Abstract
The aim of this study was to evaluate the anti-biofilm and anti-virulence properties of petroselinic acid (PSA) against the environmental pathogen Serratia marcescens. PSA significantly inhibited the quorum sensing (QS)-dependent virulence factors such as prodigiosin, protease productions, and biofilm formation in S. marcescens. The antibiofilm potential of PSA was also confirmed through light, confocal laser scanning, and scanning electron microscopic analyses. Furthermore, PSA effectively inhibited the biofilm-related phenomena such as exopolysaccharide production, hydrophobicity production, swimming, and swarming motility without affecting the bacterial growth. In FT-IR analysis, the PSA treated S. marcescens cells displayed a reduction in cellular components compared to the untreated controls. The real-time analysis revealed the downregulation of QS-controlled virulence genes such as bsmB, fimA, fimC, and flhD in S. marcescens on treatment with PSA. The obtained results strongly suggested that PSA could be further explored as an antipathogenic drug to treat QS-mediated infections caused by S. marcescens.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The Gram-negative bacterium Serratia marcescens is an environmental pathogen that infects a wide range of hosts, including plants, invertebrates, and vertebrates. In infants and immunocompromised patients, S. marcescens causes respiratory infections, urinary tract infections, meningitis, and sepsis (Hejazi and Falkiner 1997). S. marcescens is also a potent insecticidal bacterium that causes bacteremia in the insect hemolymph and results in rapid insect death (Chadwick et al. 1990). The infections caused by S. marcescens are challenging to treat as they show high resistance to a widespread variety of antibiotics including imipenem, aztreonam, and cephalosporins (Hejazi and Falkiner 1997). An important characteristic of S. marcescens is its ability to secrete a beta-lactamase enzyme, which confers resistance to a broad spectrum of beta-lactam antibiotics and complicates treatment (Yoon et al. 2005).
Biofilms are a self-secreted hydrated matrix made up of bacterial cells and biomolecules such as polysaccharides, nucleic acids, lipids, and proteins. Within a biofilm, bacterial cells are assembled into a mass and enclosed by a surrounding material referred to as extracellular polymeric substances (EPS) (Flemming and Wingender 2010). Also, growth in a biofilm enables bacterial pathogens to survive better in the presence of antibiotics or a hostile immune system (Fux et al. 2005). Development of a biofilm affords multilevel defense to bacterial pathogens against antibiotics, because of the slower growth rate and altered metabolism of bacterial pathogens in biofilms. The ability of S. marcescens to adhere to the surfaces of medical devices is enhanced by its biofilm formation. The quorum sensing (QS) mechanism plays a vital role in regulating biofilm formation in S. marcescens (Labbate et al. 2007). Similarly, the QS system regulates production of various virulence factors such as prodigiosin, protease, nuclease, lipase, hemolysin productions, and motility in S. marcescens (Labbate et al. 2007; Hejazi and Falkiner 1997). Therefore, the disruption of QS mechanism in S. marcescens possibly attenuates the virulence and protects the host against infections (Hentzer and Givskov 2003; March and Bentley 2004).
Petroselinic acid (PSA) is an uncommon fatty acid with unsaturation in the 6, 7-position, and it is a positional isomer of oleic acid (Cahoon et al. 1994). It is present in high quantities in plant seed oils belonging to the Apiaceae family (Placek 1963). In the fruits and vegetable oil of Coriandrum sativum, the quantity of PSA differs by 31–75% and it is considered as one of the maximum augmented sources of PSA (Uitterhaegen et al. 2015). PSA is an important ingredient in cosmetic preparations as a skin-irritation reducing agent, moisturizing agent, and anti-aging agent (Alaluf et al. 1999; Alaluf et al. 2002; Weinkauf et al. 1998). Also, it has substantial antimicrobial activity against several bacterial and fungal pathogens (Placek 1963). Despite its pharmacological importance, to the best of our knowledge no investigations have been made to reveal its anti-QS activity against bacterial pathogens. Therefore, the present research was performed to understand the effect of PSA on QS controlled biofilm and virulence genes expression in S. marcescens.
Materials and methods
Ethical statement
In the current study, human blood was collected from healthy donors by a technically trained person for research purposes. Written consent was obtained from donors for blood collection. The usage of healthy human blood was approved by Alagappa University Institutional Ethics Committee under the No. IEC/AU/2016/1/7.
Bacterial strains and growth conditions
S. marcescens reference strain (ATCC 14756) and clinical isolate (CI) (GenBank accession no. FJ584421) were used in this study. Both the strains were cultivated in Luria–Bertani (LB) medium (pH 7.0) at 28 °C overnight. For the experimental analyses, the S. marcescens strains were subcultured in LB medium till it reached 0.4 OD at 600 nm (1 × 108 CFU ml−1).
Compound preparation
Fifty milligram of petroselinic acid (Catalogue no. P8750, Sigma-Aldrich, St. Louis, MO, USA) was dissolved in 1 ml of methanol as stock solution and stored at 4 °C till further use.
Determination of effective QS inhibitory concentration of PSA against protease production in S. marcescens
QS inhibitory potential of PSA was assessed against protease production in S. marcescens by following the method of Srinivasan et al. (2016). After 18 h of growth at 28 °C, the PSA (100, 200, 300, 400, and 500 μg ml−1) and 1% methanol (Negative control) treated and untreated S. marcescens cultures were centrifuged at 16,770×g for 10 min and the cell-free culture supernatants (CFCS) were collected to quantify the protease production. 75 μl of PSA treated and untreated CFCS were added to 125 μl of 2% of azocasein buffered substrate in 1 M Tris–HCl (pH 8.0) and incubated for 30 min at 37 °C. Afterward, 600 μl of 10% trichloroacetic acid was added to all the tubes to dismiss the reaction and incubated for 20 min at − 20 °C. The tubes were then centrifuged for 10 min at 16,770×g and the supernatant (600 μl) was separated to which 700 μl of 1 M NaOH was added. The absorbance was taken at 440 nm in a UV–visible spectrophotometer.
Effect of PSA on the growth of S. marcescens
To examine the effect of PSA on the growth of S. marcescens, the bacterial cells were grown in LB medium in the absence and presence of PSA (200 and 400 µg ml−1) and 0.8% methanol (Negative control) at 28 °C for 18 h. After incubation, the cell density was measured using a UV–visible spectrophotometer at 600 nm.
Evaluation of cellular viability by XTT reduction assay
The cellular viability of PSA (200 and 400 µg ml−1) and 0.8% methanol (Negative control) treated and untreated S. marcescens were assessed by XTT (2,3-bis (2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide) reduction assay (Kannappan et al. 2017b). Stock solutions of XTT and menadione were prepared at 0.2 mg ml−1 in 1× PBS and 0.172 mg ml−1 in acetone, respectively. XTT/menadione solution was prepared freshly prior to experimentation at the ratio of 12.5:1 of XTT and menadione, respectively. S. marcescens cells treated with and without PSA (200 and 400 µg ml−1) were washed twice with sterile PBS (pH 7.4) and resuspended in 175 µl of the same buffer. Then, the cell suspensions were incubated for 2 h at 37 °C in the dark along with XTT/menadione solution. After incubation, the cellular viability was measured at OD 490 nm using a UV–visible spectrophotometer.
Prodigiosin inhibition assay
The effect of PSA on the production of prodigiosin pigment was assessed by the method of Morohoshi et al. (2007). 10 μl of S. marcescens cell suspensions (1 × 108 CFU ml−1) were inoculated in LB medium along with the presence (200 and 400 μg ml−1) and absence of PSA and incubated at 28 °C for 18 h. After incubation, the cultures from each well were collected in 2 ml sterile microfuge tubes and centrifuged (16,770×g for 10 min) to harvest the bacterial cells along with prodigiosin. Prodigiosin from the S. marcescens cell pellet was extracted with 1 ml of acidified ethanol (4% 1 M HCl in ethanol) and the OD of the extracted prodigiosin was measured at 534 nm.
Lipolytic assay
The lipolytic activity of PSA treated and untreated S. marcescens was estimated using p-nitrophenyl palmitate as the substrate by following the method of Gupta et al. (2002) with slight modifications. Briefly, S. marcescens was grown at 28 °C for 18 h in the presence and absence of PSA (200 and 400 µg ml−1). After incubation, the culture was centrifuged at 16,770×g for 10 min. 100 μl of CFCS was added to 900 μl of buffered substrate having 1 volume of 0.3% p-nitrophenyl palmitate in isopropanol, 9 volumes of 0.2% sodium deoxycholate, and 0.1% gummi arabicum in 50 mM Na2PO4 buffer (pH 8.0) and incubated at room temperature for 1 h. After incubation, the reaction was stopped by adding 1 ml of 1 M Na2CO3 following which the mixture was centrifuged at 16,770×g for 10 min. Then, the absorbance of the clear solution was measured spectrophotometrically at 410 nm.
Biofilm biomass quantification assay
The effect of PSA on biofilm formation of S. marcescens was analyzed by measuring the biofilm biomass through micro titer plate (MTP) assay. Briefly, 1% of S. marcescens culture was added to 1 ml of LB medium with the presence (200 and 400 µg ml−1) and absence of PSA in polystyrene 24 well MTP (Flat Bottom, Nest Scientific, USA) and incubated statically for 24 h at 28 °C. After incubation, the MTP wells were washed twice with distilled water to remove the planktonic cells. Then, the surface adhered cells were stained with 1 ml of 0.2% crystal violet (CV) solution (HiMedia, India) for 2 min. The surplus dye was removed by washing with distilled water and the CV attached cells were solubilized with the addition of 1 ml of 20% glacial acetic acid. Then, the biofilm biomass was measured by quantifying the intensity of crystal violet at OD 570 nm (Rama Devi et al. 2016).
Observation of S. marcescens biofilm formation through microscopic analyses
For visualization of biofilm by light microscopy, S. marcescens were allowed to grow on glass slides (1 × 1 cm) placed in polystyrene 24 well MTP with and without PSA (200 and 400 µg ml−1) at 28 °C for 24 h. Then, the glass slides were washed twice with distilled water and stained with 0.2% crystal violet. The biofilm in the stained glass slides were inspected by light microscope (Nikon Eclipse Ti 100, Japan) at ×400 magnification (Srinivasan et al. 2017).
Another set of glass slides with S. marcescens grown as above was washed twice with distilled water, stained with 0.1% acridine orange, and visualized using a confocal laser scanning microscope (CLSM) (Zeiss LSM 710, Carl Zeiss, Germany).
For scanning electron microscopic (SEM) analysis, the S. marcescens was allowed to form biofilm on the glass slides together with the presence and absence of PSA (400 µg ml−1) as described above. After 24 h of incubation, the glass slides were washed twice with distilled water to remove nonadherent cells. The biofilms on the glass slides were fixed with 2.5% glutaraldehyde for 3 h. Then, the glass slides were washed in distilled water and dehydrated by increasing concentrations of ethanol (50, 60, 70, 80, 90, and 100) for 30 s each. After critical-point drying and gold sputtering, the samples were examined using a SEM (VEGA 3 TESCAN, Czech Republic) (Sivaranjani et al. 2016).
Disintegration of preformed biofilm
To assess the effect of PSA on preformed biofilm, the mature biofilm on the cover glasses of S. marcescens was incubated with and without PSA (200 and 400 µg ml−1) at 28 °C for 5 h. After incubation, the biofilm biomass quantification and light microscopic analysis were performed as described above.
Exopolysaccharides quantification assay
The total exopolysaccharides (EPS) production was measured by carbohydrate assay. PSA (200 and 400 µg ml−1) treated and untreated S. marcescens cells in polystyrene 24 well MTP wells were washed with distilled water to remove the planktonic cells. The biofilm cells in wells were washed with 0.5 ml of 0.9% NaCl and an equal volume of 5% phenol and 5 volumes of concentrated H2SO4 were added. Then, the mixture was incubated in the dark for 1 h and EPS was quantified by measuring the intensity at 490 nm (Favre-Bonté et al. 2003).
Determination of hydrophobicity index
The effect of PSA on hydrophobicity of S. marcescens was measured by following the method of Courtney et al. (2009). Briefly, 1 ml of S. marcescens culture (OD 530 = 1.0) was taken in glass tubes and an equal volume of toluene was added. The mixture was vortexed for 3 min and incubated for 15 min at room temperature for phase separation. Then, the OD of the aqueous phase was measured using a spectrophotometer at 530 nm. The percentage of hydrophobicity was calculated by the formula.
Swimming and swarming assays
The swimming and swarming motility assays were performed by following the method of Abraham et al. (2012). In swimming assay, 3 μl (OD adjusted to 0.4 at 600 nm) of S. marcescens culture was stab inoculated at the center of the swimming agar medium consisting of 1% peptone, 0.5% NaCl, and 0.3% agar supplemented with and without PSA (200 and 400 µg ml−1). For swarming assay, 5 μl of S. marcescens was inoculated at the center of the swarming agar medium consisting of 1% peptone, 0.5% NaCl, 0.5% agar, and 0.5% of filter sterilized d-glucose with and without PSA (200 and 400 µg ml−1). Then, the plates were incubated at 28 °C for 16 h and observed for reduction in swimming and swarming migration zones.
FT-IR analysis
S. marcescens cells treated with and without PSA (400 µg ml−1) were washed twice with PBS (pH 7.4). Subsequently, the cell pellets were vacuum dried and 50 mg of potassium bromide (KBr) with 1 mg of bacterial cell pellets were taken to make KBr pellets. Then, the KBr pellet was analyzed by FT-IR (Nicolet iS5, Thermo Scientific, U.S.A) spectroscopy. A total of 64 scans were taken with 4 cm−1 resolution and the spectrum was scanned in the range of 400–4000 cm−1. The IR spectra were plotted as absorbance and analyzed by the OMNIC software (Kannappan et al. 2017a).
Total RNA extraction and quantitative real-time PCR analysis
To assess the effect of PSA, the expression levels of QS genes were estimated. The total RNA was extracted from PSA treated (400 µg ml−1) and untreated S. marcescens using TRIzol reagent (Sigma-Aldrich, Switzerland) (Supplementary Fig. 1B). Then, the RNA was reverse transcribed into cDNA using the Superscript III cDNA synthesis kit (Invitrogen Inc., USA) following the manufacturer’s directions. The qPCR reactions were performed using a real-time PCR system (7500 Sequence Detection System, Applied Biosystems Inc. Foster, CA, USA). The details of the gene-specific primers used for this study are given in Supplementary Table 1 and the efficacies of gene-specific primers were confirmed by 1.5% agarose gel electrophoresis (Supplementary Fig. 1A). The samples were analyzed in triplicate and the rplU gene was used as a stable internal control for stabilization.
Cytotoxicity assay
The cytotoxicity of PSA on human peripheral blood mononuclear cells (PBMC) was evaluated by MTT assay using the method of Nazarpour et al. (2012) with slight modifications. Briefly, 5 ml of blood was collected from healthy humans and then diluted with an equal volume of RPMI 1640 (Roswell Park Memorial Institute) medium (Himedia, India). To the diluted blood, an equal amount of lymphocyte separation medium (Sigma, USA) was added and centrifuged at 547 × g for 15 min in order to isolate PBMC. An intermittently formed white layer was taken and washed by using RPMI 1640 at 547 × g for 15 min. The cell pellet was then resuspended in complete medium containing RPMI 1640, 1 × penicillin and 1 × streptomycin (Himedia, India), 10% fetal bovine serum and then the final cell count was adjusted to 1 × 106 cells ml−1 and the cell suspension was incubated with increasing concentrations of PSA (200, 400, 600, 800 and 1000 µg ml−1) and 2% methanol (vehicle control) at 37 °C with 5% CO2 incubator for 24 h. After the incubation, the cells were centrifuged at 547 × g for 10 min and washed with PBS (pH 7.4) for twice. Then, the cells were incubated with MTT (1 mg ml−1) for 3 h and absorbance was taken at 570 nm. The morphology and integrity of the cell membrane of PSA treated and untreated PBMC was also evaluated by viewing the cells under a light microscope at ×20 magnification with the help of trypan blue dye.
Statistical analysis
All the results were reported as mean ± standard deviation and the statistical analyses were done using the Statistical Package for the Social Sciences (SPSS) (version 17.0, SPSS Inc., San Rafael, CA). Dunnett’s-One-way analysis of variance (one-way ANOVA) and Student-t Test were used to compare the differences among tests and controls.
Results and discussion
Fatty acids are considered as potential antibacterial agents due to their effectiveness, broad range of antibacterial activity, and the lack of resistance mechanisms for different bacterial pathogens. In spite of their well-known antimicrobial potential, fatty acids have long been used in commercial products (Desbois 2012; Desbois and Smith 2010). On the other hand, the molecular structures of most fatty acids resemble the side chain structure of autoinducer molecules, which can act as analog compounds for various bacterial QS receptor molecules. Therefore, several reports have revealed the anti-QS potential of fatty acids against different bacterial pathogens. Abd-Alla and Bashandy (2012) stated that the production of linolenic and myristic acid in onion bulbs inhibited the QS regulated virulence factors productions in Pseudomonas aeruginosa. Widmer et al. (2007) reported the anti-QS potential of long-chain fatty acids isolated from poultry meat against the AI-2 QS system. However, to the best of our knowledge, PSA has not been studied for its anti-QS and antibiofilm activity against any bacterial pathogens. Therefore, the present study focused primarily on the anti-QS and antibiofilm potential of PSA against the environmental pathogen S. marcescens.
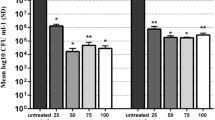
The effective QS inhibitory effect of PSA was first determined by protease inhibition assay. Proteases are extracellular virulence factors and their production is controlled by QS. The results obtained showed a concentration-dependent protease inhibition in S. marcescens on treatment with PSA (Fig. 1). The PSA exhibited 34 and 59% protease inhibition in S. marcescens ATCC and 30 and 45% protease inhibition in S. marcescens CI at 200 and 400 μg ml−1 concentrations, respectively. Hence, these dose-dependent concentrations (200 and 400 μg ml−1) alone were selected for further QS and biofilm-related assays in S. marcescens. No such protease inhibitory activity was found with 1% methanol, the negative control.
The anti-QS and antibiofilm compounds should not have inhibitory effects as this may lead to the development of resistance in bacterial pathogens. In order to assess the effect of PSA (200 and 400 μg ml−1), the growth and metabolic activity of S. marcescens was determined by growth inhibition and XTT reduction assays. The growth inhibition assay indicated that there was no variation in the growth of PSA and 0.8% methanol (Negative control) treated and untreated S. marcescens. Moreover, the XTT reduction assay confirmed the results of growth inhibition assay with no indication of adverse effects on the cellular viability (Fig. 2). Therefore, the results obtained from growth inhibition and XTT reduction assays clearly indicated that there was no growth inhibitory effect of PSA against S. marcescens.
Many bacterial pathogens including S. marcescens, P. aeruginosa, and Staphylococcus aureus are known to produce pigments that are considered to be virulence factors. These bacterial pigments hinder the host immune responses and are cytotoxic to host cells (Liu and Nizet 2009; Lau et al. 2004; Sethupathy et al. 2017). Often, the pigments produced by these bacterial pathogens are essential for their survival, invasion, or other characteristics related to pathogenicity (Liu and Nizet 2009). S. marcescens produces a prominent red pigment named prodigiosin under the control of QS mechanism. The prodigiosin inhibition assay result showed a concentration-dependent inhibition in the prodigiosin pigment by PSA (Fig. 3). The prodigiosin pigment production was significantly (p ≤ 0.05) inhibited to the level of 44 and 67% in S. marcescens ATCC and 61% and 75% in S. marcescens CI at 200 and 400 μg ml−1 concentrations, respectively. A similar impact on the pyocyanin pigment inhibition in P. aeruginosa has been reported for the fatty acid compound Cis-2-dodecenoic acid by Deng et al. (2013).
The pathogenic capability of S. marcescens has been related to the production of extracellular enzymes such as proteases, chitinases, phospholipases, and lipases (Hejazi and Falkiner 1997; Givskov et al. 1997). Thus, the effectiveness of PSA in inhibiting lipase enzyme production in S. marcescens was studied. No substantial inhibition in lipase production was observed in S. marcescens on treatment with 200 and 400 μg ml−1 of PSA (Fig. 4a). However, PSA showed significant inhibition in protease production in both S. marcescens strains (Fig. 1). Our results support the findings of Abd-Alla and Bashandy (2012), who have reported that the myrstic acid isolated from infected onion bulb significantly inhibited the protease production in P. aeruginosa.
The capabilities of S. marcescens to cause infections and endure in the environment are linked to its ability to form biofilms i.e. organized community of surface adherent bacterial microcolonies enclosed in self-produced EPS (Costerton et al. 1999). It is well established that QS plays a main role in regulating many aspects of the biofilm life cycle including initial attachment, formation of microcolonies, and three-dimensional architecture in S. marcescens (Rice et al. 2005). Therefore, we tested the effect of PSA on the S. marcescens biofilm formation by crystal violet assay. At 200 and 400 μg ml−1, PSA significantly (p ≤ 0.05) inhibited S. marcescens biofilm formation to the level of 49 and 68% and S. marcescens CI biofilm formation to the level of 47 and 72%, respectively (Fig. 3). In order to confirm the antibiofilm potential of PSA on S. marcescens biofilm, microscopic observations were carried out. In light microscopic examination, both S. marcescens ATCC and CI strains showed a thick coating of biofilm on control slides. In contrast, a visible reduction in biofilm formation was observed in S. marcescens on treatment with PSA (Fig. 5a). In order to gain deeper insight into the surface topology of biofilm architecture, the PSA treated and untreated S. marcescens biofilms were examined under CLSM. The obtained results revealed that the thickness of the biofilm is reduced in PSA treated samples compared to that of untreated controls (Fig. 5b). Likewise, the SEM images of untreated controls clearly depicted the intricate biofilm architecture, while the PSA treated samples (400 μg ml−1) showed a clear disintegration of biofilm (Fig. 6). Consistent with our findings, oleic and undecanoic acids have been shown to hinder the biofilms of S. aureus and S. marcescens, respectively (Stenz et al. 2008; Salini et al. 2015).
To be an effective antibiofilm compound, it should disrupt preformed biofilms. In order to assess the effects of PSA on the preformed biofilm, the mature biofilm disruption assay was done. The obtained result showed that the 400 µg ml−1 of PSA significantly inhibited the preformed biofilm to the level of 59 and 52% in S. marcescens ATCC and CI, compared to their controls (Fig. 7a). Furthermore, the light microscopic analysis validated the preformed biofilm inhibition efficacy of PSA, in which the PSA treatment noticeably disturbed the preformed biofilm, compared to their control slides (Fig. 7b). The outcome of the present study supports the previous study made by Sepehr et al. (2014), who have reported that the unsaturated fatty acid, cis-2-decenoic acid disturbed the preformed biofilm of Escherichia coli.
The effect of PSA on biofilm-associated virulence factors such as EPS production, HI production, swarming, and swimming motility were also assessed in this study. Many bacterial pathogens in their natural milieus are surrounded by a coating of EPS, which plays a major role in adhesion and defense against hostile environmental situations (Fett and Dunn 1989). EPS was extracted from PSA treated and untreated cultures of S. marcescens and the spectrometric results revealed that the concentration of EPS decreased with increasing concentrations of PSA. The PSA at 400 μg ml−1 caused a 33 and 46% decrease in EPS production of S. marcescens ATCC and CI, respectively (Fig. 3). The hydrophobicity of bacterial pathogens contributes to their adherence, which is responsible for biofilm formation and adhesion to epithelial cells. The adherence of bacterial cells to the epithelial surface is considered to be an important factor factors in the development of infection (Wojnicz and Jankowski 2007). The effect of PSA on hydrophobicity of S. marcescens was assessed and the hydrophobicity index (HI) of S. marcescens ATCC and CI without PSA were found to be 69 and 64%, respectively. A significant (p ≤ 0.05) level of HI reduction was observed in S. marcescens on treatment with PSA (Fig. 4b). Swarming and swimming motilities are a distinctive bacterial surface migration behaviour driven by flagella (Kearns 2010). Also, the swarming and swimming behaviour are connected to bacterial cytotoxicity (Kirov 2003). Therefore, an effort was made to observe the effect of PSA on swimming and swarming motility of S. marcescens. In the obtained results a considerable level of swimming and swarming motility inhibition was observed in PSA treated S. marcescens ATCC compared to the untreated control (Fig. 7a) and no such inhibition was observed in S. marcescens CI on treatment with PSA (Fig. 7b). However, the PSA treatment showed prodigiosin inhibition in S. marcescens CI motility plates. The reduction in biofilm formation, adhesion phenomena such as motility and HI following treatment with PSA supports the anti-biofilm potential of PSA against S. marcescens. The results of the present study are in agreement with the previous reports of Inoue et al. (2008) and Kenny et al. (2009), who have reported that the oleic and linoleic fatty acids inhibited the swarming motility and HI productions in P. aeruginosa and S. aureus, respectively.
FT-IR spectroscopy has significant advantages in understanding the functional groups of biological materials, because it depends on the individual absorbance of the corresponding molecular vibrations in the biological materials. The structural and functional groups of cellular components in bacterial pathogens are alike and stretch the same signal, although the distribution and quantity of functional groups differ significantly among bacterial pathogens on treatment with bioactive compounds due to the alteration in cellular components (Schmitt and Flemming 1998). The FT-IR spectra revealed differences between the PSA treated and untreated S. marcescens cells (Fig. 8a, b). Noticeable variations were observed in the regions of 1000–1500, 1500–1800, 2700–3100, and 3100–3500 cm−1, which represent a mixed region of proteins and fatty acids, amide linkage from proteins and peptides, fatty acids in bacterial cell membrane, and hydration of bacterial cells, respectively.
Moreover, in order to determine the in vitro results and to understand the anti-QS and antibiofilm potential of PSA at the molecular level, a real-time PCR analysis was done. Rice et al. (2005) reported that a transposon mutant bearing insertions in a C4-HSL-mediated bsmB gene was a poor biofilm former. Similarly, a study done by Labbate et al. (2007) stated that the bsmB mutant strain lacked lipase, serralysin protease, and S-layer protein production. Thus the bsmB gene plays a major role in S. marcescens virulence factor production. Therefore, the impact of PSA on the bsmB gene expression level was tested and the obtained real-time data showed a significant downregulation of bsmB gene in both the S. marcescens strains. Yamamoto et al. (1985) detected fimbriae production in S. marcescens strains isolated from patients with respiratory and urinary tract infections. Fimbriae play a major role in S. marcescens adherence and colonization to biotic and abiotic surfaces. The fimA and fimC genes encode the main structural elements for fimbriae production in S. marcescens. On treatment with PSA, the fimA and fimC genes were downregulated in both the S. marcescens ATCC and CI strains. The products of the flhDC master operon, flhC and flhD control the cell differentiation, cell division, swimming, and swarming motility in S. marcescens. Stella et al. (2008) suggested that the deficiency of flhD expression is the reason for the loss of flagella-based motility and flagellum production in S. marcescens. PSA treatment decreases the expression level of flhD up to 0.8 fold in S. marcescens ATCC, whereas a slight upregulation in flhD gene expression was observed in S. marcescens CI (Fig. 9). The real-time data obtained are in accordance with the findings of previous studies, wherein the ethyl acetate extract of Piper betle (Srinivasan et al. 2016) and methanolic extract of Anethum graveolens (Salini and Pandian 2015) down-regulated the QS-regulated virulence genes in S. marcescens (Fig. 10).
FT-IR spectra of PSA treated and untreated S. marcescens ATCC (A) and CI (B) cells. FT-IR spectra show differences in regions such as (1) mixed region, proteins, and fatty acids (1000–1500 cm−1), (2) amide linkage from proteins and peptides (1500–1800 cm−1), (3) fatty acids in bacterial cell membrane (2700–3100 cm−1), and (4) hydration of bacterial cells (3100–3500 cm−1)
A compound should be non-toxic for its application in clinical sceneries. Therefore, the effect of PSA on the viability of human PBMC was evaluated by MTT assay. No notable change was found in the viability of PSA treated and vehicle control PBMC, when compared to the control at the tested concentrations (Fig. 11a). In the positive control (H2O2), only 20% of cell viability was observed in PBMC. Light microscopic images of PBMC cells showed undamaged morphology without any change in the membrane integrity in PSA treated and vehicle control groups, whereas the positive control cells showed membrane damage (Fig. 11b). The result of MTT assay and light microscopic images indicated that PSA did not have any toxic effect on PBMC.
Conclusion
The present study supports the anti-QS and antibiofilm potential of PSA against S. marcescens through a series of virulence inhibition assays such as prodigiosin, protease, biofilm, EPS, HI, swimming, and swarming motility. The significant reduction in the expression of QS candidate genes involved in extracellular virulence enzyme production, initial adherence, biofilm formation, and motility is in good agreement with the anti-QS and antibiofilm action of PSA in vitro. More, the MTT assay revealed the non-toxic nature of PSA on PBMC at the tested concentrations. It is also notable that PSA is well known for its anti-aging, antimicrobial, and skin-irritation reducing properties; in addition the current study highlights its anti-QS and antibiofilm potential with satisfactory safety.
References
Abd-Alla MH, Bashandy SR (2012) Production of Quorum Sensing Inhibitors in Growing Onion Bulbs Infected with Pseudomonas aeruginosa E (HQ324110). ISRN Microbiol. https://doi.org/10.5402/2012/161890
Abraham I, Vasantha S, Priya S, Pandian SK, Ravi AV (2012) Inhibition of biofilm development of uropathogens by curcumin - An anti-quorum sensing agent from Curcuma longa. Food Chem 148:453–460. https://doi.org/10.1016/j.foodchem.2012.08.002
Alaluf S, Hu H-L, Green MR, Powell JR, Rawlings AV, Rogers JS, Watkinson A, Cain FW (1999) Cosmetic use of petroselinic acid. EP 1:178
Alaluf S, Green MR, Powell JR, Rogers JS, Watkinson A, Cain FW, Hu HL, Rawlings AV (2002) U.S. Patent No. 6,365,175. Washington, DC: U.S. Patent and Trademark Office
Cahoon EB, Dörmann P, Ohlrogge JB (1994) Petroselinic acid biosynthesis and production in transgenic plants. Prog Lipid Res 33:155–163. https://doi.org/10.1016/0163-7827(94)90018-3
Chadwick JS, Caldwell SS, Chadwick P (1990) Adherence patterns and virulence for Galleria mellonella larvae of isolates of Serratia marcescens. J Invertebr Pathol 55(1):133–134. https://doi.org/10.1016/0022-2011(90)90044-7
Costerton JW, Stewart PS, Greenberg EP (1999) Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322. https://doi.org/10.1126/science.284.5418.1318
Courtney HS, Ofek I, Penfound T, Nizet V, Pence MA, Kreikemeyer B, Podbielbski A, Hasty DL, Dale JB (2009) Relationship between expression of the family of M proteins and lipoteichoic acid to hydrophobicity and biofilm formation in Streptococcus pyogenes. PLoS ONE. https://doi.org/10.1371/journal.pone.0004166
Deng Y, Boon C, Chen S, Lim A, Zhang LH (2013) Cis-2-dodecenoic acid signal modulates virulence of Pseudomonas aeruginosa through interference with quorum sensing systems and T3SS. BMC Microbiol 13(1):231. https://doi.org/10.1186/1471-2180-13-231
Desbois AP (2012) Potential applications of antimicrobial fatty acids in medicine, agriculture and other industries. Recent Pat Antiinfect Drug Discov 7:111–122. https://doi.org/10.2174/157489112801619728
Desbois AP, Smith VJ (2010) Antibacterial free fatty acids: activities, mechanisms of action and biotechnological potential. Appl Microbiol Biotechnol 85:1629–1642. https://doi.org/10.1007/s00253-009-2355-3
Favre-Bonté S, Köhler T, Van Delden C (2003) Biofilm formation by Pseudomonas aeruginosa: role of the C4-HSL cell-to-cell signal and inhibition by azithromycin. J Antimicrob Chemother 52:598–604. https://doi.org/10.1093/jac/dkg397
Fett WF, Dunn MF (1989) Exopolysaccharides produced by phytopathogenic Pseudomonas syringae pathovars in infected leaves of susceptible hosts. Plant Physiol 89:5–9. https://doi.org/10.1104/pp.89.1.5
Flemming H, Wingender J (2010) The biofilm matrix. Nat Rev Microbiol 8:623–633. https://doi.org/10.1038/nrmicro2415
Fux CA, Costerton JW, Stewart PS, Stoodley P (2005) Survival strategies of infectious biofilms. Trends Microbiol 13:34–40. https://doi.org/10.1016/j.tim.2004.11.010
Givskov M, Eberl L, Molin S (1997) Control of exoenzyme production, motility and cell differentiation in Serratia liquefaciens. FEMS Microbiol Lett 148:115–122. https://doi.org/10.1111/j.1574-6968.1997.tb10276.x
Gupta N, Rathi P, Gupta R (2002) Simplified para-nitrophenyl palmitate assay for lipases and esterases. Anal Biochem 311:98–99. https://doi.org/10.1016/S0003-2697(02)00379-2
Hejazi A, Falkiner FR (1997) Serratia marcescens. J Med Microbiol 46:903–912. https://doi.org/10.1099/00222615-46-11-903
Hentzer M, Givskov M (2003) Pharmacological inhibition of quorum sensing for the treatment of chronic bacterial infections. J Clin Investig 112:1300–1307. https://doi.org/10.1172/JCI200320074
Inoue T, Shingaki R, Fukui K (2008) Inhibition of swarming motility of Pseudomonas aeruginosa by branched-chain fatty acids. FEMS Microbiol Lett 281(1):81–86. https://doi.org/10.1111/j.1574-6968.2008.01089.x
Kannappan A, Gowrishankar S, Srinivasan R, Pandian SK, Ravi AV (2017a) Antibiofilm activity of Vetiveria zizanioides root extract against methicillin-resistant Staphylococcus aureus. Microb Pathog 110:313–324. https://doi.org/10.1016/j.micpath.2017.07.016
Kannappan A, Sivaranjani M, Srinivasan R, Rathna J, Pandian SK, Ravi AV (2017b) Inhibitory efficacy of geraniol on biofilm formation and development of adaptive resistance in Staphylococcus epidermidis RP62A. J Med Microbiol. https://doi.org/10.1099/jmm.0.000570
Kearns DB (2010) A field guide to bacterial swarming motility. Nature Rev Microbiol 8(9):634–644. https://doi.org/10.1038/nrmicro2405
Kenny JG, Ward D, Josefsson E, Jonsson M, Hinds J, Rees HH, Lindsay JA, Tarkowski A, Horsburgh MJ (2009) The Staphylococcus aureus response to unsaturated long chain free fatty acids: survival mechanisms and virulence implications. PLoS ONE 4(2):e4344. https://doi.org/10.1371/journal.pone.0004344
Kirov SM (2003) Bacteria that express lateral flagella enable dissection of the multifunctional roles of flagella in pathogenesis. FEMS Microbiol Lett 224:151–159. https://doi.org/10.1016/S0378-1097(03)00445-2
Labbate M, Zhu H, Thung L, Bandara R, Larsen MR, Willcox MDP, Givskov M, Rice SA, Kjelleberg S (2007) Quorum-sensing regulation of adhesion in Serratia marcescens MG1 is surface dependent. J Bacteriol 189:2702–2711. https://doi.org/10.1128/JB.01582-06
Lau GW, Hassett DJ, Ran H, Kong F (2004) The role of pyocyanin in Pseudomonas aeruginosa infection. Trends Mol Med 10(12):599–606. https://doi.org/10.1016/j.molmed.2004.10.002
Liu GY, Nizet V (2009) Color me bad: microbial pigments as virulence factors. Trends Microbiol 17:406–413. https://doi.org/10.1016/j.tim.2009.06.006
March JC, Bentley WE (2004) Quorum sensing and bacterial cross-talk in biotechnology. Curr Opin Biotechnol 15:495–502. https://doi.org/10.1016/j.copbio.2004.08.013
Morohoshi T, Shiono T, Takidouchi K, Kato M, Kato N, Kato J, Ikeda T (2007) Inhibition of quorum sensing in Serratia marcescens AS-1 by synthetic analogs of N-acylhomoserine lactone. Appl Environ Microbiol 73:6339–6344. https://doi.org/10.1128/AEM.00593-07
Nazarpour R, Zabihi E, Alijanpour E, Abedian Z, Mehdizadeh H, Rahimi F (2012) Optimization of human peripheral blood mononuclear cells (PBMCs) cryopreservation. Int J Mol Cell Med. 1(2):88
Placek LL (1963) A review on petroselinic acid and its derivatives. J Am Oil Chem Soc 40:319–329. https://doi.org/10.1007/BF02631548
Rama Devi K, Srinivasan R, Kannappan A, Santhakumari S, Bhuvaneswari M, Rajasekar P, Prabhu NM, Veera Ravi A (2016) In vitro and in vivo efficacy of rosmarinic acid on quorum sensing mediated biofilm formation and virulence factor production in Aeromonas hydrophila. Biofouling 32:1171–1183. https://doi.org/10.1080/08927014.2016.1237220
Rice SA, Koh KS, Queck SY, Labbate M, Lam KW, Kjelleberg S (2005) Biofilm formation and sloughing in Serratia marcescens are controlled by quorum sensing and nutrient cues. J Bacteriol 187:3477–3485. https://doi.org/10.1128/JB.187.10.3477-3485.2005
Salini R, Pandian SK (2015) Interference of quorum sensing in urinary pathogen Serratia marcescens by Anethum graveolens. Pathog Dis. https://doi.org/10.1093/femspd/ftv038
Salini R, Sindhulakshmi M, Poongothai T, Pandian SK (2015) Inhibition of quorum sensing mediated biofilm development and virulence in uropathogens by Hyptis suaveolens. Antonie Leeuwenhoek 107(4):1095–1106. https://doi.org/10.1007/s10482-015-0402-x
Schmitt J, Flemming HC (1998) FTIR-spectroscopy in microbial and material analysis. Int Biodeterior Biodegrad 41:1–11. https://doi.org/10.1016/S0964-8305(98)80002-4
Sepehr S, Rahmani-Badi A, Babaie-Naiej H, Soudi MR (2014) Unsaturated fatty acid, cis-2-decenoic acid, in combination with disinfectants or antibiotics removes pre-established biofilms formed by food-related bacteria. PLoS ONE 9(7):e101677. https://doi.org/10.1371/journal.pone.0101677
Sethupathy S, Vigneshwari L, Valliammai A, Balamurugan K, Pandian SK (2017) L-Ascorbyl 2,6-dipalmitate inhibits biofilm formation and virulence in methicillin-resistant Staphylococcus aureus and prevents triacylglyceride accumulation in Caenorhabditis elegans. RSC Adv 7:23392–23406
Sivaranjani M, Gowrishankar S, Kamaladevi A, Pandian SK, Balamurugan K, Ravi AV (2016) Morin inhibits biofilm production and reduces the virulence of Listeria monocytogenes—An in vitro and in vivo approach. Int J Food Microbiol 237:73–82. https://doi.org/10.1016/j.ijfoodmicro.2016.08.021
Srinivasan R, Devi KR, Kannappan A, Pandian SK, Ravi AV (2016) Piper betle and its bioactive metabolite phytol mitigates quorum sensing mediated virulence factors and biofilm of nosocomial pathogen Serratia marcescens in vitro. J Ethnopharmacol 193:592–603. https://doi.org/10.1016/j.jep.2016.10.017
Srinivasan R, Santhakumari S, Ravi AV (2017) In vitro antibiofilm efficacy of Piper betle against quorum sensing mediated biofilm formation of luminescent Vibrio harveyi. Microb Pathog 110:232–239. https://doi.org/10.1016/j.micpath.2017.07.001
Stella NA, Kalivoda EJ, O’Dee DM, Nau GJ, Shanks RM (2008) Catabolite repression control of flagellum production by Serratia marcescens. Res Microbiol 159(7):562–568. https://doi.org/10.1016/j.resmic.2008.07.003
Stenz L, François P, Fischer A, Huyghe A, Tangomo M, Hernandez D, Cassat J, Linder P, Schrenzel J (2008) Impact of oleic acid (cis-9-octadecenoic acid) on bacterial viability and biofilm production in Staphylococcus aureus. FEMS Microbiol Lett 287:149–155. https://doi.org/10.1111/j.1574-6968.2008.01316.x
Uitterhaegen E, Nguyen QH, Sampaio KA, Stevens CV, Merah O, Talou T, Rigal L, Evon P (2015) Extraction of coriander oil using twin-screw extrusion: feasibility study and potential press cake applications. J Am Oil Chem Soc 92(8):1219–1233. https://doi.org/10.1007/s11746-015-2678-4
Weinkauf R, Santhanam U, Palanker LR, Januario TG, Brinker A (1998) Petroselinic acid as an anti-irritant in compositions containing alpha hydroxy acids. US Pat 6(022):896
Widmer KW, Soni KA, Hume ME, Beier RC, Jesudhasan P, Pillai SD (2007) Identification of poultry meat-derived fatty acids functioning as quorum sensing signal inhibitors to autoinducer-2 (AI-2). J Food Sci 72:363–368. https://doi.org/10.1111/j.1750-3841.2007.00527.x
Wojnicz D, Jankowski S (2007) Effects of subinhibitory concentrations of amikacin and ciprofloxacin on the hydrophobicity and adherence to epithelial cells of uropathogenic Escherichia coli strains. Int J Antimicrob Agents 29:700–704. https://doi.org/10.1016/j.ijantimicag.2007.01.007
Yamamoto T, Ariyoshi A, Amako K (1985) Fimbria-mediated adherence of serratia marcescens strain US5 to human urinary bladder surface. Microbiol Immun. 29(7):677–681. https://doi.org/10.1111/j.1348-0421.1985.tb00871.x
Yoon HJ, Choi JY, Park YS, Kim CO, Kim JM, Yong DE, Lee KW, Song YG (2005) Outbreaks of Serratia marcescens bacteriuria in a neurosurgical intensive care unit of a tertiary care teaching hospital: a clinical, epidemiologic, and laboratory perspective. Am J Infect Control 33:595–601. https://doi.org/10.1016/j.ajic.2005.01.010
Acknowledgements
The authors acknowledge the Department of Biotechnology, Government of India, for providing Bioinformatics Infrastructure Facility (Grant No. BT/BI/25/015/2012). Financial assistance rendered to R. Srinivasan in the form of UGC-BSR fellowship by the University Grants Commission, New Delhi [F.4-1/2006(BSR)/7-326/2011(BSR)] is thankfully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest with this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
10482_2017_971_MOESM1_ESM.docx
Supplementary Table 1. Nucleotide sequences of S. marcescens primers used in this study. Supplementary material 1 (DOCX 14 kb)
10482_2017_971_MOESM2_ESM.jpg
Supplementary Fig. 1. PCR amplification for the detection of genes responsible for biofilm formation and virulence factors production in (A) S. marcescens and (B) gel image represents RNA isolated from PSA treated and untreated S. marcescens for real-time PCR analysis. Supplementary material 2 (JPEG 854 kb)
Rights and permissions
About this article
Cite this article
Ramanathan, S., Ravindran, D., Arunachalam, K. et al. Inhibition of quorum sensing-dependent biofilm and virulence genes expression in environmental pathogen Serratia marcescens by petroselinic acid. Antonie van Leeuwenhoek 111, 501–515 (2018). https://doi.org/10.1007/s10482-017-0971-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-017-0971-y