and Wang 2002; McArdle and Prescott 2005; van den Oord, 2001). Fifth, we provide the sophisticated user with considerable flexibility by defining flags for certain data problems (e.g., inconsistencies that cannot be resolved). One of these flags notes potential for unreliability within the responses themselves. When respondent or maternal reports are simply in error (because of duplicity, confusion, or incorrect information), no amount of logical data management can correct this problem; however, certain data patterns imply incorrect responses, and we identify those for use by researchers.
Some of our modern validity studies are similar to the original methods used in the 1990’s. We still use height, weight, body-mass-index (BMI), and other phenotypes in ACE models, and compare the results to meta-analyses and other research studies (e.g., Visscher 2008). The current results for the updated NLSY79 and NLSYC data show high concurrent validity in relation to other studies. Rodgers et al. (2007, p. 349) listed a number of past kinship link findings along with results from other research teams and data sources that matched; phenotypes included age at first intercourse, birth weight and gestational length, delinquency, and education/IQ/age at first birth. To add to published validity studies from the past, we present validation results for NLSY79 and NLSY-C/YA height later in this paper.
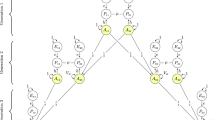
Many performance metrics informed the linking algorithm’s development. One newly-developed approach resembles receiver-operator characteristic (ROC) methodology, in which agreements and disagreements between different classification methods are monitored (for discussion of ROC methodology, see Green and Swets 1966; Lynn and Barrett 2014). We were most interested in agreement between the explicit and implicit assignments. In Fig. 2, each point along the line indicates the performance of a successive version of the linking algorithm. The left-hand figure shows agreements plotted against disagreements in the same metric. That the points are all in the upper left-hand corner indicates that there were many more agreements than disagreements. This portrayal is expanded using different scales on the right hand side.
Using ROC curve logic, the performance of each algorithm version was monitored to inform future versions. The tension between the implicit and explicit assignments among Gen2 pairs eventually arrived at a 21:1 agreement-to-disagreement ratio. The axes represent the counts of agreement/disagreement, and the text indicates the algorithm version (which progresses from version 1 to version 40). The right panel zooms on the relevant movement
To illustrate interpretation of Fig. 2: Our 1st algorithm version had 6,101 Gen 2 sibling pairs whose implicit assignment perfectly agreed with the explicit assignment, balanced against 417 disagreements (a 15:1 ratio). The second version, achieved by making a change in the algorithm, barely improved the number of agreements, but substantially reduced the number of disagreements. By the 40th version, there were 7492 agreements and 355 disagreements (a 21:1 ratio). Figure 2‘s progression is not always smooth, reflecting how we experimented and used many different approaches in an effort to create more agreements and fewer disagreements. The initial versions targeted easily-classified pairs. Later versions targeted more difficult pairs; these algorithmic decisions typically used many NLSY items simultaneously. In defining new algorithms, we balanced face validity against methods that empirically drove the ROC curves further toward the upper left-hand corner of Fig. 2.
Other ROC-like comparisons were monitored besides the ones in Fig. 2. These include curves comparing the success of current links against previously-released links, Gen1 implicit against Gen1 explicit assignments, Gen1 explicit assignment against the Gen1 1979 household roster, and the Gen1 implicit assignment against the Gen1 1979 household roster. Ideally, the ROC points for later versions would move toward the top left corner (i.e., high agreement and low disagreement), but trade-off decisions were necessary in some cases and so the movement was not always systematic (see Fig. 2, versions 5 through 9). Further, some performance metrics could not be expressed in ROC terms, such as the total count of Gen 1 pairs linked (because many pairs had only the implicit items, and no definitive assignment from the explicit items or 1979 roster).
In summary, our goal within this section has been to provide background and support for the quality of both the NLSY79 and NLSYC kinship links. Besides the overall quality of these links—which has improved substantially from earlier ones developed in the 1990’s and early 2000’s—there are other features of the NLSY data to recommend to researchers. Among those are the broad array of outcomes that can be used to construct phenotype scores for many different variables of interest to BG researchers, the longitudinal structure for BG researchers who study development and aging, the broad age range that now exists to support research from infancy through the mid-50s (and consistently increasing), and the fact that the original NLSY79 links were obtained within the context of a national household probability sample. The NLSY97 links, once developed, will have many of the same important features.
It is worth emphasizing that many researchers contributing to the corpus of BG literature have consistently sacrificed external validity for high internal validity (see Mook 1983, for expansive justification of this decision). Especially in using recruited twin samples and many adoption designs, the focus of biometrical research has been on internal validity, and issues of generalizability have often been ignored. The NLSY as a national household probability sample (and whose data release includes complex sample weight calculation and availability) provides a relatively high level of external validity (in combination with potentially high internal validity). Only a few other such datasets (e.g., the Add Health data) support biometrical research with high levels of both internal and external validity and longitudinal structure. In addition, there also exist national registry-based twin samples (e.g., in the U.S., Denmark, Sweden, and Australia, among others) that provide large sample sizes and excellent levels of external validity.
Publications
A catalogue of journal articles, book chapters, technical reports, and dissertations/theses that have used the NLSY kinship links is posted online on our kinship link website: http://liveoak.github.io/NlsyLinks/. Many of these publications emerged from work by Rodgers, Rowe, and their collaborators; a number of other research teams have used the kinship links as well. As of January, 2016, the site above lists a total of 61 research contributions that have used the kinship links: 47 research articles (44 refereed journal articles and 3 book chapters), 4 internal publications (including documentation of the NLSY kinship links), and 10 dissertations/theses that have used the NLSY kinship links. Of the 47 research articles, 14 exclusively used the NLSY79 kinship links, 26 exclusively used the NLSY-C/YA kinship links, and 7 used both the NLSY79 and NLSY-C/YA. (We note that there are likely several published kinship link articles that we have not identified.) The phenotypes that have been investigated include an extensive set of studies of fertility outcomes and precursors, including pubertal development, adolescent sexual behavior, and fertility outcomes; a number of studies of childhood and adolescent problem behaviors and conduct disorder; studies of smoking and drinking, both by NLSY respondents and their parents; several studies of intelligence/cognitive development; studies of educational outcomes; studies of health and physical development; a study of childhood maladjustment; a study of the quality of the child’s home environment; and several studies that have used combinations of these phenotypes.
Two general methodologies are represented in past NLSY kinship link studies. First, a number of biometrical studies are represented. Some of these are standard heritability studies fitting ACE models to univariate and multivariate outcomes. Others are more complex biometrical designs using Cholesky models, competing pathway models, multivariate models, or other design structures developed for particular purposes (e.g., see the Mother–Daughter–Aunt–Niece, or MDAN, design presented in Rodgers et al. 2007). Prototypes of biometrical research on intelligence and cognitive functioning include Rodgers et al. (1994b), Rowe et al. (1995), Rowe and Cleveland (1996), and van den Oord and Rowe (1997). Examples of biometrical studies of problem behaviors include Rodgers et al. (1994a), Cleveland et al. (2000), (Rodgers et al. 2001), Van Hulle et al. (2009), and Connelly and Beaver (2014). Examples of biometrical studies of pubertal development, sexuality, and fertility include (Rodgers et al. 1999), Doughty and Rodgers (2000), Neiss et al. (2002); Rodgers et al. (2007), and Miller et al. (2010).
The second general methodology has used the relatedness indicators from the kinship links within design structures to control for family background selection factors to assess causal influences on various child outcomes, including sexuality, cognitive development, and problem behaviors. Most of these designs are designated as “children-of-sibling” designs (or, sometimes, “discordant sibling designs”), slightly weaker but often more practical than children-of-twin designs. Some of those studies have also been biometrically informed (e.g., D’Onofrio et al. 2008; Rodgers et al. 2008), but the major innovation is the ability to control family background by linking sisters from the NLSY79 dataset to study child outcomes in the NLSYC dataset. Prototypes include Lahey et al. (2009), Mendle et al. (2009), D’Onofrio et al. (2009), Jaffee et al. (2011), Goodnight et al. (2012), D’Onofrio et al. (2012), and Cheung et al. (2014). In some studies, the family has been used as a unit, and results compared to those obtained from cross-sectional studies, providing the same kind of within-family control (e.g., Rodgers et al. 2000; Wichman et al. 2006); these have used the NLSY sibling data, but have not needed the kinship links themselves.
In the sections that follow, we separately assess the NLSY79, the NLSYC, and cross-generational file structures in terms of their utility for fitting biometrical models. Included within these are validity analyses, which illustrate at a basic level some of the principles involved in using the NLSY kinship links. We then emphasize the potential for cross-dataset research, using the “super-file” of kinship links that combine over 42,000 links from two of the NLSY datasets. We conclude with general discussion of the kinship links, including suggestions for biometrical work with all three NLSY data sources.
Using the NLSY79 kinship links
In the updated NLSY79 kinship links released in November, 2013, we have classified 5038 links out of a possible 5302 links, a 95 % classification rate (compare to the 63 % classification rate from the earlier mid-1990’s NLSY79 files used in previous studies using the NLSY79 kinship links). Table 1 presents the different kinship categories available for classification, with sample sizes for each category. It is notable that there are many more full siblings than the combination of all other categories, which may appear problematic at first glance. We note, though, that many past successful studies have been conducted using smaller NSLY79 sample sizes with the same unbalanced patterns. Further, these sample sizes reflect the distribution of those kinship categories in households in the December, 1978 U.S. population. The earlier 1996 NLSY79 kinship links had a much smaller denominator of kinship pairs to classify, because only sibling links involving the oldest NLSY79 sibling in the household were submitted to the linking algorithm. The 2012 NLSY79 links were not based on a larger sample—the NLSY79 is the same dataset as the original (except for loss at later rounds due to mortality, attrition, and design adjustments; see the overall summary above). However, all pairs of within-family and cousin pairs were submitted to the linking algorithm to create the most recent links. This adjustment explains the difference between the 3890 potential links in 1996 and the 5302 in 2012. Obviously, not only did the number of potential links increase substantially, but also the success rate increased (from 63 to 95 %) because of the availability of the explicit indicators collected in the 2006 survey, used in combination with the earlier implicit links.
We used a similar validity analysis for the 2012 NLSY79 kinship links to the one we developed for the 1996 links (the original was documented in Rodgers 1996). The original NLSY79 validity study followed a similar strategy to the one used for the 1994 NLSY-C/YA links. Both relied on several mechanisms, the primary of which was ACE modeling (Neale and Cardon 1992) of adult height and weight data, which have high and carefully documented heritabilities, with a focus on height (which has higher heritability than weight).
Johnson et al. (2010) specified height heritabilities between .70 and .95, with adult height h2 generally around .80. Visscher et al. (2006) found adult height h2 = .80 in Australian Twin Registry data, and Silverntoinen et al. (2000) found male h2 = .78 and female h2 = .75 in a Finnish twin cohort. Furthermore, past research has noted race differences in height heritability, with lower h2 values for African-Americans and especially Hispanics (e.g., Lai 2006; Roberts et al. 1978). Estimated c2 values for adult height are consistently around zero.
We used gender-standardized and single-entered adult height measures contained within the NLSY79 files. We estimated an ACE model using both OpenMx (Neale et al. 2015) and lavaan; we report lavaan results, in Table 2. (We report the lavaan results, because that is the structural equation modeling program that runs in the background of our online vignettes; reporting lavaan results provides compatibility for users who wish to use the vignettes in their research). Using the NLSY79 sibling-based kinship pairs (excluding cousins, adoptive siblings, and ambiguous siblings, because of low sample sizes and questions of reliability), h2 = .70, c2 = .11, and e2 = .18. Patterns by race and gender are also presented, and these approximately match what is expected based on results above.
Using the NLSY-children/young adult kinship links
We released a complete NLSY-C/YA kinship link dataset in May, 2012. In November, 2013, we released updated and final versions of three datasets: NLSY79, NLSY-C/YA, and a “super-dataset” combining both within- and cross-generational links. The NLSY-C/YA links were slightly updated in the 2013 release compared to the 2012, and we report sample sizes from the most recent (2013) release.
The 2013 NLSY-C/YA kinship links file contained 16,083 kinship pairs with identified links (100 % of the total); see Table 3. If we exclude the ambiguous sibling and ambiguous twin categories, the sibling classification rate (i.e., not counting cousins) is 10,466 links out of a possible 11,088 sibling links, a 94 % classification rate (compared to the 75 % classification rate from the 2005 NLSY-C/YA links). Within the 2013 release, there are 27 MZ twin pairs; 83 DZ twin pairs; 6914 full sibling (non-DZ twin) pairs; and 3,442 half sibling pairs; these can be considered approximately representative of the distribution of siblings produced by the NLSY79 female cohort. Remaining are 610 sibling kinship pairs who our algorithm could not classify (but who are definitely either full or half siblings; this “ambiguous sibling” category was reduced from 2,636 ambiguous pairs in the 2005 kinship links to the current 610, and can be used in some research studies as their own category with R = .375), and 12 ambiguous twin pairs who are definitely same-sex twins, but cannot be distinguished by our algorithm as either MZ or DZ twins (and who can also be used in some studies by assigning R = .75). In addition, there are 4995 cousin pairs.
There are many categories of cousins, because there are many categories of relatedness among the mothers/aunts of cousins. NLSY-C/YA cousin pairs whose mothers are MZ twins are genetically equivalent to half siblings (R = .25); there are 18 “half-sibling cousins.” Pairs whose mothers are full siblings are full cousins (R = .125); there are 3,941 full cousins identified in the 2013 dataset. Pairs whose mothers are R = .375 have an estimated cousin genetic coefficient of R = .09375; there are 12 such identified cousin pairs. Pairs whose mothers are half-siblings (R = .25) are half-cousins with a genetic coefficient of R = .0625; there are 309 such half-cousin pairs. There are several other cousin categories as well, including 131 cousin pairs whose mothers are of entirely unknown relatedness. The overall classification rate for cousins is 97 %. In Table 4, we identify all of the possible cousin pair categories in the NLSY-C/YA dataset, and provide sample sizes for each. These large and diverse cousin categories support the kind of cousin studies suggested by Fisher (1918) and discussed earlier in this paper. There is potential to explore the “cousin anomaly,” often noted by NLSY researchers (including our own research, in a number of different studies). The anomaly is that cousin correlations are often too high on a number of different phenotypes to be consistent with other biometrical patterns.
The 3276 mothers (females from the NLSY79) whose children contribute sibling pairs to the NLSY-C/YA dataset include 1521 mothers with one kinship pair (i.e., mothers with two NLSY-C/YA children), 94 mothers with two kinship pairs (mothers with three or more children, but missing data for some of the kinship pairs), 858 mothers with three kinship pairs (almost all of whom have three children, a few of whom have more than three children but with missing data for some of their children), 120 mothers with four kinship pairs, 7 mothers with five kinship pairs, 366 mothers with six kinship pairs (almost all of whom had four children, resulting in six pairs of children), and 310 mothers with more than six kinship pairs (ranging from seven to 58 pairs).
We fit the NLSY-C/YA kinship pairs using ACE models to measures of adult height. Adult height was measured in inches in the NLSY-YA survey between ages 19–25. We used gender-standardized measures of adult height to adjust for gender differences. Data were used in single-entry form, ambiguous siblings and twins were deleted, and models were fit using DF Analysis (DeFries and Fulker 1985; Rodgers and Kohler 2005), OpenMx (Neale et al. 2015), and lavaan; results were highly similar across the software procedures; we report lavaan results. As noted above, meta-analyses show adult height h2 is generally in the range of .70–.90, with c2 around 0. ACE model results are presented in Table 5. These estimates are highly consistent with meta-analyses and studies cited above. For the overall sample, height heritability was h2 = .79, with c2 = .03. Heritabilities for African-American and Hispanic samples are smaller, as expected. We note that Tables 2 and 5 are of identical structure, and each can be considered a replication of the other; the potential for this kind of replication is one of the many strong features of the NLSY files, with equivalent instrumentation in cross-generational data.
Though the height results are the ones we use as an example, and the ones to which we give greatest credence, we ran other similar validity analyses using different phenotypes. Those included adult weight, BMI, female age at menarche, female age at first intercourse, and eye color, all used to estimate biometrical models. These results will be presented in future research, with papers devoted to the several separate topics (e.g., pubertal development; fertility/reproduction; physical development, etc.).
Using the NLSY79 and NLSY-children/young adult “super file”
With the final release of the NLSY79 and NLSY-C/YA kinship links in November 2013, we also released a master file that has great potential to support interesting and innovative BG (and many other types of) research in the future. We combined 5302 NLSY79 kinship pairs, 16,083 NLSY-C/YA kinship pairs, and 21,388 cross-generational kinship pairs to create a “super file” with 42,773 kinship pairs. The cross-generational kinship pairs consist of mother–daughter pairs, aunt–niece/aunt–nephew pairs, and uncle–niece/uncle–nephew pairs.Footnote 4 Few datasets exist with the breadth of kinship pairs available within this “NLSY Super-File,” and those that do have provided extensive empirical support for a broad range of past research. One example is the “VA 30,000 Study,” a large dataset of almost 15,000 twin pairs from Virginia that has similar cross-generational features (e.g., D’Onofrio et al. 2007; Eaves et al. 1999).
The NLSY super-file has data structure that can be used for new and innovative research studies based on ideas such as the following. First, because in many cases instrumentation is identical across the two generations, analyses can be run with relatively little concern for instrumentation bias (except that the meaning of some words can change over time). Second, invariance studies can be conducted to determine if the status of the measurement process is fixed, or has changed, over the two different generations. In cases where it has not (and in those where it has with adjustments), the two generations can be combined in a remarkably large biometrically-informed dataset. Third, certain assumptions can be investigated within this dataset. For example, the equal environments assumption, which has been carefully studied within the BG community and the cause of much criticism from outside, can be studied from a unique perspective, because the nature of the shared environment is different across generations (who never shared a childhood rearing environment) compared to within-generation. If results converge between the cross-generational data and the within-generational data, this convergence would provide at least partial support for the legitimacy of the equal environments assumption in relation to a given phenotype. Fourth, the NLSY97 kinship links can eventually be added to this super-file to support broader research across time and across all three NLSY datasets.
Conclusion
In conclusion, we recommend the three NLSY datasets to researchers doing biometrical research (and also to other broader developmental/family studies researchers). We especially recommend the kinship links to graduate students planning thesis or dissertation research, and to early-career faculty. These recommendations are based on the ease and accessibility of the NLSY data and kinship links, the broad array of research topics that can be developed, and the potential for doing unique and methodologically sophisticated research using these data resources. A publication history based on over 60 articles, book chapters, technical reports, and dissertations/theses supports the viability of the use of the NLSY kinship links for productive research, and can be consulted for research ideas. Note that this earlier publication stream was based on earlier versions of the kinship links, which were of lower quality (reliability and validity) and reduced sample sizes compared to those recently released and described in detail within the current paper. Further, extensive online support and a User Service research support function (available by contacting any of the first three authors) is provided by the current NLSY kinship research team to assist with software and modeling issues associated with using the kinship links. The NLSY datasets are remarkable resources, even without accounting for their biometrical structure. Combining the kinship link information with the long-term longitudinal structure, the cross-generational design, the thousands of phenotypes that can be constructed, and the probability sample origin of the original NLSY79 creates a highly unique and valuable data resource.
Notes
David Rowe, Rodgers’ original collaborator on the kinship linking projects, passed away in February, 2003.
In September, 1996, Baydar and Rodgers/Rowe compared kinship links, and found that their independent efforts produced highly similar linking results, a rate of agreement of 94.3 %.
Rodgers et al. (2008) presented the Mother–Daughter–Aunt–Niece (MDAN) design, in which correlations from mother to daughter pairs are compared biometrically to those from aunt to niece pairs. Mothers and aunts are often either the same person (i.e., a mother of one or more NLSYC daughters also was the aunt of one or more NLSYC nieces), or are related as sisters or half-sisters, providing substantial control over unobserved heterogeneity due to family background.
An interesting challenge involves verbal identification of certain cross-generational pairs, because of implied gender. We specify Parent–Child kinship links simply as “ParentChild” links. But to separately identify aunt–niece, aunt–nephew, uncle–niece, and uncle–nephew files would require gender identification of members of the kinship links (creating inconsistency compared to other links such as full sibling, half sibling, twin, and parent–children, identified without gender specification). We have searched the literature/internet for resolutions of this problem; an interesting solution refers to non-gendered Nieces or Nephews as “Niblings,” the counterpart to non-gendered “Siblings.” We created a new name for non-gendered Uncle/Aunts of “Niblings;” we use “AUncle” for this category. Thus, an Uncle/Aunt in Gen1 paired with Niblings in Gen2 are designated as “AUncleNibling” pairs within our files.
References
Baydar N, Greek A (2001) Analysis of data from related individuals. Working paper, Battell Centers for Public Health Research and Evaluation, Seattle
Beaver KM, Connolly EJ, Schwartz JA, Al-Ghamdi AS, Kobeisy AN (2013) Genetic and environmental contributions to stability and change in levels of self-control. J Crim Justice 41:300–308
Chase-Lansdale PL, Mott FL, Brooks-Gunn J, Phillips DA (1991) Children of the National Longitudinal Survey of Youth: a unique research opportunity. Dev Psychol 27:918–931
Cheung AK, Harden KP, Tucker-Drob EM (2014) Gene x Environment interactions in early externalizing behaviors: parental emotional support and socioeconomic context as moderators of genetic influences? Behav Genet 44(5):468–486
Cleveland HH, Wiebe RP, van den Oord EJCG, Rowe DC (2000) Behavior problems among children from different family structures: the influence of genetic self-selection. Child Dev 71:733–751
Connolly EJ, Beaver KM (2014) Examining the genetic and environmental influences on self-control and delinquency: results from a genetically informative analysis of sibling pairs. J Interpers Violence 29:707–735
D’Onofrio BM, Turkheimer E, Emery RE, Maes HH, Eaves LJ (2007) A children of twins study of parental divorce and offspring psychopathology. J Child Psychol Psychiatry 48:667–675
D’Onofrio BM, Van Hulle CA, Waldman ID, Rodgers JL, Harden KP, Rathouz PJ, Lahey BB (2008) Smoking during pregnancy and offspring externalizing problems: an exploration of genetic and environmental confounds. Dev Psychopathol 20:139–164
D’Onofrio BM, Goodnight JA, Van Hulle CA, Waldman ID, Rodgers JL, Rathouz PJ, Lahey BB (2009) Maternal age at childbirth and offspring disruptive behavior: testing the causal hypothesis. J Child Psychol Psychiatry 50:1018–1028
D’Onofrio BM, Rickert ME, Langstrom N, Donahue KL, Coyne CA, Larsson H, Ellingson JM, Van Hulle CA, Iliadou AN, Rathouz PJ, Lahey BB, Lichtenstein P (2012) Familial confounding of the association between maternal smoking during pregnancy and offspring substance use and problems: converging evidence across samples and measures. Arch Gen Psychiatry 69:1140–1150
DeFries JC, Fulker D (1985) Multiple regression analysis of twin data. Behav Genet 15:467–473
Doughty D, Rodgers JL (2000) Behavior genetic modeling of menarche in U.S. females. In: Rodgers JL, Rowe DC, Miller WB (eds) Genetic influences on fertility and sexuality. Kluwer Academic Press, Boston
Eaves L, Heath A, Martin N, Maes H, Neale M, Kendler K, Kirk K, Corey L (1999) Comparing the biological and cultural inheritance of personality and social attitudes in the Virginia 30,000 study of twins and their relatives. Twin Res 2:62–80
Fisher RA (1918) The correlation between relatives on the supposition of Mendelian inheritance. Philos Trans R Soc Edinburgh 52:399–433
Goodnight JA, Lahey BB, Van Hulle CA, Rodgers JL, Rathouz PJ, Waldman ID, D’Onofrio BM (2012) A quasi-experimental analysis of the influence of neighborhood disadvantage on child and adolescent conduct problems. J Abnorm Psychol 121:95–108
Goodnight JA, D’Onofrio BM, Cherlin AJ, Emery RE, Van Hulle CA, Lahey BB (2013) Effects of multiple maternal relationship transitions on offspring antisocial behavior in childhood and adolescence: a cousin-comparison analysis. J Abnorm Child Psychol 41:185–198
Green DM, Swets JA (1966) Signal detection theory and psychophysics. Wiley, New York
Guo G, Wang JM (2002) The mixed or multilevel models for behavior genetic analysis. Behav Genet 32:37–49
Harris KM, Halpern CT, Smolen A, Haberstick B (2006) The national longitudinal study of adolescent health (Add Health) twin data. Twin Res Hum Genet 9:988–997
Harris KM, Halpern CT, Haberstick BC, Smolen A (2013) The national longitudinal study of adolescent health (Add Health) sibling pairs data. Twin Res Hum Genet 16:391–398
Hart SA, Petrill SA, Kamp Dush CM (2010) Genetic influences on language, reading, and mathematics skills in a national sample: an analysis using the National Longitudinal Survey of Youth. Lang Speech Hear Serv Sch 41:118
Jaffee S, Van Hulle C, Rodgers JL (2011) Effects of non-maternal care in the first three years on children’s academic skills and behavioral functioning in childhood and early adolescence: a sibling comparison study. Child Dev 84:1076–1081
Johnson W, Turkheimer E, Gottesman I, Bouchard TJ (2010) Beyond heritability: twin studies in behavioral research. Curr Dir Psychol Sci 18:217–220
Lahey BB, Van Hulle CA, Rathouz PJ, Rodgers JL, D’Onofrio BM, Waldman ID (2009) Are oppositional-defiant and hyperactive-inattentive symptoms developmental precursors to conduct problems in late childhood?: genetic and environmental links. J Abnorm Child Psychol 37:45–58
Lai C-Q (2006) How much of human height is genetic and much is due to nutrition? Scientific American
Lynn SK, Barrett LF (2014) “Utilizing” signal detection theory. Psychol Sci 25:1663–1673
McArdle JJ, Prescott CA (2005) Mixed-effects variance components models for biometrical family analyses. Behav Genet 35:631–652
Mendle J, Harden KP, Turkheimer E, Van Hulle CA, D’Onofrio BM, Brooks-Bunn J, Rodgers JL, Emery RE, Lahey BB (2009) Associations between father absence and age of first sexual intercourse. Child Dev 80:1463–1480
Miller WB, Bard DE, Pasta DJ, Rodgers JL (2010) Biodemographic modeling of the links between fertility motivation and fertility outcomes in the NLSY79. Demography 47:393–414
Mook DG (1983) In defense of external invalidity. Am Psychol 38:379–387
Neale MC, Cardon LR (1992) Methodology for genetic studies of twins and families. Springer, New York
Neale MC, Hunter MD, Pritikin JN, Zahery M, Brick TR, Kickpatrick RM, Estabrook R, Bates TC, Maes HH, Boker SM (2015) OpenMx 2.0: extended structural equation and statistical modeling. Psychometrika. 10.1007/s11336-014-9435-8
Neiss M, Rowe DC, Rodgers JL (2002) Does education mediate the relationship between IQ and age of first birth? A behavior genetic analysis. J Biosoc Sci 34:259–275
Roberts DF, Billewicz WZ, McGregor IA (1978) Heritability of stature in a West African population. Ann Hum Genet 42:15–24
Rodgers JL (1996) NLSY Youth linking algorithm. Unpublished manuscript, Department of Psychology, University of Oklahoma
Rodgers JL, Kohler H-P (2005) Reformulating and simplifying the DF analysis model. Behav Genet 35:211–217
Rodgers JL, Rowe DC, Li C (1994a) Beyond nature versus nurture: DF analysis of nonshared influences on problem behaviors. Dev Psychol 30:374–384
Rodgers JL, Rowe DC, May K (1994b) DF analysis of NLSY IQ/achievement data: nonshared environmental influences. Intelligence 19:157–177
Rodgers JL, Rowe DC, Buster M (1999) Nature, nurture, and first sexual intercourse in the USA: fitting behavioral genetic models to NLSY kinship data. J Biosoc Sci 31:29–41
Rodgers JL, Cleveland HH, van den Oord EJCG, Rowe DC (2000) Resolving the debate over birth order, family size, and intelligence. Am Psychol 55:599–612
Rodgers JL, Buster M, Rowe DC (2001) Genetic and environmental influences on delinquency: DF analysis of NLSY kinship data. J Quant Criminol 17:145–168
Rodgers JL, Bard DE, Miller WB (2007) Multivariate Cholesky models of human female fertility patterns in the NLSY. Behav Genet 37:345–361
Rodgers JL, Bard DE, Johnson AB, D’Onofrio BM, Miller WB (2008) The cross-generational mother-daughter-aunt-niece design: establishing validity of the MDAN design with NLSY fertility variables. Behav Genet 38:567–578
Rodgers JL, Van Hulle C, D’Onofrio BM, Rathouz PJ, Beasley WH, Johnson AB, Waldman ID, Lahey BB (2015) Behavior problems and timing of menarche: a developmental longitudinal biometrical analysis using the NLSY-Children data. Behav Genet 45:51–70
Rowe DC, Cleveland HH (1996) Academic achievement in African-Americans and Whites: are the developmental processes similar? Intelligence 23:205–228
Rowe DC, Vaszonyi AT, Flannery DJ (1995) Ethnic and racial similarity in developmental process: a study of academic achievement. Psychol Sci 6:33–38
Salsberry PJ, Reagan PB (2010) Effects of heritability, shared environment and non-shared intrauterine conditions on child and adolescent BMI. Obesity 18(9):1775–1780
Shadish WR, Cook TD, Campbell DT (2002) Experimental and quasi-experimental designs for generalized causal inference. Houghton Mifflin, Boston
Silverntoinen K, Kaprio J, Kahelma E (2000) Genetic and environmental contribgutions to the association etween body height and educational attainment: a study of adult Finnish twins. Behav Genet 30:477–485
van den Oord EJCG (2001) Estimating effects of latent and measured genotypes in multilevel models. Stat Methods Med Res 10:393–407
van den Oord EJCG, Rowe DC (1997) An examination of genotype-environment interactions for academic achievement in an U.S. national longitudinal survey. Intelligence 25:205–228
van den Oord EJCG, Rowe DC (1999) A cousin study of associations between family demographic characteristics and children’s intellectual ability. Intelligence 27:251–266
Van Hulle CA, Waldman ID, D’Onofrio BM, Rodgers JL, Rathouz PJ, Lahey BB (2009) Developmental structure of genetic influences on antisocial behavior across childhood and adolescence. J Abnorm Psychol 118:711–721
Visscher PM (2008) Sizing up human height variation. Nat Genet 40:489–490
Visscher PM, Medland SE, Ferreira MAR, Morley KI, Zhu G, Cornes BK, Montgomery GW, Martin NG (2006) Assumption-free estimation of heritability from genome-wide identity-by-descent sharing between full siblings. PLoS Genet 2(e41):316–325
Wichman A, Rodgers JL, MacCallum RC (2006) A multilevel approach to the relationship between birth order and intelligence. Pers Soc Psychol Bull 32:117–127
Acknowledgments
Three grants from the National Institutes of Health (NICHD) have supported the work described within this paper: in the 1990s, R01-HD21973; in 2003–2007 RO1-HD043265; in 2012–2014, R01-HD065865 (Joseph Lee Rodgers was PI on each grant). The authors express appreciation to the Bureau of Labor Statistics and the National Opinion Research Center for longstanding expertise in conceptualizing, funding, and collecting the NLSY data. Particular appreciation is expressed to many individuals at the Center for Human Resource Research at Ohio State for expert management of the NLSY data, and more specifically for conceptual, statistical, and data management support of past NLSY kinship linking efforts. Those individuals at CHRR include Randy Olsen, Elizabeth Cooksey, Frank Mott, Paula Baker, Steven McClaskie, and Karima Nagy. The co-authors on this paper are those who have contributed to coding or conceptualizing the NLSY kinship links since the original linking projects in the early 1990s; with the exception of van den Oord, who developed his own code for NLSYC kinship links, each co-author has been financially supported on one or more of the NIH grants listed above.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by the any of the authors.
Additional information
Edited by Brian D’Onofrio.
David C. Rowe—deceased.
Rights and permissions
About this article
Cite this article
Rodgers, J.L., Beasley, W.H., Bard, D.E. et al. The NLSY Kinship Links: Using the NLSY79 and NLSY-Children Data to Conduct Genetically-Informed and Family-Oriented Research. Behav Genet 46, 538–551 (2016). https://doi.org/10.1007/s10519-016-9785-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10519-016-9785-3