Abstract
The evolutionary interrelationships between the archaeal organisms which comprise the class Halobacteria have proven difficult to elucidate using traditional phylogenetic tools. The class currently contains three orders. However, little is known about the family level relationships within these orders. In this work, we have completed a comprehensive comparative analysis of 129 sequenced genomes from members of the class Halobacteria in order to identify shared molecular characteristics, in the forms of conserved signature insertions/deletions (CSIs) and conserved signature proteins (CSPs), which can provide reliable evidence, independent of phylogenetic trees, that the species from the groups in which they are found are specifically related to each other due to common ancestry. Here we present 20 CSIs and 31 CSPs which are unique characteristics of infra-order level groups of genera within the class Halobacteria. We also present 40 CSIs and 234 CSPs which are characteristic of Haloarcula, Halococcus, Haloferax, or Halorubrum. Importantly, the CSIs and CSPs identified here provide evidence that the order Haloferacales contains two main groups, one consisting of Haloferax and related genera supported by four CSIs and five CSPs and the other consisting of Halorubrum and related genera supported by four CSPs. We have also identified molecular characteristics that suggest that the polyphyletic order Halobacteriales contains at least two large monophyletic clusters of organisms in addition to the polyphyletic members of the order, one cluster consisting of Haloarcula and related genera supported by ten CSIs and nineteen CSPs and the other group consisting of the members of the genus Halococcus supported by nine CSIs and 23 CSPs. We have also produced a highly robust phylogenetic tree based on the concatenated sequences of 766 proteins which provide additional support for the relationships identified by the CSIs and CSPs. On the basis of the phylogenetic analyses and the identified conserved molecular characteristics presented here, we propose a division of the order Haloferacales into two families, an emended family Haloferacaceae and Halorubraceae fam. nov. and a division of the order Halobacteriales into three families, an emended family Halobacteriaceae, Haloarculaceae fam. nov., and Halococcaceae fam. nov.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The class Halobacteria consists of a large and phenotypically heterogeneous assemblage of halophilic archaea within the phylum Euryarchaeota (Grant et al. 2001; Oren 2012). The class Halobacteria currently contains over 175 species placed into fifty distinct genera (Parte 2013; NamesforLife 2015). Our understanding of interrelationships of the genera within the class Halobacteria was previously based on chemotaxonomic characteristics and analysis of the 16S rRNA gene (Grant et al. 2001; Oren 2006; Wright 2006; Oren 2012). However, chemotaxonomic properties have not proven useful for classification above the genus level (Oren 2006, 2012) and the presence of multiple highly divergent copies of the 16S rRNA gene in many members of the class Halobacteria has limited the inferences that can be drawn from its analysis (Mylvaganam and Dennis 1992; Vreeland et al. 2002; Boucher et al. 2004; Cui et al. 2009; Oren 2012). Analyses of other individual gene/protein sequences, such as rpoB’/rpoC, have also thus far proven of limited value in elucidating the evolutionary relationships of the members of the class Halobacteria due to concerns regarding recombination events and lateral gene transfers (Walsh et al. 2004; Enache et al. 2007; Minegishi et al. 2010b; Naor et al. 2012; Williams et al. 2012).
The use of a concatenated set of unlinked and conserved loci in phylogenetic reconstruction is able to mitigate the effects of any instances of recombination or lateral gene transfer and provide greater resolving power than trees based on single genes/proteins (Rokas et al. 2003; Ciccarelli et al. 2006; Wu et al. 2009). The advent of widely available genome sequencing technology has provided taxonomists with a wealth of data from which to elucidate the relationships between various prokaryotic groups (Gao and Gupta 2012a; Zhi et al. 2012; Oren and Garrity 2014). A genome-centric, polyphasic approach to taxonomy, in which categorisation is driven primarily by inferences drawn from the genome sequence data and secondarily by molecular, biochemical, and phenotypic traits is now the recommended approach for prokaryotic taxonomy (Klenk and Goker 2010; Oren and Garrity 2014; Rossello-Mora and Amann 2015; Sutcliffe 2015; Whitman 2015).
The increasing availability of genome sequencing technology has provided us with genome sequence data from 35 genera within the class Halobacteria, covering a majority of the diversity within the group (NCBI 2015). This genome sequence data has allowed for robust and in-depth phylogenetic reconstructions of the sequenced Halobacteria species based on multiple concatenated gene and protein sequences (Papke et al. 2011; Andam et al. 2012; Williams et al. 2012; Soucy et al. 2014; Gupta et al. 2015). The most comprehensive phylogenetic reconstructions to date have been based on the sequences of thirty-two concatenated housekeeping proteins from 98 Halobacteria genomes (Gupta et al. 2015) and fifty-five concatenated ribosomal proteins from 118 Halobacteria genomes (Soucy et al. 2014). This genome sequence data is also enabling the detection of conserved molecular characteristics shared by evolutionarily related groups of organisms. In particular, two classes of conserved molecular characteristics have recently been utilised in prokaryotic taxonomy (Bhandari and Gupta 2014; Gupta 2014; Naushad et al. 2014; Gupta et al. 2015): conserved signature insertions/deletions (CSIs), which are insertions or deletions (indels) that are present only in a related group of organisms, and conserved signature proteins (CSPs), which are whole proteins that are present only in a related group of organisms. Both classes of molecular characteristics represent synapomorphic characteristics and provide reliable evidence, independent of phylogenetic trees, that the species from the groups in which they are found are specifically related to each other due to common ancestry. Recently, the class Halobacteria, which previously contained a single order (Halobacteriales), was divided into three orders (Halobacteriales, Haloferacales, and Natrialbales) on the basis of CSIs and CSPs (Gupta et al. 2015). However, due to the size of the class Halobacteria, the previous analysis only focused on the higher level divisions within the class Halobacteria, placing a single family within each of the three orders despite the size, diversity, and, in the case of the order Halobacteriales, the polyphyly of the identified groups (Gupta et al. 2015).
In this work, we have employed the whole genome sequences of 129 genome sequenced members of the class Halobacteria to reconstruct a highly robust phylogenetic tree based on 766 shared proteins and to identify conserved molecular characteristics that can be used to determine the interrelationships of the halobacterial genera within the three orders. We present 20 CSIs and 31 CSPs which are unique characteristics of infra-order level groups of genera within the class Halobacteria. Additionally, we present 40 CSIs and 234 CSPs that are characteristic of Haloarcula, Halococcus, Haloferax, or Halorubrum. Importantly, the order Haloferacales has been found to contain two main groups, a group containing the genus Haloferax and related genera, which is supported by four CSIs and five CSPs and a group containing the genus Halorubrum and related genera, which is supported by four CSPs. We have also identified molecular characteristics that suggest that the polyphyletic order Halobacteriales contains at least two large monophyletic clusters of organisms in addition to the polyphyletic members of the order, one cluster containing the genus Haloarcula and related genera supported by ten CSIs and nineteen CSPs and the other containing the members of the genus Halococcus which is supported by nine CSIs and 23 CSPs. On the basis of the phylogenetic analyses and the identified conserved molecular characteristics presented here, we propose a division of the order Haloferacales into two families, an emended family Haloferacaceae and Halorubraceae fam nov. and a division of the order Halobacteriales into three families, an emended family Halobacteriaceae, Haloarculaceae fam. nov., and Halococcaceae fam. nov.
Methods
Phylogenetic analyses
A phylogenetic tree was produced based on the concatenated sequences of 766 proteins obtained from 129 genome sequenced members of the class Halobacteria. The protein families used in this phylogeny were identified using the UCLUST algorithm (Edgar 2010) to identify proteins families present in at least 80 % of the input genomes which shared at least 50 % sequence identity and 50 % sequence length. Input genomes that were not annotated had all of their open reading frames translated from nucleotide sequences to amino acid sequences using USEARCH 8 (Edgar 2010). Each identified protein family was individually aligned using Clustal Omega (Sievers et al. 2011) and trimmed using Gblocks 0.91b (Castresana 2000) with relaxed parameters (Talavera and Castresana 2007). The concatenated dataset of the trimmed sequence alignments contained 212,988 aligned amino acid residues. A maximum-likelihood tree based on this alignment was constructed using FastTree 2 (Price et al. 2010) employing the Whelan and Goldman model of protein sequence evolution (Whelan and Goldman 2001) and RAxML 8 (Stamatakis 2014) using the Le and Gascuel model of protein sequence evolution (Le and Gascuel 2008). SH-like statistical support values (Guindon et al. 2010) for each branch node in the final phylogenetic tree were calculated using RAxML 8 (Stamatakis 2014). The resultant phylogenetic tree was drawn and artificially rooted on the midpoint using MEGA 6 (Tamura et al. 2013). This process was completed using an internally developed software pipeline. A manuscript for this pipeline is currently under preparation and the pipeline will be available for public use on Gleans.net once released.
In parallel, a phylogenetic tree based on the 16S rRNA gene sequences of type strains covering all validly named genera within the class Halobacteria was also constructed. The 16S rRNA sequences were retrieved from Ribosomal Database Project (Cole et al. 2014) and aligned using the SINA aligner (Pruesse et al. 2012) to form a multiple sequence alignment that was 1604 nucleotides long with common gaps removed. A maximum-likelihood phylogenetic tree based on this multiple sequence alignment was created using in MEGA 6 (Tamura et al. 2013) employing the General Time-Reversible model of sequence evolution (Tavaré 1986) with branch support based on 1000 bootstrap replicates.
Identification of conserved signature indels
Conserved signature indels were identified as detailed by Gupta (2014). In summary, BLASTp (Altschul et al. 1997) searches were performed on each protein in the genome of Halobacterium salinarum R1 (Pfeiffer et al. 2008) against all available sequences in the GenBank non-redundant database. Multiple sequence alignments were then created using ClustalX (Jeanmougin et al. 1998) for proteins that returned high scoring matches from Halobacteria and other prokaryotes. The alignments were then visually inspected for the presence of insertions or deletions that were flanked on both sides by at least 5–6 conserved amino acid residues in the neighbouring 30–40 amino acids. Detailed BLASTp searches were then carried out on short sequence segments containing the indel and the flanking conserved regions (60–100 amino acids long) to determine the specificity of the indels. SIG_CREATE and SIG_STYLE (available on Gleans.net) were then used to create Signature files for CSIs that were specific to Halobacteria subgroups as described by Gupta (2014). Due to the large number of genome sequences available for Halobacteria, the sequence alignment files presented here contain sequence information for only a limited number of species (generally only the type species from different genera). However, unless otherwise indicated, all members of the specified groups displayed similar sequence characteristics.
Identification of conserved signature proteins
Identification of conserved signature proteins for different Halobacteria subgroups was carried out by completing BLASTp (Altschul et al. 1997) searches using all proteins in the genomes of Halobacterium salinarum R1, Haloarcula marismortui ATCC 43049, Halococcus thailandensis JCM 13552, Haloferax volcanii DS2 and Halorubrum lacusprofundi ATCC 49239 (Baliga et al. 2004; Pfeiffer et al. 2008; Hartman et al. 2010) as query sequences. BLASTp searches were performed against all available sequences in the GenBank non-redundant sequence database. The results of the BLAST searches were then manually inspected for proteins for which all significant hits were from well-defined groups within the class Halobacteria or proteins for which there was a large increase in E value from the last hit belonging to a particular group of organism within the class Halobacteria and the first hit from an organism for any other bacterial group and the E value for the latter hits were >1 × 10−4 (Gao and Gupta 2007; Gupta and Mok 2007; Naushad et al. 2014).
Results
Phylogenetic analysis of the class Halobacteria based on concatenated protein sequences
In this work, we have produced a phylogenetic tree containing 129 genome sequenced members of the class Halobacteria using a concatenated set of seven hundred and sixty-six proteins (Fig. 1). The phylogenetic reconstruction produced for this work is the most comprehensive phylogenetic analysis of the genome sequenced Halobacteria completed to date. The branching patterns in this concatenated protein based phylogenetic tree largely reflect those seen in prior publications (Soucy et al. 2014; Gupta et al. 2015). In this concatenated protein based phylogenetic tree, the orders Natrialbales and Haloferacales form monophyletic groups with strong statistical support. The order Haloferacales can be further divided into two smaller groups; one containing the genera Haloferax and other closely related Halobacteria and the other containing Halorubrum and its relatives, labelled HF1 and HF2, respectively, in Fig. 1. The order Halobacteriales is currently a polyphyletic assemblage of halobacterial groups which do not show strong affinity to either the order Natrialbales or Haloferacales (Gupta et al. 2015). In our concatenated protein tree, the order Halobacteriales spans at least three subgroups. Two of these subgroups consist of distinct pairings of genera (viz. Halobacterium/Halarchaeum, Haladaptus/Halalkalicoccus). In addition to these smaller subgroups, two large neighbouring clusters of Halobacteriales, one consisting of Haloarcula, Halomicrobium, Halorhabdus, Halosimplex, and Natronomonas (labelled HB1) and the other consisting of the members of the genus Halococcus (labelled HB2), are also observed in the phylogenetic tree.
A maximum likelihood phylogenetic tree based on the concatenated sequences of 766 proteins obtained from 129 genome sequenced members of the class Halobacteria. The members of orders Natrialbales, Haloferacales, and Halobacteriales are highlighted in green, blue, and red, respectively. Major clades are labelled. SH-like statistical support values are shown at branch nodes
In this work, we have also produced a 16S rRNA gene sequence based phylogenetic tree containing all named Halobacteria species (Fig. 2). The branching patterns observed in the 16S rRNA based phylogenetic tree are similar to those observed in the concatenated protein tree. As observed in the concatenated protein tree, the orders Natrialbales and Haloferacales are monophyletic and well-separated entities in the 16 rRNA gene based tree. Within the order Natrialbales, the members of the genus Halopiger form a monophyletic grouping that is not observed in the concatenated protein tree. We have further examined the significance of the monophyletic grouping of the Halopiger in the 16S rRNA tree by creating individual and concatenated phylogenetic trees for the five multi-locus sequence analysis proteins (viz. atpB, EF-2, radA, rpoB’, and secY) for the class Halobacteria proposed by Papke et al. (2011) (Supplemental Figs. 54–59). A monophyletic grouping of the members of the genus Halopiger was not observed in any of these phylogenetic trees and there was stochasticity in their branching patterns, indicating a high level of genetic heterogeneity within the members of this genus. In phylogenetic trees based on different protein sequences, Natrinema altuense 1AG-DGR consistently shows a longer branch in comparison to the other Natrinema spp., but its significance at present is unclear. Within the order Haloferacales in the 16S rRNA tree, the clades HF1 (Haloferax and related genera) and HF2 (Halorubrum and related genera) are identifiable and well separated with the sole exception of the members of the genus Haloplanus which branched within the Haloferacales adjacent to the clades HF1 and HF2. The order Halobacteriales weakly supported monophyletic branching in the 16S rRNA based phylogenetic tree. Within this weakly supported grouping of the Halobacteriales, there are nine distinct and well-supported subgroups. Importantly, the large clusters of Halobacteriales species observed in the concatenated protein tree (clades HB1 and HB2) are also identifiable in the 16S rRNA tree. However, the genus Halococcus (clade HB2) did not branch with clade HB1 in the 16S rRNA tree, instead Halococcus branched with the genus Halalkalicoccus alongside the genera Haladaptus, Halorussus, and Halorubellus. The genus Halosimplex did not branch within clade HB1 in the 16S rRNA tree. The single member of the genus Halosimplex formed a cluster with the lone member of the genus Halovenus which branched in the vicinity of clade HB1 and other Halobacteriales.
A maximum likelihood phylogenetic tree based on 144 16S rRNA gene sequences from members of the class Halobacteria covering all known species. The members of orders Natrialbales, Haloferacales, and Halobacteriales are highlighted in green, blue, and red, respectively. Major clades are labelled. Bootstrap support values above 50 % are shown at branch nodes
Molecular characteristics distinguishing the two main groups within the order Haloferacales
Molecular characteristics, such as CSIs and CSPs, that are uniquely found in a well-defined group of organisms are powerful tools for evolutionary studies (Rokas and Holland 2000; Gao and Gupta 2012a; Jones 2012; Gupta 2014). Recently, CSIs and CSPs have been used to revise the taxonomy of the class Halobacteria and a large number of other prokaryotic groups at varying taxonomic depths (Sawana et al. 2014; Gupta et al. 2015; Naushad et al. 2015). However, the previous analysis of the class Halobacteria focused only on the higher level divisions within the class. In this work we have identified four CSIs and five CSPs which are shared by clade HF1 (Haloferax and related genera). An example of one such CSI, consisting of a one amino acid insertion in the members of clade HF1 located in a conserved region of a hypothetical protein, is shown in Fig. 3. This insertion is uniquely found in all members of clade HF1 and absent in the members of clade HF2 and all other members of the class Halobacteria. The sequence alignments for three additional CSIs which are specific for clade HF1 are shown in Supplemental Figs. 1–3 and their properties are briefly summarised in Table 1. The second group within the order Haloferacales, clade HF2 (Halorubrum and related genera), is characterised by four identified CSPs. GenInfo Identifier numbers (GI Numbers) for the five CSPs specific for clade HF1 and the four CSPs specific for clade HF2 are provided in Table 3A, B, respectively.
A partial sequence alignment of a hypothetical protein showing a 1 amino acid insertion (boxed) that is characteristic of the members of clade HF1. Sequence information for a limited number of Halobacteria and other archaea are shown here, but unless otherwise indicated similar CSIs were detected in all members of the indicated group and not detected in any other species in the top 250 Blastp hits. The dashes (-) in the alignments indicate identity with the residue in the top sequence. GenInfo identification (GI) numbers for each sequence are indicated in the second column
Molecular characteristics which provide evolutionary insights for the order Halobacteriales
In this work, we have also identified a number of CSPs and CSIs that provide novel insights into the interrelationships within the polyphyletic order Halobacteriales. Most importantly, we have identified three CSIs and four CSPs which are shared by clade HB1 as seen in the concatenated protein based phylogenetic tree (viz. the genera Haloarcula, Halomicrobium, Halorhabdus, Halosimplex, and Natronomonas) and seven CSIs and fifteen CSPs which are unique characteristics of all clade HB1 members except the genus Natronomonas, which forms the outermost branch of clade HB1. One example each of these two types of CSI are shown in Fig. 4. The first CSI, consisting of a one amino acid insertion in ATP-dependent DNA helicase (Fig. 4a), is uniquely found in the members of clade HB1 while the second CSI, consisting of a one amino acid deletion in a conserved region of the protein acetylglutamate kinase (Fig. 4b), is uniquely found in all members of clade HB1 except the genus Natronomonas. Neither of the CSIs shown in Fig. 4, or any of the other CSIs indicated to be specific for clade HB1, are found in the members of clade HB2 or other members of the class Halobacteria. The sequence alignments for eight additional CSIs which are specific for clade HB1 are shown in Supplemental Figs. 4–11 and their properties are briefly summarized in Table 2. In addition, we have also identified nineteen CSPs which are also specifically found in members of clade HB1. Identification numbers for these nineteen clade HB1-specific CSPs are provided in Table 3C. These CSIs and CSPs provide strong support that the genus Halosimplex shares an evolutionary lineage with the members of clade HB1 despite not branching within the clade in our 16S rRNA tree (Fig. 2).
Partial sequence alignments of a a ATP-dependent DNA helicase showing a one amino acid insertion deletion that is characteristic of the members of clade HB1, b acetylglutamate kinase showing a 1 amino acid deletion that is characteristic of the members of clade HB1 except the genus Natronomonas. Sequence information for a limited number of Halobacteria and other archaea are shown here, but the indicated CSIs were detected only in members of the indicated group and not detected in any other species in the top 250 Blastp hits. The dashes (-) in the alignments indicate identity with the residue in the top sequence. GenInfo identification (GI) numbers for each sequence are indicated in the second column
We have also identified a number of CSIs and CSPs that are uniquely shared by groups within the order Halobacteriales that show inconsistent branching affinity in phylogenetic trees. An association between the members of the genera Halococcus and Halalkalicoccus is supported by two CSIs and one CSP identified in this work (Tables 1, 3D). Additionally, Halococcus and Halalkalicoccus, cluster together in the 16S rRNA sequence based phylogenetic tree alongside the genera Haladaptus, Halorussus, and Halrubellus. However, there is no association observed between Halococcus and Halalkalicoccus in the concatenated protein tree and no CSIs or CSPs are identified that are specific to a grouping of the sequenced members of the genus Halococcus and associated genera identified in the 16S rRNA tree (viz. Halalkalicoccus and Haladaptus). Thus, clade HB2 is currently limited to the members of the genus Halococcus until the affinity between the genus Halococcus and the genera Halalkalicoccus, Haladaptus, Halorussus, and Halrubellus can be determined. Within the order Halobacteriales, we have also identified phylogenetic and molecular support for a supergeneric relationship between the genera Halobacterium and Halarchaeum. The genera Halobacterium and Halarchaeum cluster together in the concatenated protein and in the 16S rRNA gene based phylogenetic trees along with the genus Salarchaeum. A relationship between Halobacterium and Halarchaeum is further supported by four CSIs (Table 1) and one CSP (GI: 169236474) identified in this work which are uniquely shared by these two genera. We have also identified some CSIs that support a relationship Halobacterium and Haladaptus, the significance of which is currently unclear. The sequence alignments for the CSIs specific to either Halococcus and Halalkalicoccus or Halobacterium and Halarchaeum are shown in Supplemental Figs. 12–17 and their properties are briefly summarised in Table 2.
Molecular characteristics which characterise important genus level groups within the class Halobacteria
In addition to the CSIs and CSPs identified for groups of multiple genera within the class Halobacteria, we have also identified a number of CSIs and CSPs which are uniquely found within members of specific genera within the class. Within the order Haloferacales, we have identified seven CSIs and 38 CSPs which are restricted to the members of the genus Haloferax, and seven CSIs and 60 CSPs which are restricted to the members of the genus Halorubrum. Examples of CSIs which are uniquely found in members of the genera Haloferax and Halorubrum, respectively, are shown in Fig. 5. A one amino acid insertion that is specific for members of the genus Haloferax found in a hypothetical protein is shown in Fig. 5a whereas a one amino acid insertion that is specific for members of the genus Halorubrum which is present in the 50S ribosomal protein L19 is shown in Fig. 5b. In both cases, these inserts are unique characteristics of these genera and are not found in any other member of the class Halobacteria. The sequence alignments for the other identified Haloferax and Halorubrum CSIs are presented in Supplemental Figs. 18–29 and their properties are summarised in Table 2. The GI numbers for the seventeen CSPs specific for the genus Haloferax and the seven CSPs specific for the genus Halorubrum are provided in Table 3E, F, respectively.
Partial sequence alignments of a a hypothetical protein showing a one amino acid insertion (boxed) identified in all sequenced members of the genus Haloferax, b 50S ribosomal protein L19 showing a one amino acid insertion (boxed) identified in all sequenced members of the genus Halorubrum. Sequence information for a limited number of Halobacteria and other archaea are shown here, but unless otherwise indicated similar CSIs were detected in all members of the indicated group and not detected in any other species in the top 250 Blastp hits. The characteristics of all identified CSIs specific for the genera Haloferax and Halorubrum are summarized in Table 2
We have also identified nine CSIs and twenty-three CSPs which are restricted to the large assemblage of sequenced species within the genus Halococcus and seventeen CSIs and 113 CSPs that are restricted to the genus Haloarcula, both of which are found within the order Halobacteriales. An example of a CSI which is specific for the genus Halococcus and another CSI which is specific for the genus Haloarcula are shown in Fig. 6. The first CSI, shown in Fig. 6a, consists of a two amino acid insertion in DNA gyrase subunit B that is uniquely found in the genus Halococcus whereas the second CSI, shown in Fig. 6b, consists of a five amino acid insertion in a gufA family protein that is uniquely found in the members of the genus Haloarcula. The sequence alignments for eight additional CSIs which are specific for the genus Halococcus and sixteen additional CSIs which are specific for the genus Haloarcula are shown in Supplemental Figs. 30–53 and their properties are briefly summarised in Table 2. The identification numbers for the 23 CSPs which are specific for the genus Halococcus and the 113 CSPs specific for the genus Haloarcula are provided in Table 3D, G, respectively.
Partial sequence alignments of a a gufA family protein showing a five amino acid insertion (boxed) identified in all sequenced members of the genus Haloarcula, b DNA gyrase subunit B showing a two amino acid insertion (boxed) identified in all sequenced members of the genus Halococcus. Sequence information for a limited number of Halobacteria and other archaea are shown here, but unless otherwise indicated similar CSIs were detected in all members of the indicated group and not detected in any other species in the top 250 Blastp hits. The characteristics of all identified CSIs specific for the genera Haloarcula and Halococcus are summarized in Table 2
Discussion
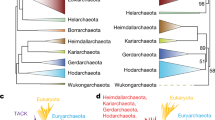
The analyses presented here, in the form of a highly robust phylogenetic tree based on 266 proteins obtained from 129 genome sequenced members of the class Halobacteria (Fig. 1), the 20 identified CSIs (Table 1), and the 31 identified CSPs (Table 3) provide a reliable basis for understanding the infra-order level relationships of genera within the class Halobacteria. These analyses have identified a number of family level clusters within the orders Haloferacales and Halobacteriales which are supported by a large number of CSIs and CSPs (Fig. 7). The result of phylogenomic studies and identified molecular characteristics provide strong evidence that the order Haloferacales contains two main groups, one consisting of Haloferax and related genera and the other consisting of Halorubrum and related genera, each of which is supported by multiple lines of evidence.
A summary diagram depicting the distribution of identified CSIs and CSPs within the class Halobacteria and the proposed families described in this study. The genera within each family are listed under the corresponding family name. The superscript letter T beside a genus indicates that it is the type genus of the family
The order Halobacteriales, as currently described (Gupta et al. 2015), is a polyphyletic assemblage of members of the class Halobacteria that do not fall into the monophyletic orders Haloferacales and Natrialbales. The present analysis has identified one large and consistently observed group of multiple genera within the order Halobacteriales, referred to as clade HB1, and a second clade consisting of the large assemblage of species which make up the genus Halococcus, referred to as clade HB2, which are both supported by a large number of identified CSIs and CSPs (Fig. 7). Prior research on the interrelationships of the species within the genus Halococcus has noted that the members of the genus are more genetically diverse than other genera within the class Halobacteria and that the genus Halococcus contains two phylotypes which may eventually be separated into two genera (Montero et al. 1993; Goh et al. 2006). It should be mentioned that in the rRNA tree, the genera Halalkalicoccus, Haladaptus, Halorussus and Halrubellus also group together with the Halococcus spp. (Clade HB2). An affinity of Halococcus and Halalkalicoccus is also supported by a number of identified CSIs and CSPs. Although we are presently limiting the clade HB2 to the genus Halococcus, with some additional research it is likely that this group will be expanded to include the genera Halalkalicoccus, Haladaptus, Halorussus and Halrubellus, which show some affinity to this clade. Further work is also required to clearly delimit the groups within the genus Halococcus.
The phylogenetic trees and molecular characteristics presented here also provide strong evidence for the separation of the genera Halobacterium, Halarchaeum, and Salarchaeum from the other members of the order Halobacteriales as a novel family level taxa. However, the Bacteriological Code (Lapage et al. 1992) states that a family consisting of the genera Halobacterium, Halarchaeum, and Salarchaeum must retain the name Halobacteriaceae precluding any reclassification of this group. Thus, an important task for the future will be to identify reliable, genome-derived characteristics, utilising novel techniques and the increasing wealth of genomic data, which can be used to classify the other members of the order Halobacteriales as members of distinct clades/families or clearly establish their close affinity to the grouping of the genera Halobacterium, Halarchaeum, and Salarchaeum.
The analyses presented here have also lead to the identification of 31 CSIs and 211 CSPs which are characteristic of Haloferax, Halorubrum, and Haloarcula (Table 2, 3E–G). Based upon these molecular characteristics, it is now possible to differentiate these groups of species from all other Halobacteria on the basis of the presence or absence of unique molecular features. Earlier work by our group on other groups of prokaryotes has shown that these CSIs and CSPs have strong predictive value and will likely be found in other members of these groups as more members are sequenced (Gao and Gupta 2012b; Gupta and Lali 2013; Bhandari and Gupta 2014; Howard-Azzeh et al. 2014). Additionally, the conserved nature of these CSIs and CSPs make them promising targets for the development of diagnostic assays that can be used to identify novel members of these genera from isolates or environmental samples (Ahmod et al. 2011; Wong et al. 2014). Further analyses of these genus specific molecular characteristics should also lead to the discovery of novel functions in these organisms mediated by CSIs and CSPs which may provide important insights into the physiology, evolution, and novel adaptations of these groups of Halobacteria.
Overall, the results presented here provide strong support for the presence of two family-level groups within the order Haloferacales and for two novel family level clusters within the polyphyletic order Halobacteriales. Based on the phylogenetic analyses, the identified CSIs and the identified CSPs presented in this work, we propose a division of the order Haloferacales into two families, an emended family Haloferacaceae (clade HF1) and Halorubraceae fam nov. (clade HF2) and a division of the order Halobacteriales into three families, Haloarculaceae fam. nov. (clade HB1), Halococcaceae fam. nov. (clade HB2), and an emended family Halobacteriaceae, containing all members of the order Halobacteriales that do not fall into clade HB1 or HB2. Descriptions of the new and emended families are provided below.
Emended description of the family Halobacteriaceae Gibbons 1974 (approved lists 1980)
The family Halobacteriaceae contains the type genus Halobacterium (Oren et al. 2009) and the genera Haladaptatus (Cui et al. 2010d), Halalkalicoccus (Xue et al. 2005), Halarchaeum (Minegishi et al. 2010a), Haloarchaeobius (Yuan et al. 2015), Halomarina (Inoue et al. 2011), Halorubellus (Cui et al. 2012), Halorussus (Cui et al. 2010c), Halovenus (Makhdoumi-Kakhki et al. 2012), Natronoarchaeum (Shimane et al. 2010), Natronomonas (Burns et al. 2010b) and Salarchaeum (Shimane et al. 2011). The description of the family is the same as that of the order Halobacteriales given by Gupta et al. (2015) with the following modifications: The members of this order also lack the CSIs and CSPs that are specific for the families Haloarculaceae and Halococcaceae.
Description of Haloarculaceae fam. nov.
Haloarculaceae (Ha.lo.ar.cu.la.ce’ae. N.L. fem. n. Haloarcula type genus of the family; -aceae ending to denote a family; N.L. fem. pl. n. Haloarculaceae the family whose nomenclatural type is the genus Haloarcula).
The family Haloarculaceae contains the type genus Haloarcula (Oren et al. 2009) and the genera Halapricum (Song et al. 2014), Halomicroarcula (Echigo et al. 2013), Halomicrobium (Oren et al. 2002), Halorhabdus (Antunes et al. 2008), Halorientalis (Amoozegar et al. 2014), and Halosimplex (Han and Cui 2014). The description of the family is the same as that of the order Halobacteriales given by Gupta et al. (2015) with the following modifications: the members of the family Haloarculaceae can be distinguished from the Halobacteriaceae and all other archaea by the ten CSIs listed in Table 1 and by the twenty CSPs listed in Table 3C.
Description of Halococcaceae fam. nov.
Halococcaceae (Ha.lo.coc.ca.ce’ae.N.L. masc. n. Halococcus type genus of the family; -aceae ending to denote a family; N.L. masc. pl. n. Halococcaceae the family whose nomenclatural type is the genus Halococcus).
The family Halococcaceae contains only the type genus Halococcus (Oren et al. 2009). The description of the family is the same as that of the order Halobacteriales given by Gupta et al. (2015) with the following modifications: the members of the family Halococcaceae can be distinguished from the Halobacteriaceae and all other archaea by the nine CSIs listed in Table 2 and by 23 CSPs CSPs listed in Table 3D.
Emended description of the family Haloferacaceae Gupta 2015
The family Haloferacaceae contains the type genus Haloferax (Oren et al. 2009) and the genera Halobellus (Cui et al. 2011b), Halogeometricum (Cui et al. 2010e), Halogranum (Cui et al. 2011c), Halopelagius (Zhang et al. 2013), Haloplanus (Qiu et al. 2013), Haloquadratum (Burns et al. 2007), and Halosarcina (Cui et al. 2010a). The description of the family is the same as that of the order Haloferacales given by Gupta et al. (2015) with the following modifications: the members of the family Haloferacaceae can be distinguished from the Halorubraceae and all other archaea by their branching in phylogenetic trees and by the four CSIs listed in Table 1 and by the five CSPs listed in Table 2.
Description of Halorubraceae fam. nov.
Halorubraceae (Ha.lo.ru.bra.ce’ae. N.L. neut. n. Halorubrum type genus of the family; -aceae ending to denote a family; N.L. neut. pl. n. Halorubraceae the family whose nomenclatural type is the genus Halorubrum).
The family Halorubraceae contains the type genus Halorubrum (Oren et al. 2009) and the genera Halobaculum (Oren et al. 1995), Halogranum (Cui et al. 2010b), Halohasta (Mou et al. 2012), Halolamina (Cui et al. 2011a), Halonotius (Burns et al. 2010a), Halopenitus (Amoozegar et al. 2012), and Salinigranum (Cui and Zhang 2014). The description of the family is the same as that of the order Haloferacales given by Gupta et al. (2015) with the following modifications: the members of the family Halorubraceae can be distinguished from the Haloferacaceae and all other archaea by their branching in phylogenetic trees and by the four CSPs listed in Table 2.
References
Ahmod NZ, Gupta RS, Shah HN (2011) Identification of a Bacillus anthracis specific indel in the yeaC gene and development of a rapid pyrosequencing assay for distinguishing B. anthracis from the B. cereus group. J Microbiol Methods 87(3):278–285
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25(17):3389–3402
Amoozegar MA, Makhdoumi-Kakhki A, Shahzadeh Fazeli SA, Azarbaijani R, Ventosa A (2012) Halopenitus persicus gen. nov., sp. nov., an archaeon from an inland salt lake. Int J Syst Evol Microbiol 62(Pt 8):1932–1936
Amoozegar MA, Makhdoumi-Kakhki A, Mehrshad M, Fazeli SA, Sproer C, Ventosa A (2014) Halorientalis persicus sp. nov., an extremely halophilic archaeon isolated from a salt lake and emended description of the genus Halorientalis. Int J Syst Evol Microbiol 64(Pt 3):940–944
Andam CP, Harlow TJ, Papke RT, Gogarten JP (2012) Ancient origin of the divergent forms of leucyl-tRNA synthetases in the Halobacteriales. BMC Evol Biol 12:85
Antunes A, Taborda M, Huber R, Moissl C, Nobre MF, da Costa MS (2008) Halorhabdus tiamatea sp. nov., a non-pigmented, extremely halophilic archaeon from a deep-sea, hypersaline anoxic basin of the Red Sea, and emended description of the genus Halorhabdus. Int J Syst Evol Microbiol 58(Pt 1):215–220
Baliga NS, Bonneau R, Facciotti MT, Pan M, Glusman G, Deutsch EW, Shannon P, Chiu Y, Weng RS, Gan RR et al (2004) Genome sequence of Haloarcula marismortui: a halophilic archaeon from the Dead Sea. Genome Res 14(11):2221–2234
Bhandari V, Gupta RS (2014) Molecular signatures for the phylum (class) Thermotogae and a proposal for its division into three orders (Thermotogales, Kosmotogales ord. nov. and Petrotogales ord. nov.) containing four families (Thermotogaceae, Fervidobacteriaceae fam. nov., Kosmotogaceae fam. nov. and Petrotogaceae fam. nov.) and a new genus Pseudothermotoga gen. nov. with five new combinations. Anton Leeuw Int J G 105(1):143–168
Boucher Y, Douady CJ, Sharma AK, Kamekura M, Doolittle WF (2004) Intragenomic heterogeneity and intergenomic recombination among haloarchaeal rRNA genes. J Bacteriol 186(12):3980–3990
Burns DG, Janssen PH, Itoh T, Kamekura M, Li Z, Jensen G, Rodriguez-Valera F, Bolhuis H, Dyall-Smith ML (2007) Haloquadratum walsbyi gen. nov., sp. nov., the square haloarchaeon of Walsby, isolated from saltern crystallizers in Australia and Spain. Int J Syst Evol Microbiol 57(Pt 2):387–392
Burns DG, Janssen PH, Itoh T, Kamekura M, Echigo A, Dyall-Smith ML (2010a) Halonotius pteroides gen. nov., sp. nov., an extremely halophilic archaeon recovered from a saltern crystallizer. Int J Syst Evol Microbiol 60(Pt 5):1196–1199
Burns DG, Janssen PH, Itoh T, Minegishi H, Usami R, Kamekura M, Dyall-Smith ML (2010b) Natronomonas moolapensis sp. nov., non-alkaliphilic isolates recovered from a solar saltern crystallizer pond, and emended description of the genus Natronomonas. Int J Syst Evol Microbiol 60(Pt 5):1173–1176
Castresana J (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17(4):540–552
Ciccarelli FD, Doerks T, Von Mering C, Creevey CJ, Snel B, Bork P (2006) Toward automatic reconstruction of a highly resolved tree of life. Science 311(5765):1283–1287
Cole J, Wang Q, Fish J, Chai B, McGarrell D, Sun Y, Brown C, Porras-Alfaro A, Kuske C, Tiedje J (2014) Ribosomal database project: data and tools for high throughput rRNA analysis. Nucleic Acids Res 42(1):D633
Cui HL, Zhang WJ (2014) Salinigranum rubrum gen. nov., sp. nov., a member of the family Halobacteriaceae isolated from a marine solar saltern. Int J Syst Evol Microbiol 64(Pt 6):2029–2033
Cui HL, Zhou PJ, Oren A, Liu SJ (2009) Intraspecific polymorphism of 16S rRNA genes in two halophilic archaeal genera, Haloarcula and Halomicrobium. Extremophiles 13(1):31–37
Cui HL, Gao X, Li XY, Xu XW, Zhou YG, Liu HC, Zhou PJ (2010a) Halosarcina limi sp. nov., a halophilic archaeon from a marine solar saltern, and emended description of the genus Halosarcina. Int J Syst Evol Microbiol 60(Pt 10):2462–3466
Cui HL, Gao X, Sun FF, Dong Y, Xu XW, Zhou YG, Liu HC, Oren A, Zhou PJ (2010b) Halogranum rubrum gen. nov., sp. nov., a halophilic archaeon isolated from a marine solar saltern. Int J Syst Evol Microbiol 60(Pt 6):1366–1371
Cui HL, Gao X, Yang X, Xu XW (2010c) Halorussus rarus gen. nov., sp. nov., a new member of the family Halobacteriaceae isolated from a marine solar saltern. Extremophiles 14(6):493–499
Cui HL, Sun FF, Gao X, Dong Y, Xu XW, Zhou YG, Liu HC, Oren A, Zhou PJ (2010d) Haladaptatus litoreus sp. nov., an extremely halophilic archaeon from a marine solar saltern, and emended description of the genus Haladaptatus. Int J Syst Evol Microbiol 60(Pt 5):1085–1089
Cui HL, Yang X, Gao X, Li XY, Xu XW, Zhou YG, Liu HC, Zhou PJ (2010e) Halogeometricum rufum sp. nov., a halophilic archaeon from a marine solar saltern, and emended description of the genus Halogeometricum. Int J Syst Evol Microbiol 60(Pt 11):2613–2617
Cui HL, Gao X, Yang X, Xu XW (2011a) Halolamina pelagica gen. nov., sp. nov., a new member of the family Halobacteriaceae. Int J Syst Evol Microbiol 61(Pt 7):1617–1621
Cui HL, Yang X, Gao X, Xu XW (2011b) Halobellus clavatus gen. nov., sp. nov. and Halorientalis regularis gen. nov., sp. nov., two new members of the family Halobacteriaceae. Int J Syst Evol Microbiol 61(Pt 11):2682–2689
Cui HL, Yang X, Gao X, Xu XW (2011c) Halogranum gelatinilyticum sp. nov. and Halogranum amylolyticum sp. nov., isolated from a marine solar saltern, and emended description of the genus Halogranum. Int J Syst Evol Microbiol 61(Pt 4):911–915
Cui HL, Mou YZ, Yang X, Zhou YG, Liu HC, Zhou PJ (2012) Halorubellus salinus gen. nov., sp. nov. and Halorubellus litoreus sp. nov., novel halophilic archaea isolated from a marine solar saltern. Syst Appl Microbiol 35(1):30–34
Echigo A, Minegishi H, Shimane Y, Kamekura M, Itoh T, Usami R (2013) Halomicroarcula pellucida gen. nov., sp. nov., a non-pigmented, transparent-colony-forming, halophilic archaeon isolated from solar salt. Int J Syst Evol Microbiol 63(Pt 10):3556–3562
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26(19):2460–2461
Enache M, Itoh T, Fukushima T, Usami R, Dumitru L, Kamekura M (2007) Phylogenetic relationships within the family Halobacteriaceae inferred from rpoB ‘ gene and protein sequences. Int J Syst Evol Microbiol 57:2289–2295
Gao B, Gupta R (2007) Phylogenomic analysis of proteins that are distinctive of Archaea and its main subgroups and the origin of methanogenesis. BMC Genomics 8(1):86
Gao B, Gupta RS (2012a) Microbial systematics in the post-genomics era. Anton Leeuw Int J G 101(1):45–54
Gao B, Gupta RS (2012b) Phylogenetic framework and molecular signatures for the main clades of the phylum Actinobacteria. Microbiol Mol Biol Rev 76(1):66–112
Goh F, Leuko S, Allen MA, Bowman JP, Kamekura M, Neilan BA, Burns BP (2006) Halococcus hamelinensis sp. nov., a novel halophilic archaeon isolated from stromatolites in Shark Bay, Australia. Int J Syst Evol Microbiol 56(6):1323–1329
Grant WD, Kamekura M, McGenity TJ, Ventosa A (2001) Class III. Halobacteria class. nov. In: Boone, Castenholz RW, Garrity GM, Bergey DH (eds) Bergey’s manual of systematic bacteriology, vol 1, 2nd edn. Springer, New York
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59(3):307–321
Gupta RS (2014) Identification of conserved indels that are useful for classification and evolutionary studies methods in microbiology, vol 41. Academic Press, Oxford
Gupta RS, Lali R (2013) Molecular signatures for the phylum Aquificae and its different clades: proposal for division of the phylum Aquificae into the emended order Aquificales, containing the families Aquificaceae and Hydrogenothermaceae, and a new order Desulfurobacteriales ord. nov., containing the family Desulfurobacteriaceae. Anton Leeuw Int J G 104(3):349–368
Gupta RS, Mok A (2007) Phylogenomics and signature proteins for the Alphaproteobacteria and its main groups. BMC Microbiol 7(1):106
Gupta RS, Naushad S, Baker S (2015) Phylogenomic analyses and molecular signatures for the class Halobacteria and its two major clades: a proposal for division of the class Halobacteria into an emended order Halobacteriales and two new orders, Haloferacales ord. nov. and Natrialbales ord. nov., containing the novel families Haloferacaceae fam. nov. and Natrialbaceae fam. nov. Int J Syst Evol Microbiol 65(Pt 3):1050–1069
Han D, Cui HL (2014) Halosimplex pelagicum sp. nov. and Halosimplex rubrum sp. nov., isolated from salted brown alga Laminaria, and emended description of the genus Halosimplex. Int J Syst Evol Microbiol 64(Pt 1):169–173
Hartman AL, Norais C, Badger JH, Delmas S, Haldenby S, Madupu R, Robinson J, Khouri H, Ren Q, Lowe TM et al (2010) The complete genome sequence of Haloferax volcanii DS2, a model archaeon. PLoS One 5(3):e9605
Howard-Azzeh M, Shamseer L, Schellhorn HE, Gupta RS (2014) Phylogenetic analysis and molecular signatures defining a monophyletic clade of heterocystous cyanobacteria and identifying its closest relatives. Photosynth Res 122(2):171–185
Inoue K, Itoh T, Ohkuma M, Kogure K (2011) Halomarina oriensis gen. nov., sp. nov., a halophilic archaeon isolated from a seawater aquarium. Int J Syst Evol Microbiol 61(Pt 4):942–946
Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ (1998) Multiple sequence alignment with Clustal X. Trends Biochem Sci 23(10):403
Jones AL (2012) The future of taxonomy. In: Gadd GM, Sariaslani S (eds) Advances in applied microbiology, vol 80, 1st edn. Academic Press Inc, San Diego, pp 23–35
Klenk HP, Goker M (2010) En route to a genome-based classification of Archaea and Bacteria? Syst Appl Microbiol 33(4):175–182
Lapage SP, Sneath PHA, Lessel EF, Skerman VBD, Seeliger HPR, Clark WA (1992) International code of nomenclature of bacteria: bacteriological code, 1990 revision. ASM Press International Union of Microbiological Societies, Washington, DC
Le SQ, Gascuel O (2008) An improved general amino acid replacement matrix. Mol Biol Evol 25(7):1307–1320
Makhdoumi-Kakhki A, Amoozegar MA, Ventosa A (2012) Halovenus aranensis gen. nov., sp. nov., an extremely halophilic archaeon from Aran-Bidgol salt lake. Int J Syst Evol Microbiol 62(Pt 6):1331–1336
Minegishi H, Echigo A, Nagaoka S, Kamekura M, Usami R (2010a) Halarchaeum acidiphilum gen. nov., sp. nov., a moderately acidophilic haloarchaeon isolated from commercial solar salt. Int J Syst Evol Microbiol 60(Pt 11):2513–2516
Minegishi H, Kamekura M, Itoh T, Echigo A, Usami R, Hashimoto T (2010b) Further refinement of the phylogeny of the Halobacteriaceae based on the full-length RNA polymerase subunit B ‘ (rpoB ‘) gene. Int J Syst Evol Microbiol 60:2398–2408
Montero CG, Klenk H-P, Nieto JJ, Ventosa A (1993) DNA-rRNA hybridization studies on Halococcus saccharolyticus and other halobacteria. FEMS Microbiol Lett 111(1):69–72
Mou YZ, Qiu XX, Zhao ML, Cui HL, Oh D, Dyall-Smith ML (2012) Halohasta litorea gen. nov. sp. nov., and Halohasta litchfieldiae sp. nov., isolated from the Daliang aquaculture farm, China and from Deep Lake, Antarctica, respectively. Extremophiles 16(6):895–901
Mylvaganam S, Dennis PP (1992) Sequence heterogeneity between the two genes encoding 16S rRNA from the halophilic archaebacterium Haloarcula marismortui. Genetics 130(3):399–410
NamesforLife (2015) NamesforLife. http://www.namesforlife.com/
Naor A, Lapierre P, Mevarech M, Papke RT, Gophna U (2012) Low species barriers in halophilic archaea and the formation of recombinant hybrids. Curr Biol 22(15):1444–1448
Naushad HS, Lee B, Gupta RS (2014) Conserved signature indels and signature proteins as novel tools for understanding microbial phylogeny and systematics: identification of molecular signatures that are specific for the phytopathogenic genera Dickeya, Pectobacterium and Brenneria. Int J Syst Evol Microbiol 64(2):366–383
Naushad S, Adeolu M, Wong S, Sohail M, Schellhorn HE, Gupta RS (2015) A phylogenomic and molecular marker based taxonomic framework for the order Xanthomonadales: proposal to transfer the families Algiphilaceae and Solimonadaceae to the order Nevskiales ord. nov. and to create a new family within the order Xanthomonadales, the family Rhodanobacteraceae fam. nov., containing the genus Rhodanobacter and its closest relatives. Antonie Van Leeuwenhoek 107(2):467–485
NCBI (2015) NCBI genome database. http://www.ncbi.nlm.nih.gov/genome/
Oren A (2006) The order Halobacteriales. In: Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E (eds) The prokaryotes. Springer, New York, pp 113–164
Oren A (2012) Taxonomy of the family Halobacteriaceae: a paradigm for changing concepts in prokaryote systematics. Int J Syst Evol Microbiol 62(Pt 2):263–271
Oren A, Garrity GM (2014) Then and now: a systematic review of the systematics of prokaryotes in the last 80 years. Antonie Van Leeuwenhoek 106(1):43–56
Oren A, Gurevich P, Gemmell RT, Teske A (1995) Halobaculum gomorrense gen. nov., sp. nov., a novel extremely halophilic archaeon from the Dead Sea. Int J Syst Bacteriol 45(4):747–754
Oren A, Elevi R, Watanabe S, Ihara K, Corcelli A (2002) Halomicrobium mukohataei gen. nov., comb. nov., and emended description of Halomicrobium mukohataei. Int J Syst Evol Microbiol 52(Pt 5):1831–1835
Oren A, Arahal DR, Ventosa A (2009) Emended descriptions of genera of the family Halobacteriaceae. Int J Syst Evol Microbiol 59(Pt 3):637–642
Papke RT, White E, Reddy P, Weigel G, Kamekura M, Minegishi H, Usami R, Ventosa A (2011) A multilocus sequence analysis approach to the phylogeny and taxonomy of the Halobacteriales. Int J Syst Evol Microbiol 61(Pt 12):2984–2995
Parte AC (2013) LPSN-list of prokaryotic names with standing in nomenclature. Nucleic Acids Res 42:D613–D616
Pfeiffer F, Schuster SC, Broicher A, Falb M, Palm P, Rodewald K, Ruepp A, Soppa J, Tittor J, Oesterhelt D (2008) Evolution in the laboratory: the genome of Halobacterium salinarum strain R1 compared to that of strain NRC-1. Genomics 91(4):335–346
Price MN, Dehal PS, Arkin AP (2010) FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One 5(3):e9490
Pruesse E, Peplies J, Glockner FO (2012) SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28(14):1823–1829
Qiu XX, Zhao ML, Han D, Zhang WJ, Cui HL (2013) Haloplanus salinus sp. nov., an extremely halophilic archaeon from a Chinese marine solar saltern. Arch Microbiol 195(12):799–803
Rokas A, Holland PWH (2000) Rare genomic changes as a tool for phylogenetics. Trends Ecol Evol 15(11):454–459
Rokas A, Williams BL, King N, Carroll SB (2003) Genome-scale approaches to resolving incongruence in molecular phylogenies. Nature 425(6960):798–804
Rossello-Mora R, Amann R (2015) Past and future species definitions for Bacteria and Archaea. Syst Appl Microbiol 38(4):209–216
Sawana A, Adeolu M, Gupta RS (2014) Molecular signatures and phylogenomic analysis of the genus Burkholderia: proposal for division of this genus into the emended genus Burkholderia containing pathogenic organisms and a new genus Paraburkholderia gen. nov. harboring environmental species. Front Genet 5:429
Shimane Y, Hatada Y, Minegishi H, Mizuki T, Echigo A, Miyazaki M, Ohta Y, Usami R, Grant WD, Horikoshi K (2010) Natronoarchaeum mannanilyticum gen. nov., sp. nov., an aerobic, extremely halophilic archaeon isolated from commercial salt. Int J Syst Evol Microbiol 60(Pt 11):2529–2534
Shimane Y, Hatada Y, Minegishi H, Echigo A, Nagaoka S, Miyazaki M, Ohta Y, Maruyama T, Usami R, Grant WD et al (2011) Salarchaeum japonicum gen. nov., sp. nov., an aerobic, extremely halophilic member of the Archaea isolated from commercial salt. Int J Syst Evol Microbiol 61(Pt 9):2266–2270
Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J et al (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7(1):539
Song HS, Cha IT, Yim KJ, Lee HW, Hyun DW, Lee SJ, Rhee SK, Kim KN, Kim D, Choi JS et al (2014) Halapricum salinum gen. nov., sp. nov., an extremely halophilic archaeon isolated from non-purified solar salt. Antonie Van Leeuwenhoek 105(5):979–986
Soucy SM, Fullmer MS, Papke RT, Gogarten JP (2014) Inteins as indicators of gene flow in the halobacteria. Front Microbiol, 5
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313
Sutcliffe IC (2015) Challenging the anthropocentric emphasis on phenotypic testing in prokaryotic species descriptions: rip it up and start again. Front Genet 6:218
Talavera G, Castresana J (2007) Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol 56(4):564–577
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30(12):2725–2729
Tavaré S (1986) Some probabilistic and statistical problems in the analysis of DNA sequences. In: Miura RM (ed) Lectures on mathematics in the life sciences, 17th edn. American Mathematical Society, Providence, RI, pp 57–86
Vreeland RH, Straight S, Krammes J, Dougherty K, Rosenzweig WD, Kamekura M (2002) Halosimplex carlsbadense gen. nov., sp. nov., a unique halophilic archaeon, with three 16S rRNA genes, that grows only in defined medium with glycerol and acetate or pyruvate. Extremophiles 6(6):445–452
Walsh DA, Bapteste E, Kamekura M, Doolittle WF (2004) Evolution of the RNA polymerase B’ subunit gene (rpoB’) in Halobacteriales: a complementary molecular marker to the SSU rRNA gene. Mol Biol Evol 21(12):2340–2351
Whelan S, Goldman N (2001) A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol 18(5):691–699
Whitman WB (2015) Genome sequences as the type material for taxonomic descriptions of prokaryotes. Syst Appl Microbiol 38(4):217–222
Williams D, Gogarten JP, Papke RT (2012) Quantifying homologous replacement of loci between haloarchaeal species. Genome Biol Evol 4(12):1223–1244
Wong SY, Paschos A, Gupta RS, Schellhorn HE (2014) Insertion/deletion-based approach for the detection of Escherichia coli O157:H7 in freshwater environments. Environ Sci Technol 48(19):11462–11470
Wright AD (2006) Phylogenetic relationships within the order Halobacteriales inferred from 16S rRNA gene sequences. Int J Syst Evol Microbiol 56(Pt 6):1223–1227
Wu D, Hugenholtz P, Mavromatis K, Pukall R, Dalin E, Ivanova NN, Kunin V, Goodwin L, Wu M, Tindall BJ (2009) A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea. Nature 462(7276):1056–1060
Xue Y, Fan H, Ventosa A, Grant WD, Jones BE, Cowan DA, Ma Y (2005) Halalkalicoccus tibetensis gen. nov., sp. nov., representing a novel genus of haloalkaliphilic archaea. Int J Syst Evol Microbiol 55(Pt 6):2501–2505
Yuan PP, Zhang WJ, Han D, Cui HL (2015) Haloarchaeobius salinus sp. nov., isolated from an inland salt lake, and emended description of the genus Haloarchaeobius. Int J Syst Evol Microbiol 65(Pt 3):910–914
Zhang X, Zhang WY, Shen AH, Huo YY, Zhu XF, Wu M (2013) Halopelagius longus sp. nov., a member of the family Halobacteriaceae isolated from a salt mine, and emended description of the genus Halopelagius. Int J Syst Evol Microbiol 63(Pt 10):3585–3590
Zhi X-Y, Zhao W, Li W-J, Zhao G-P (2012) Prokaryotic systematics in the genomics era. Anton Leeuw Int J G 101(1):21–34
Author information
Authors and Affiliations
Corresponding author
Additional information
An erratum to this article can be found at http://dx.doi.org/10.1007/s10482-016-0765-7.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Gupta, R.S., Naushad, S., Fabros, R. et al. A phylogenomic reappraisal of family-level divisions within the class Halobacteria: proposal to divide the order Halobacteriales into the families Halobacteriaceae, Haloarculaceae fam. nov., and Halococcaceae fam. nov., and the order Haloferacales into the families, Haloferacaceae and Halorubraceae fam nov.. Antonie van Leeuwenhoek 109, 565–587 (2016). https://doi.org/10.1007/s10482-016-0660-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-016-0660-2