Abstract
The current taxonomy of the order Xanthomonadales is highly problematic and no comprehensive phylogenomic studies have been completed that include the most divergent members within the order. In this work, we have completed a phylogenomic analysis of a wide range of genomes, five of which were sequenced for the first time for this work, representing the vast majority of the diversity within the order Xanthomonadales. Using comparative genomic techniques, we have identified a large number of conserved signature inserts/deletions (CSIs) that are specifically found in different groups of related organisms, at different taxonomic levels, within the order. Our phylogenetic analyses do not support a monophyletic grouping of the members of the order Xanthomonadales and no CSIs were identified which are uniquely shared by all sequenced species within this order. However, our work has identified 10 CSIs which are specific to all members of the family Xanthomonadaceae and an additional 10 and 11 CSIs that are specific to one of two phylogenetically well-defined clades within the family Xanthomonadaceae. On the basis of the identified CSIs and the results of phylogenomic analyses, we propose a new taxonomic framework for the order Xanthomonadales. In this proposal, the families Algiphilaceae and Solimonadaceae (Nevskiaceae), which do not branch with the other members of the order Xanthomonadales, are transferred into the order Nevskiales ord. nov. The remaining members of the order Xanthomonadales are divided into two families: the family Xanthomonadaceae, containing the genus Xanthomonas and its closest relatives, and a new family, Rhodanobacteraceae fam. nov., containing the genus Rhodanobacter and its closest relatives. Additionally, we have also emended descriptions of the order Lysobacterales, the family Lysobacteraceae, and the family Nevskiaceae to indicate that they are earlier synonyms of the order Xanthomonadales, the family Xanthomonadaceae, and the family Solimonadaceae, respectively.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The order Xanthomonadales is an early diverging group of bacteria within the class Gammaproteobacteria (Cutino-Jimenez et al. 2010; Williams et al. 2010; Naushad and Gupta 2013). The order Xanthomonadales currently contains 5 families (viz. Algiphilaceae, Nevskiaceae, Sinobacteraceae, Solimonadaceae, and Xanthomonadaceae) which contain 30 genera encompassing a large number of species that possess a diverse range of phenotypic and biochemical characteristics (Saddler and Bradbury 2005a; Parte 2013). The members of this order include a number of major plant pathogens that have significant economic and agricultural impact. Members of the genera Xylella and Xanthomonas, in particular, are major phytopathogens which cause a wide variety of serious diseases in more than 400 agriculturally important plants including tomatoes, bananas, citrus plants, rice, and coffee plants (da Silva et al. 2002; Van Sluys et al. 2003; Lee et al. 2005; Chatterjee et al. 2008; Salzberg et al. 2008; Ryan et al. 2011). The order also contains the genus Stenotrophomonas which harbours a number of increasingly important multidrug resistant opportunistic pathogens that are responsible for hospital-acquired infections in immunodeficient patients (Crossman et al. 2008; Looney et al. 2009). Despite the important plant and human pathogens present within this order, the taxonomy of this group is highly problematic and no comprehensive phylogenetic studies have been completed that focus specifically on the interrelationships of the different members within the order Xanthomonadales (Gao et al. 2009; Cutino-Jimenez et al. 2010; Williams et al. 2010; Naushad and Gupta 2013; Tindall 2014b).
The current taxonomy of the order Xanthomonadales is largely based on 16S rRNA sequence analysis (Saddler and Bradbury 2005a; Gutierrez et al. 2012; Losey et al. 2013). However, the 16S rRNA gene sequence has shown limited ability to resolve the branching and relationships of organisms within the order Xanthomonadales (Zhou et al. 2008; Cutino-Jimenez et al. 2010; Yilmaz et al. 2013). Phylogenetic trees based on the 16S rRNA gene sequence often do not resolve a monophyletic cluster of all Xanthomonadales; the most divergent members of the order often branch separately from the majority of the species within the group (Yilmaz et al. 2013). Apart from the 16S rRNA sequence, no biochemical, morphological or physiological characteristics are known which distinguish the order Xanthomonadales from all other bacteria or the families and major phylogenetic clusters within the order from each other (Saddler and Bradbury 2005a; Gutierrez et al. 2012; Losey et al. 2013). Thus, it is of interest to identify shared characteristics that can clearly elucidate the evolutionary relationships within this highly diverse group of organisms and form the basis for a coherent taxonomic framework of the order.
Whole genome sequences for members of the order Xanthomonadales provide a rich resource for the discovery of molecular characteristics which are unique to evolutionarily related organisms (Gao et al. 2009; Cutino-Jimenez et al. 2010; Naushad and Gupta 2013). One useful type of shared molecular characteristic that has been a focus of recent research are Conserved Signature Indels (CSIs), which are insertions/deletions uniquely present in protein sequences from a group of evolutionarily related organisms (Gupta 2010; Gao and Gupta 2012b; Gupta and Lali 2013; Gupta et al. 2013; Gupta 2014). Due to the specificity of CSIs for particular groups of bacteria, they represent molecular synapomorphies (markers of common evolutionary decent) which can be used to identify and demarcate specific bacterial groups in clear molecular terms (Gupta 1998, 2010). We have previously carried out comparative genomic analysis of a limited number of members from the order Xanthomonadales in which we identified a large number of CSIs in diverse proteins that were uniquely present in all analyzed members of the order or a subgroup of the Xanthomonadales (Naushad and Gupta 2013). In this work, we have extended these studies, by carrying out detailed phylogenomic and comparative genomic analyses on a greatly expanded dataset on members of the order Xanthomonadales which includes 43 genomes from the NCBI, JGI, and EzBioCloud genome databases and 5 additional Xanthomonadales genomes, which we have sequenced de novo, representing 2 families, 20 genera, and 42 named species. Our analyses have identified no phylogenetic support for a monophyletic grouping of all sequenced members the order Xanthomonadales and no CSIs were identified which are uniquely shared by all sequenced species within the order Xanthomonadales suggesting that the order Xanthomonadales does not represent a single monophyletic lineage. Additionally, we have identified 31 CSIs which are either specific to the family Xanthomonadaceae or to one of its subgroups which demarcate these groups in molecular terms. On the basis of the identified CSIs and the results of phylogenomic analyses, we propose a new taxonomic framework for the order Xanthomonadales. In this proposal, the families Algiphilaceae and Solimonadaceae (Nevskiaceae), which do not branch with the other members of the order Xanthomonadales, are transferred into the order Nevskiales ord. nov, along with their closest evolutionary neighbour, the family Salinisphaeraceae fam. nov. The remaining members of the order Xanthomonadales are divided into two families: the family Xanthomonadaceae, containing the genus Xanthomonas and its closest relatives, and a new family, Rhodanobacteraceae fam. nov., containing the genus Rhodanobacter and its closest relatives. Lastly, the descriptions of the order Lysobacterales, the family Lysobacteraceae, and the family Nevskiaceae are emended to indicate that they are earlier synonyms of the order Xanthomonadales, the family Xanthomonadaceae, and the families Solimonadaceae and Sinobacteraceae, respectively.
Methods
DNA extraction and genome sequencing
Five Xanthomonadales isolates were sequenced de novo in this study; Dyella japonica DSM 16301T (Genbank accession number JPLA00000000), Luteibacter rhizovicinus DSM 16549T (JPLB00000000), Thermomonas brevis DSM 15422T (JPLC00000000), Xanthomonas hyacinthi DSM 19077T (JPLD00000000), and Xanthomonas pisi DSM 18956T (JPLE00000000). The isolates were obtained from the German Collection of Microorganisms and Cell Cultures (Leibniz-Institut DSMZ). The isolates were grown for 24 h under the growth conditions described in Supplemental Table 1. Genomic DNA was extracted using a CTAB based DNA extraction methodology (Wilson 1987) with specific modifications for Xanthomonadales described by Jaufeerally-Fakim and Dookun (2000). The DNA samples were diluted to 0.2 ng/µl and standard Illumina multiplex libraries were generated using the Nextera XT DNA Sample Prep Kit. The fragment size distribution of each library was verified using the Agilent High Sensitivity DNA Kit. Sequencing was performed using an Illumina HiSeq 2000 and 150 bp paired end reads were generated. Genomes were assembled using CLC Genomics Workbench 7.0.4 with default de novo assembly parameters and trimmed for contamination using the UniVec vector database (Build 8.0) (Table 1).
Phylogenetic sequence analysis
Phylogenetic analysis was performed on a concatenated sequence alignment of 15 highly conserved housekeeping proteins (viz. dimethyladenosine transferase, alanyl-tRNA synthetase, arginyl-tRNA synthetase, chaperone protein DnaK, signal recognition particle-docking protein FtsY, chaperonin GroL, DNA gyrase subunit A, DNA gyrase subunit B, ATP-dependent DNA helicase UvrD, valyl-tRNA synthetase, Isoleucyl-tRNA synthetase, DNA polymerase I, SecA, RpoB, and RpoC) which have been widely used for phylogenetic analysis (Kyrpides et al. 1999; Charlebois and Doolittle 2004; Ciccarelli et al. 2006). Sequences for these proteins were obtained from the NCBI and JGI-IMG genome databases for strains of all Xanthomonadales and a representative selection of outgroup Gammaproteobacteria (which included members from the orders Aeromonadales, Alteromonadales, Cardiobacteriales, Chromatiales, “Enterobacteriales”, Legionellales, Methylococcales, Oceanospirillales, Pasteurellales, Pseudomonadales, “Salinisphaerales”, Thiotrichales, and “Vibrionales”) and Betaproteobacteria. Sequences for these proteins were also obtained from the five Xanthomonadales genomes which we have sequenced in this work and the genome of Riemerella anatipestifer, which was used to root the tree. Multiple sequence alignments for these proteins were created using Clustal_X 1.83 (Jeanmougin et al. 1998) and concatenated into a single alignment file. Poorly aligned regions from this alignment file were removed using Gblocks 0.92 (Castresana 2000). The resulting alignment, which contained 6995 aligned amino acids, was used for phylogenetic analysis. The maximum-likelihood tree based on 100 bootstrap replicates of this alignment was constructed using MEGA 5.2 (Tamura et al. 2011) employing the Whelan and Goldman substitution model.
A 16S rRNA gene sequence based phylogenetic tree was also created based on 197 sequences that included representative strains of all cultured Xanthomonadales genera. 16S rRNA gene sequences larger than 1,200 bp were obtained for all strains used in our concatenated protein based phylogenetic tree and all type strains classified under the order Xanthomonadales in the Ribosomal Database Project (Cole et al. 2014). A maximum-likelihood tree based on these sequences was created using 100 bootstrap replicates of the 16S rRNA sequence alignments in MEGA 5.2 (Tamura et al. 2011) employing the General Time-Reversible (Tavaré 1986) substitution model.
Identification and assessment of specificity of conserved signature indels
Identification of CSIs that are commonly shared by members of the Xanthomonadaceae was carried out as described by Naushad and Gupta (2013). Briefly, for the identification of CSIs, BLASTp searches were performed on each protein in the genome of Rhodanobacter fulvus Jip2. These searches were performed using the default BLAST parameters against all available sequences in the GenBank non-redundant database. For those proteins for which high scoring homologs (E values < 1e−20) were present in other species from the Xanthomonadales, multiple sequence alignments were created using the Clustal_X 1.83 program (Jeanmougin et al. 1998). These alignments were visually inspected for the presence of insertions or deletions that were flanked on both sides by at least 5–6 conserved amino acid residues in the neighbouring 30–40 amino acids. Indels that were not flanked by conserved regions were not further considered, as they do not provide useful molecular markers. To assess the specificity of the indels we identified here and to reassess the specificity of the indels identified in our previous work, we carried out detailed BLASTp and tBLASTn searches against both the NCBI and JGI-IMG genome databases using as query short sequence segments containing the indel and the flanking conserved regions (60–100 amino acids long). Local tBLASTn searches were also completed on the indel containing regions for genomes of Xanthomonadales organisms missing from the NCBI and JGI-IMG genome databases. To ensure that the identified signatures are only present in Xanthomonadales homologues, the 250 BLAST hits with the highest similarity to the query sequence were examined for the presence or absence of these CSIs. Signature files were created and formatted using the programs Sig_Create and Sig_Style (accessible from Gleans.net) as described by Gupta (2014). In this work, we report the results of CSIs that are specific for different groups within the Xanthomonadales and where similar CSIs were not observed in any other bacteria in the top 250 BLAST hits. Due to space constraints, the sequence alignment files presented here contain sequence information for a limited number of species within the order Xanthomonadales and a representative selection of outgroup species. However, in each case, all members of the order and outgroups exhibited similar sequence characteristics to the representatives.
Results
Phylogenetic analysis
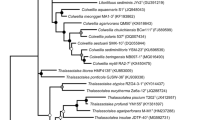
The current understanding of the evolutionary relationships of the Xanthomonadales is based largely on analyses of the 16S rRNA gene (Saddler and Bradbury 2005a; Gutierrez et al. 2012; Losey et al. 2013). In past studies, the 16S rRNA gene sequence has shown limited ability to resolve some of the phylogenetic relationships of organisms within the order Xanthomonadales (Zhou et al. 2008; Yilmaz et al. 2013). Phylogenetic trees based on multiple conserved genes/proteins have been shown to provide greater resolving power than those based on any single gene or protein (Rokas et al. 2003; Wu et al. 2009). Thus, we have constructed a highly resolved phylogenetic tree of the Xanthomonadales based on a concatenated set of 15 housekeeping and ribosomal proteins (Fig. 1). In this concatenated protein based phylogenetic tree a majority of the members of the Xanthomonadales formed a well-supported monophyletic clade which branched as an outgroup of the other members of the Gammaproteobacteria. The members of the order Xanthomonadales formed two distinct and well-supported main monophyletic clades: one clade consisting of members of the family Xanthomonadaceae and another clade consisting of the family Solimonadaceae (including the genera Nevskia and Hydrocarboniphaga) and the species Salinisphaera shabanensis, a member of the family “Salinisphaeraceae”. The Xanthomonadaceae clade contained two smaller clades that were well-supported by bootstrap analysis. The first of these clades, contained the genera Xanthomonas, Xylella, Stenotrophomonas, Lutimonas, Lysobacter and their relatives (Clade 1) while the second clade contained the genera Rudaea, Dylella, Luteibacter, Rhodanobacter and Frateuria (Clade 2). Two members of the Xanthomonadales, Wohlfahrtiimonas chitiniclastica and Ignatzschineria larvae, branched separately from the rest of the order, in a well-supported clade with members of the order Cardiobacteriales, another early diverging group within the class Gammaproteobacteria.
A maximum-likelihood phylogenetic tree of the order Xanthomonadales, other Gammaproteobacteria, and Betaproteobacteria based on the concatenated amino acid sequences of 25 conserved proteins. Bootstrap values are shown at branch nodes. The major groups within the order Xanthomonadales as well as the related taxa, Salinisphaeraceae and Cardiobacteriales, are indicated
We have also produced a phylogenetic tree based on the 16S rRNA gene which contains representative species of Xanthomonadales that encompass all of the currently named genera (Fig. 2). The 16S rRNA based phylogenetic tree exhibited broadly similar branching to our concatenated protein based phylogenetic tree. In the 16S rRNA gene tree, the families Xanthomonadaceae and Solimonadaceae did not form a monophyletic clade and were separated by a large number of organisms. The family Xanthomonadaceae was divided into two well-supported clades which were analogous to the clades found in our concatenated protein based phylogenetic tree (Clades 1 and 2). In the 16S rRNA gene tree, the family Solimonadaceae (including the genera Nevskia, Hydrocarboniphaga, and Alkanibacter) branched with the family Algiphilaceae, another disparate group within the Xanthomonadales, and the genus Steroidobacter, which is currently recognized as a member of the Xanthomonadaceae. As in our concatenated protein based phylogenetic tree, Solimonadaceae and the other disparate members of the Xanthomonadales showed an association with the members of the genus Salinisphaera, the sole members of the family “Salinisphaeraceae”. Additionally, in the 16S rRNA based phylogenetic tree, the genera Wohlfahrtiimonas and Ignatzschineria, which branched with the order Cardiobacteriales in our concatenated protein based phylogenetic tree, formed a weakly supported monophyletic group with the other members of the family Xanthomonadaceae. However, Wohlfahrtiimonas and Ignatzschineria were well separated from the other Xanthomonadaceae by a long branch.
Conserved signature indels
CSIs that are restricted to a group of related species are a novel class of molecular marker with high utility for evolutionary studies (Gupta 1998; Rokas and Holland 2000; Gupta 2010; Gao and Gupta 2012a; Gupta 2014). Recently, CSIs have been used to define novel taxonomic groups and to propose important taxonomic changes for various groups of bacteria (viz. Spirochaetes, Aquificae, Neisseriales, and Bacillus) at different taxonomic ranks (Adeolu and Gupta 2013; Bhandari et al. 2013; Gupta and Lali 2013; Gupta et al. 2013). We have recently reported a comparative genomic analysis on a limited number of members of the order Xanthomonadales in which we identified a large number of CSIs in diverse proteins that were uniquely present in all available members of the order or different phylogenetic groups within the order and absent in homologs from all other bacterial groups (Naushad and Gupta 2013). However, the genomes analyzed in our previous study were all from members of one family within the Xanthomonadales, the family Xanthomonadaceae, and did not include any of the more divergent species within the order whose phylogenetic placement is less clear. In this work, we have reassessed the specificity of these previously identified CSIs for a large number of additional Xanthomonadales, including five strains which we have sequenced, de novo, covering a vast majority of the diversity within the order and thereby have identified 31 CSIs which are either specific to the family Xanthomonadaceae or to one of its subgroups and absent in all other sequenced bacterial groups.
Of the 31 CSIs described in this work, none were present in all members of the order Xanthomonadales. All of the CSIs identified in our previous study of the Xanthomonadales (Naushad and Gupta 2013) were found to be specific to only the family Xanthomonadaceae or one of its subgroups. Of the 31 CSIs identified, 10 were uniquely found in all or most members of the Xanthomonadaceae, except Wohlfahrtiimonas and Ignatzschineria, and absent in organisms from all other sequenced bacterial groups. One example of a CSI uniquely present in members of the Xanthomonadace is shown in Fig. 3. In the example, an 18 aa insertion in a conserved region of glutaminyl t-RNA synthetase is uniquely present in all members of the Xanthomonadace, except Wohlfahrtiimonas and Ignatzschineria, but it is not found in sequences from any other bacterial group. Sequence information for the 9 other CSIs specific for all members of the Xanthomonadace, except Wohlfahrtiimonas and Ignatzschineria are presented in Supplemental Figs. 1–9 and a summary of all 10 Xanthomonadace specific CSIs is presented in Table 2A. Our analyses have also identified 10 CSIs which were found to be unique molecular characteristics of most members of Clade 1 of the Xanthomonadaceae. Two examples of such CSIs are presented in Fig. 4. One CSI, a 4 aa insert in DNA polymerase III subunit alpha, is present in all members of Clade 1 of the Xanthomonadaceae except Silanimonas lenta (Fig. 4a), while the other CSI, a 4 aa insert in the protein protoheme IX farnesyltransferase, is uniquely present in all members of Clade 1 of the Xanthomonadaceae except the early branching genera Arenimonas and Silanimonas (Fig. 4b). Sequence information for the other identified CSIs specific to the members of Clade 1 of the Xanthomonadaceae are presented in Supplemental Figs. 10–17 and summarized in Table 2B.
A partial sequence alignment of the protein Glutaminyl t-RNA synthetase, showing a CSI (boxed) that is uniquely present in all members of the order Xanthomonadales. Sequence information for only representative Xanthomonadales and a limited number other bacteria is shown here. However, unless otherwise indicated, similar CSIs were present in all members of the indicated group and not detected in any other bacterial species in the top 250 BLAST hits. The dashes (-) in the alignments indicate identity with the residue in the top sequence. GenBank identification (GI) numbers for each sequence are indicated in the second column. Sequence information for 10 other CSIs that are specific for all sequenced Xanthomonadales is provided in Supplemental Figs. 1–9 and Table 2A
Partial sequence alignments of a DNA polymerase III subunit alpha showing a 4 amino acid insertion (boxed) identified in all members of Clade 1 of the Xanthomonadaceae except Silanimonas lenta b the protein Protoheme IX farnesyltransferase showing a 4 amino acid insertion (boxed) identified in all members of Clade 1 of the Xanthomonadaceae except the genera Arenimonas and Silanimonas. Due to space constraints, sequence information for only representative Xanthomonadales and a limited number other bacteria is shown here, but similar CSIs were present in all members of the indicated group and not detected in any other bacterial species in the top 250 BLAST hits. Sequence information for other CSIs showing similar group specificities are presented in Supplemental Figs. 10–17 and summarized in Tables 2B
Our analyses have also identified 11 CSIs that were specifically found in diverse proteins from members of Clade 2 of the Xanthomonadaceae, 7 of which were uniquely found in all members of Clade 2 of the Xanthomonadaceae except the early branching genus Rudaea. An example of a CSI specifically found in all members of Clade 2 of the Xanthomonadaceae is shown in Fig. 5a. In this CSI a 1 aa insert in the protein uridylyltransferase is shown to be found in all members of Clade 2 of the Xanthomonadaceae and absent in all other Xanthomonadales and all other bacterial groups. Another CSI, a 4 aa insert in the protein CDP-diacylglycerol–glycerol-3-phosphate 3-phosphatidyltransferase, specifically found in all members of Clade 2 of the Xanthomonadaceae except Rudaea cellulosilytica is shown in Fig. 5b. Sequence information for the other identified CSIs specific to the members of Clade 2 of the Xanthomonadaceae are presented in Supplemental Figs. 18–26 and summarized in Table 2C and D. Our analyses have not identified any CSIs uniquely found in all of the disparate members of the Xanthomonadales or uniquely shared by the genera Wohlfahrtiimonas and Ignatzschineria and the rest of the Xanthomonadaceae.
Partial sequence alignments of a the protein Uridylyltransferase showing a 1 amino acid insertion (boxed) identified in all members of Clade 2 of the Xanthomonadaceae b the protein CDP-diacylglycerol–glycerol-3-phosphate 3-phosphatidyltransferase showing a 4 amino acid insertion (boxed) identified in all members of Clade 2 of the Xanthomonadaceae except the early branching genus Rudaea. Sequence information for only representative Xanthomonadales and a limited number other bacteria is shown here, but similar CSIs were not detected in any other bacterial species in the top 250 BLAST hits. Sequence information for the other CSIs specific to the Clade 2 Xanthomonadaceae are presented in Supplemental Figs. 18–26 and summarized in Table 2C and D
Discussion
The current phylogeny of the order Xanthomonadales is based largely on the analysis of 16S rRNA gene sequences (Saddler and Bradbury 2005a; Gutierrez et al. 2012; Losey et al. 2013). However, the 16S rRNA gene based phylogenies exhibit limited support for a single monophyletic clade consisting of all members of the Xanthomonadales (Yilmaz et al. 2013; Fig. 2). The current taxonomy of the order Xanthomonadales is not concordant with 16S rRNA gene based phylogenies of the members of the order and the nomenclature of the order Xanthomonadales and a majority of the family names within this order are problematic and not in accordance with the International Code of Nomenclature of Bacteria (Oren 2010; Yilmaz et al. 2013; Tindall 2014a, b). However, apart from the 16S rRNA gene, no reliable morphological, biochemical, or molecular characteristics are known that are specifically shared by all members of this order or its distinct subgroups and can be used for their demarcation and classification (Saddler and Bradbury 2005a; Gutierrez et al. 2012; Losey et al. 2013). In this work, we have completed a robust phylogenetic analysis of the order Xanthomonadales and have utilized comparative genomic techniques to identify large numbers of novel molecular markers of common evolutionary decent (CSIs) shared by subgroups within the Xanthomonadales. The CSIs identified in this work both supplement gene based phylogenies and demarcate the groups within the Xanthomonadales in more definitive molecular terms. A summary diagram of the identified CSIs and the species in which they are found is shown in Fig. 6.
A summary of the evolutionary relationships of the Xanthomonadales genera based upon phylogenetic analyses and the identified CSIs. Genera with genome sequenced members are indicated with asterisks (*). The distribution of the identified CSIs and the proposed reclassification of taxonomic groups are indicated. The genera Wohlfahrtiimonas and Ignatzschineria do not branch with the members of the order Xanthomonadales and hence are regarded as order incertae sedis. The families Sinobacteraceae and Solimonadaceae are synonymous; however, only the name of the family Solimonadaceae is shown here (1). The placement of the genus Steroidobacter within the family Solimonadaceae (Nevskiaceae) is tentative until a more detailed phylogenetic analysis can be completed for this genus (2)
The results of phylogenetic analyses presented here do not support a monophyletic grouping of the members of the order Xanthomonadales and no CSI was identified that is uniquely shared by all members of this order. In phylogenetic trees, members of the order Xanthomonadales formed two main clades, one grouping together most of the members from the family Xanthomonadaceae, whereas the other clade was comprised of members from the families Algiphilaceae (containing the genus Algiphilus) and Solimonadaceae (containing the genera Fontimonas, Singularimonas and Solimonas) and genera related to these two families (viz. Alkanibacter, Hydrocarboniphaga, Nevskia and Steroidobacter). The lack of any identified CSIs or a consistent phylogenetic relationship between these two clades suggests that they may represent distinct evolutionary lineages within the Gammaproteobacteria. Additionally, in both our concatenated protein tree and the 16S rRNA gene tree, members of the clade containing Algiphilaceae and Solimonadaceae families consistently grouped with the members of the genus Salinisphaera, the sole members of the family “Salinisphaeraceae”, suggesting that the species from these groups may share a common ancestor exclusive of the Xanthomonadaceae and other Gammaproteobacteria. The genera Wohlfahrtiimonas and Ignatzschineria branch distinctly from the Xanthomonadales in a clade with members of the order Cardiobacteriales in our concatenated protein based phylogenetic tree and show limited phylogenetic association with the other members of the Xanthomonadales in our 16S rRNA tree. Due to this inconsistent branching, further research will be required to accurately assess the phylogenetic placement of the genera Wohlfahrtiimonas and Ignatzschineria, but the available data suggests that they do not belong to the order Xanthomonadales sensu stricto.
Our work has identified 10 CSIs that support a monophyletic grouping of a majority of the members of the order Xanthomonadales that are currently part of the family Xanthomonadaceae. These CSIs were initially identified in our earlier comparative genomic study (Naushad and Gupta 2013) and the sequence information for them was updated in the present work for a large number of additional Xanthomonadales, including 5 genomes which were sequenced, de novo, for this study. Our earlier work identified 13 CSIs which were specific to this group (Naushad and Gupta 2013). Of these, all but 3 CSIs were found to be still specific to the whole group, while the remaining three CSIs were found to be specific for subsets of this large group. The observed specificity of the previously identified CSIs for a distinct bacterial group despite a large increase in the number of analyzed genomes strongly indicate that they constitute reliable molecular characteristics with predictive ability for distinguishing and demarcation of evolutionarily related bacterial groups.
Our work also provides strong molecular and phylogenetic support for the existence of two distinct clades within the Xanthomonadaceae: One clade consists of the genera Xanthomonas, Xylella, Stenotrophomonas, Lutimonas, Lysobacter and their relatives (Clade 1), whereas the other clade groups together members of the genera Rudaea, Dylella, Lutibacter, Rhodanobacter and their relatives (Clade 2). The members of these monophyletic clades branch distinctly from each other with strong bootstrap support in both the concatenated protein tree as well as in the 16S rRNA gene trees. Importantly, Clade 1 and Clade 2 are also supported by 10 and 11 identified CSIs, respectively, which serve to clearly distinguish them from each other and every other bacterial group in molecular terms. This evidence suggests that the Clade 1 and Clade 2 represent two phylogenetically and molecularly distinguishable evolutionary lineages.
Taxonomic implications
Based on the branching of the members of the order Xanthomonadales in the concatenated protein and 16S rRNA gene trees and the large number of identified molecular markers (CSIs) that are specific for this group of bacteria, the following main inferences regarding the phylogeny of the Xanthomonadales can be derived.
-
(1)
The order presently designated as Xanthomonadales contains 2 highly divergent phylogenetic groups, one made up of the members of the family Xanthomonadaceae and the other made up of the members of the families Algiphilaceae, Solimonadaceae, and “Salinisphaeraceae”
-
(2)
The family presently designated as Xanthomonadaceae, which harbours a majority of the members from the order Xanthomonadales, also contains 2 distinct and distinguishable phylogenetic groups, one consisting of the genera Xanthomonas, Xylella, Stenotrophomonas, Lutimonas, Lysobacter and their relatives (Clade 1) and another clade consisting of the genera Rudaea, Dylella, Lutibacter, Rhodanobacter and their relatives (Clade 2)
Thus, the current taxonomy of the order Xanthomonadales does not accurately reflect the evolutionary histories of its members which exhibit enormous genetic diversity. In order to alleviate the taxonomic incongruences within the order Xanthomonadales, we propose that the families Algiphilaceae (containing the genus Algiphilus) and Solimonadaceae (or Nevskiaceae) (containing the genera Fontimonas, Singularimonas and Solimonas) and genera related to these two families (viz. Alkanibacter, Hydrocarboniphaga, Nevskia and Steroidobacter), which do not branch with the other members of the order Xanthomonadales, be placed within a novel order, Nevskiales ord. nov., along with their closest evolutionary relatives, the members of the family Salinisphaeraceae fam. nov. Further, to recognize the presence of two distinct groups within the family presently designated as Xanthomonadaceae, the members of this family should be divided into two families: the family Xanthomonadaceae (containing the genera Arenimonas, Luteimonas, Lysobacter, Metallibacterium, Panacagrimonas, Pseudoxanthomonas, Silanimonas, Stenotrophomonas, Thermomonas, Xanthomonas, and Xylella) and a novel family, Rhodanobacteraceae fam. nov. (containing the genera Aquimonas, Chiayiivirga, Dokdonella, Dyella, Frateuria, Fulvimonas, Luteibacter, Pseudofulvimonas, Rhodanobacter, and Rudaea). The remaining two genera, Wohlfahrtiimonas and Ignatzschineria, whose taxonomic affiliation to the above two orders is not supported should be regarded as order incertae sedis.
Additionally, the present proposal also serves to help rectify several problems associated with the nomenclature of the order Xanthomonadales, the family Xanthomonadaceae, and the family Solimonadaceae (Tindall 2014b). It has been noted previously (Oren 2010; Tindall 2014a, b) that the names of these taxa are later synonyms for the order Lysobacterales, the family Lysobacteraceae, and the family Nevskiaceae, respectively. In recognition of these nomenclatural concerns, we are providing emended descriptions of the order Lysobacterales, the family Lysobacteraceae, and the family Nevskiaceae, which indicate that they are earlier synonyms of the order Xanthomonadales, the family Xanthomonadaceae, and the family Solimonadaceae, respectively. Descriptions of Rhodanobacteraceae fam. nov., Nevskiales ord. nov., and Salinisphaeraceae fam. nov. and emended descriptions of the order Lysobacterales (Xanthomonadales), the family Lysobacteraceae (Xanthomonadaceae), and the family Nevskiaceae (Solimonadaceae) are provided below.
Emended description of the order Lysobacterales Christensen and Cook (1978) (Approved Lists 1980)
Synonym: Xanthomonadales Saddler and Bradbury (2005a, b).
The order contains two families, Lysobacteraceae and Rhodanobacteraceae. Organisms are rods, 0.2–1.8 µm in diameter and 0.8–70 µm in length. Cells are both motile and non-motile. Organisms are aerobic, or facultatively anaerobic. Organisms are chemoorganotrophic and non-spore-forming. Organisms within this order may be either positive or negative in both oxidase and catalase tests. The G + C content of the DNA is 42–75 (mol%).The type genus of the order is Lysobacter Christensen and Cook (1978) (Approved Lists 1980) (Skerman et al. 1980) emend. Park et al. (2008).
Organisms from this order are distinguished from all other bacteria examined to date by 10 conserved signature indels in Glutaminyl t-RNA synthetase, GTP-binding protein, Queuine tRNA-ribosyltransferase, Lipoyl synthase, Lysyl-tRNA synthetase, Dihydroorotate dehydrogenase, Carbamoyl phosphate synthase large subunit, Aspartate aminotransferase, DNA polymerase I, and DNA topoisomerase IV subunit B (Tables 2A).
Emended description of the family Lysobacteraceae Christensen and Cook (1978) (Approved Lists 1980)
Synonym: Xanthomonadaceae Saddler and Bradbury (2005a, b).
The family contains twelve genera, Arenimonas, Luteimonas, Lysobacter, Metallibacterium, Panacagrimonas, Pseudoxanthomonas, Silanimonas, Stenotrophomonas, Thermomonas, Xanthomonas and Xylella. Organisms are rods, 0.2–1.8 µm in diameter and 0.8–70 µm in length. Cells are both motile and non-motile. Organisms are aerobic, or facultatively anaerobic. Organisms are chemoorganotrophic and non-spore-forming. Organisms within this family may be either positive or negative in both oxidase and catalase tests. The G+C content of the DNA is 42–70 (mol%). The type genus of the family is Lysobacter Christensen and Cook (1978) (Approved Lists 1980) emend. Park et al. (2008).
Organisms from this order are distinguished from all other bacteria examined to date by 10 conserved signature indels in DNA polymerase III subunit alpha, Uroporphyrinogen decarboxylase, DNA polymerase I, Coproporphyrinogen III oxidase, tRNA isopentenyltransferase, Protoheme IX farnesyltransferase, Ribose-5-phosphate isomerase A, Aspartyl-tRNA synthetase, 2-oxoglutarate dehydrogenase E1, and Asparagine synthetase B (Tables 2B).
Description of Rhodanobacteraceae fam. nov
Rhodanobacteraceae (Rho.da.no.bac.ter.a,ce’ae N.L. masc. n. Rhodanobacter type genus of the family; -aceae ending to denote a family; N.L. fem. pl. n. Rhodanobacteraceae the family whose nomenclatural type is the genus Rhodanobacter).
The family contains nine genera, Aquimonas, Dokdonella, Dyella, Frateuria, Fulvimonas, Luteibacter, Pseudofulvimonas, Rhodanobacter and Rudaea. Organisms are rods, 0.3–0.5 µm in diameter and 1–4.5 µm in length. Cells are both motile and non-motile. Organisms are aerobic, chemoorganotrophic, and non-spore-forming. Organisms within this family may be either positive or negative in both oxidase and catalase tests. The G + C content of the DNA is 62–75 (mol%). The type genus of the family is Rhodanobacter Nalin et al. (1999).
Organisms from this order are distinguished from all other bacteria examined to date by 11 conserved signature indels in Uridylyltransferase, a xanthomonadin exporter protein, a signal peptidase, Tryptophan synthase subunit alpha, CDP-diacylglycerol–glycerol-3-phosphate 3-phosphatidyltransferase, Protease tldD, S-adenosylmethionine decarboxylase, DEAD/DEAH box helicase, F0F1 ATP synthase subunit gamma, Proline aminopeptidase P II, and Glycosyl transferase (Table 2C, D).
Description of Nevskiales ord. nov
Nevskiales (Nev.ski.a’les. N.L. fem. n. Nevskia type genus of the order; -ales ending to denote an order; N.L. fem. pl. n. Nevskiales the order whose nomenclatural type is the genus Nevskia).
The order contains three families, Algiphilaceae, Salinisphaeraceae, and Nevskiaceae. Organisms are rods and cocci, 0.6–1.3 µm in diameter and 0.4–2 µm in length. Cells are non-motile or motile by means of a one or more polar flagella. Organisms are aerobic, or facultatively anaerobic. Organisms are chemoorganotrophic and non-spore-forming. Oxidase and catalase positive. The G+C content of the DNA is 60–68 (mol%). The type genus of the order is Nevskia Famintzin 1892 (Approved Lists 1980).
Emended Description of the family Nevskiaceae Henrici and Johnson 1935 (Approved Lists 1980)
Synonyms: Sinobacteraceae Zhou et al. (2008), Solimonadaceae Losey et al. (2013).
The family contains six genera, Alkanibacter, Fontimonas, Hydrocarboniphaga, Nevskia, Solimonas and Steroidobacter.Footnote 1 Organisms are rods, 0.6–0.85 µm in diameter and 0.9–2 µm in length. Cells are non-motile or motile by means of a single polar flagellum. Organisms are aerobic, or facultatively anaerobic. Organisms are chemoorganotrophic and non-spore-forming. Oxidase and catalase positive. The G + C content of the DNA is 60–65 (mol%). The type genus of the family is Nevskia Famintzin 1892 (Approved Lists 1980).
Description of Salinisphaeraceae fam. nov
Salinisphaeraceae (Sa.li.ni.sphae.ra.ce’ae. N.L. fem. n. Salinisphaera type genus of the family; -aceae ending to denote a family; N.L. fem. pl. n. Salinisphaeraceae the family whose nomenclatural type is the genus Salinisphaera).
The family contains one genus, Salinisphaera, which is also the type genus of the family. The description of the family is the same as that of the genus Salinisphaera Antunes et al. (2003) emend. Shimane et al. (2013).
Notes
The genus Steroidobacter does not branch monophyletically with the other members of the family Nevskiaceae in 16S rRNA gene based phylogenies. However, Steroidobacter is clearly distinct from the order Xanthomonadales and family Xanthomonadaceae in which it was previously placed. Its placement within the family Nevskiaceae is tentative until more detailed phylogenetic analysis can be completed for this genus.
References
Adeolu M, Gupta RS (2013) Phylogenomics and molecular signatures for the order Neisseriales: proposal for division of the order Neisseriales into the emended family Neisseriaceae and Chromobacteriaceae fam. nov. Anton Leeuw Int J G 104(1):1–24
Antunes A, Eder W, Fareleira P, Santos H, Huber R (2003) Salinisphaera shabanensis gen. nov., sp. nov., a novel, moderately halophilic bacterium from the brine–seawater interface of the Shaban Deep, Red Sea. Extremophiles 7(1):29–34
Bhandari V, Ahmod NZ, Shah HN, Gupta RS (2013) Molecular signatures for Bacillus species: demarcation of the Bacillus subtilis and Bacillus cereus clades in molecular terms and proposal to limit the placement of new species into the genus Bacillus. Int J Syst Evol Microbiol 63(7):2712–2726
Castresana J (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17(4):540–552
Charlebois RL, Doolittle WF (2004) Computing prokaryotic gene ubiquity: Rescuing the core from extinction. Genome Res 14(12):2469–2477
Chatterjee S, Almeida RPP, Lindow S (2008) Living in two worlds: the plant and insect lifestyles of Xylella fastidiosa. Annu Rev Phytopathol 46:243–271
Chen J, Xie G, Han S, Chertkov O, Sims D, Civerolo EL (2010) Whole genome sequences of two Xylella fastidiosa strains (M12 and M23) causing almond leaf scorch disease in California. J Bacteriol 192(17):4534
Christensen P, Cook FD (1978) Lysobacter, a new genus of nonfruiting, gliding bacteria with a high base ratio. Int J Syst Bacteriol 28(3):367–393
Ciccarelli FD, Doerks T, Von Mering C, Creevey CJ, Snel B, Bork P (2006) Toward automatic reconstruction of a highly resolved tree of life. Science 311(5765):1283–1287
Cole J, Wang Q, Fish J, Chai B, McGarrell D, Sun Y, Brown C, Porras-Alfaro A, Kuske C, Tiedje J (2014) Ribosomal database project: data and tools for high throughput rRNA analysis. Nucleic Acids Res 42(1):D633
Crossman LC, Gould VC, Dow JM, Vernikos GS, Okazaki A, Sebaihia M, Saunders D, Arrowsmith C, Carver T, Peters N (2008) The complete genome, comparative and functional analysis of Stenotrophomonas maltophilia reveals an organism heavily shielded by drug resistance determinants. Genome Biol 9(4):R74
Cutino-Jimenez AM, Martins-Pinheiro M, Lima WC, Martin-Tornet A, Morales OG, Menck CFM (2010) Evolutionary placement of Xanthomonadales based on conserved protein signature sequences. Mol Phylogen Evol 54(2):524–534
da Silva AR, Ferro JA, Reinach F, Farah C, Furlan L, Quaggio R, Monteiro-Vitorello C, Van Sluys M, Almeida N, Alves L (2002) Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 417(6887):459–463
Darrasse A, Carrere S, Barbe V, Boureau T, Arrieta-Ortiz ML, Bonneau S, Briand M, Brin C, Cociancich S, Durand K et al (2013) Genome sequence of Xanthomonas fuscans subsp. fuscans strain 4834-R reveals that flagellar motility is not a general feature of xanthomonads. BMC Genomics 14:761
De Clercq D, Van Trappen S, Cleenwerck I, Ceustermans A, Swings J, Coosemans J, Ryckeboer J (2006) Rhodanobacter spathiphylli sp. nov., a gammaproteobacterium isolated from the roots of Spathiphyllum plants grown in a compost-amended potting mix. Int J Syst Evol Microbiol 56(Pt 8):1755–1759
Gao B, Gupta RS (2012a) Microbial systematics in the post-genomics era. Anton Leeuw Int J G 101(1):45–54
Gao B, Gupta RS (2012b) Phylogenetic framework and molecular signatures for the main clades of the phylum Actinobacteria. Microbiol Mol Biol Rev 76(1):66–112
Gao B, Mohan R, Gupta RS (2009) Phylogenomics and protein signatures elucidating the evolutionary relationships among the Gammaproteobacteria. Int J Syst Evol Microbiol 59(2):234–247
Gupta RS (1998) Protein phylogenies and signature sequences: a reappraisal of evolutionary relationships among archaebacteria, eubacteria, and eukaryotes. Microbiol Mol Biol Rev 62(4):1435
Gupta RS (2010) Applications of conserved indels for understanding microbial phylogeny. In: Oren A, Papke RT (eds) Molecular phylogeny of microorganisms. Caister Academic Press, Norfolk, pp 135–150
Gupta RS (2014) Identification of conserved indels that are useful for classification and evolutionary studies Methods in Microbiology, vol 41. Academic Press: 10.1016/bs.mim.2014.05.003
Gupta RS, Lali R (2013) Molecular signatures for the phylum Aquificae and its different clades: proposal for division of the phylum Aquificae into the emended order Aquificales, containing the families Aquificaceae and Hydrogenothermaceae, and a new order Desulfurobacteriales ord. nov., containing the family Desulfurobacteriaceae. Anton Leeuw Int J G 104(3):349–368
Gupta RS, Mahmood S, Adeolu M (2013) A phylogenomic and molecular signature based approach for characterization of the phylum Spirochaetes and its major clades: proposal for a taxonomic revision of the phylum. Frontiers in microbiology 4:217
Gutierrez T, Green DH, Whitman WB, Nichols PD, Semple KT, Aitken MD (2012) Algiphilus aromaticivorans gen. nov., sp. nov., an aromatic hydrocarbon-degrading bacterium isolated from a culture of the marine dinoflagellate Lingulodinium polyedrum, and proposal of Algiphilaceae fam. nov. Int J Syst Evol Microbiol 62(11):2743–2749
Henrici AT, Johnson DE (1935) Studies of Freshwater Bacteria: II. Stalked Bacteria, a New Order of Schizomycetes. J Bacteriol 30(1):61–93
Im WT, Lee ST, Yokota A (2004) Rhodanobacter fulvus sp. nov., a beta-galactosidase-producing gammaproteobacterium. J Gen Appl Microbiol 50(3):143–147
Jalan N, Kumar D, Yu F, Jones JB, Graham JH, Wang N (2013) Complete genome sequence of Xanthomonas citri subsp. citri Strain Aw12879, a restricted-host-range citrus canker-causing bacterium. Genome Announc 1(3):e00235-13
Jaufeerally-Fakim Y, Dookun A (2000) Extraction of high quality DNA from polysaccharides-secreting xanthomonads. Sci Technol Res J Univ Maurit 6:33–40
Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ (1998) Multiple sequence alignment with clustal X. Trends Biochem Sci 23(10):403
Kostka JE, Green SJ, Rishishwar L, Prakash O, Katz LS, Marino-Ramirez L, Jordan IK, Munk C, Ivanova N, Mikhailova N et al (2012) Genome sequences for six Rhodanobacter strains, isolated from soils and the terrestrial subsurface, with variable denitrification capabilities. J Bacteriol 194(16):4461–4462
Kyrpides N, Overbeek R, Ouzounis C (1999) Universal protein families and the functional content of the last universal common ancestor. J Mol Evol 49(4):413–423
Lee B-M, Park Y-J, Park D-S, Kang H-W, Kim J-G, Song E-S, Park I-C, Yoon U-H, Hahn J-H, Koo B-S (2005) The genome sequence of Xanthomonas oryzae pathovar oryzae KACC10331, the bacterial blight pathogen of rice. Nucleic Acids Res 33(2):577–586
Lee CS, Kim KK, Aslam Z, Lee ST (2007) Rhodanobacter thiooxydans sp. nov., isolated from a biofilm on sulfur particles used in an autotrophic denitrification process. Int J Syst Evol Microbiol 57(Pt 8):1775–1779
Lee SH, Jin HM, Lee HJ, Kim JM, Jeon CO (2012) Complete genome sequence of the BTEX-degrading bacterium Pseudoxanthomonas spadix BD-a59. J Bacteriol 194(2):544
Looney WJ, Narita M, Mühlemann K (2009) Stenotrophomonas maltophilia: an emerging opportunist human pathogen. Lancet Infect Dis 9(5):312–323
Losey NA, Stevenson BS, Verbarg S, Rudd S, Moore ER, Lawson PA (2013) Fontimonas thermophila gen. nov., sp. nov., a moderately thermophilic bacterium isolated from a freshwater hot spring, and proposal of Solimonadaceae fam. nov. to replace Sinobacteraceae Zhou et al. 2008. Int J Syst Evol Microbiol 63(1):254–259
Meidanis J, Braga MD, Verjovski-Almeida S (2002) Whole-genome analysis of transporters in the plant pathogen Xylella fastidiosa. Microbiol Mol Biol Rev 66(2):272–299
Nalin R, Simonet P, Vogel TM, Normand P (1999) Rhodanobacter lindaniclasticus gen. nov., sp. nov., a lindane-degrading bacterium. Int J Syst Bacteriol 49(1):19–23
Naushad HS, Gupta RS (2013) Phylogenomics and molecular signatures for species from the plant pathogen-containing order Xanthomonadales. PLoS ONE 8(2):e55216
Oren A (2010) The phyla of prokaryotes—cultured and uncultured. In: Oren A, Papke RT (eds) Molecular phylogeny of microorganisms. Caister Academic Press, Norfolk, pp 85–107
Park JH, Kim R, Aslam Z, Jeon CO, Chung YR (2008) Lysobacter capsici sp. nov., with antimicrobial activity, isolated from the rhizosphere of pepper, and emended description of the genus Lysobacter. Int J Syst Evol Microbiol 58(2):387–392
Parte AC (2013) LPSN–list of prokaryotic names with standing in nomenclature. Nucleic Acids Res 42:D613–D616
Pieretti I, Royer M, Barbe V, Carrere S, Koebnik R, Cociancich S, Couloux A, Darrasse A, Gouzy J, Jacques MA et al (2009) The complete genome sequence of Xanthomonas albilineans provides new insights into the reductive genome evolution of the xylem-limited Xanthomonadaceae. BMC Genomics 10:616
Qian W, Jia Y, Ren SX, He YQ, Feng JX, Lu LF, Sun Q, Ying G, Tang DJ, Tang H et al (2005) Comparative and functional genomic analyses of the pathogenicity of phytopathogen Xanthomonas campestris pv. campestris. Genome Res 15(6):757–767
Rokas A, Holland PWH (2000) Rare genomic changes as a tool for phylogenetics. Trends Ecol Evol 15(11):454–459
Rokas A, Williams BL, King N, Carroll SB (2003) Genome-scale approaches to resolving incongruence in molecular phylogenies. Nature 425(6960):798–804
Ryan RP, Vorhölter F-J, Potnis N, Jones JB, Van Sluys M-A, Bogdanove AJ, Dow JM (2011) Pathogenomics of Xanthomonas: understanding bacterium–plant interactions. Nat Rev Microbiol 9(5):344–355
Saddler GS, Bradbury JF (2005a) Order III. Xanthomonadales ord. nov. In: Brenner DJ, Krieg NR, Staley JT, Garrity GM, Boone, Vos P, Goodfellow M, Rainey FA, Schleifer K-H (eds) Bergey’s manual of systematic bacteriology. Springer, Austin, pp 63–122
Saddler GS, Bradbury JF (2005b) Xanthomonadaceae fam. nov Validation of publication of new names and new combinations previously effectively published outside the IJSEM, List no 106. Int J Syst Evol Microbiol 55:2235–2238
Salzberg SL, Sommer DD, Schatz MC, Phillippy AM, Rabinowicz PD, Tsuge S, Furutani A, Ochiai H, Delcher AL, Kelley D (2008) Genome sequence and rapid evolution of the rice pathogen Xanthomonas oryzae pv. oryzae PXO99A. BMC Genomics 9(1):204
Shimane Y, Tsuruwaka Y, Miyazaki M, Mori K, Minegishi H, Echigo A, Ohta Y, Maruyama T, Grant WD, Hatada Y (2013) Salinisphaera japonica sp. nov., a moderately halophilic bacterium isolated from the surface of a deep-sea fish, Malacocottus gibber, and emended description of the genus Salinisphaera. Int J Syst Evol Microbiol 63(6):2180–2185
Skerman VBD, McGowan V, Sneath PHA (1980) Approved lists of bacterial names. Int J Syst Bacteriol 30(1):225–420
Studholme DJ, Wasukira A, Paszkiewicz K, Aritua V, Thwaites R, Smith J, Grant M (2011) Draft genome sequences of Xanthomonas sacchari and two banana-associated xanthomonads reveal insights into the Xanthomonas group 1 clade. Genes 2(4):1050–1065
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Tavaré S (1986) Some probabilistic and statistical problems in the analysis of DNA sequences. In: Miura RM (ed) Lectures on mathematics in the life sciences, 17th edn. American Mathematical Society, Providence, pp 57–86
Tindall B (2014a) Names at the rank of class, subclass and order, their typification and current status: supplementary information to Opinion 79. Judicial commission of the international committee on systematics of prokaryotes. Int J Syst Evol Microbiol 64(10):3599–3602
Tindall BJ (2014b) The family name Solimonadaceae Losey et al. 2013 is illegitimate, proposals to create the names ‘Sinobacter soli’ comb. nov. and ‘Sinobacter variicoloris’ contravene the Code, the family name Xanthomonadaceae Saddler and Bradbury 2005 and the order name Xanthomonadales Saddler and Bradbury 2005 are illegitimate and notes on the application of the family names Solibacteraceae Zhou et al. 2008, Nevskiaceae Henrici and Johnson 1935 (Approved Lists 1980) and Lysobacteraceae Christensen and Cook 1978 (Approved Lists 1980) and order name Lysobacteriales Christensen and Cook 1978 (Approved Lists 1980) with respect to the classification of the corresponding type genera Solibacter Zhou et al. 2008 Nevskia Famintzin 1892 (Approved Lists 1980) and Lysobacter Christensen and Cook 1978 (Approved Lists 1980) and importance of accurately expressing the link between a taxonomic name, its authors and the corresponding description/circumscription/emendation. Int J Syst Evol Microbiol 64(1):293–297
Van Sluys MA, de Oliveira MC, Monteiro-Vitorello CB, Miyaki CY, Furlan LR, Camargo LE, da Silva AC, Moon DH, Takita MA, Lemos EG et al (2003) Comparative analyses of the complete genome sequences of Pierce’s disease and citrus variegated chlorosis strains of Xylella fastidiosa. J Bacteriol 185(3):1018–1026
Williams KP, Gillespie JJ, Sobral BW, Nordberg EK, Snyder EE, Shallom JM, Dickerman AW (2010) Phylogeny of gammaproteobacteria. J Bacteriol 192(9):2305–2314
Wilson K (1987) Preparation of genomic DNA from bacteria. In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (eds) Current protocols in molecular biology. Wiley, New York, pp 2.4.1–2.4.2
Wu D, Hugenholtz P, Mavromatis K, Pukall R, Dalin E, Ivanova NN, Kunin V, Goodwin L, Wu M, Tindall BJ (2009) A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea. Nature 462(7276):1056–1060
Yilmaz P, Parfrey LW, Yarza P, Gerken J, Pruesse E, Quast C, Schweer T, Peplies J, Ludwig W, Glöckner FO (2013) The SILVA and “all-species living tree project (LTP)” taxonomic frameworks. Nucleic Acids Res. doi:10.1093/nar/gkt1209
Zhou Y, Zhang Y-Q, Zhi X-Y, Wang X, Dong J, Chen Y, Lai R, Li W-J (2008) Description of Sinobacter flavus gen. nov., sp. nov., and proposal of Sinobacteraceae fam. nov. Int J Syst Evol Microbiol 58(1):184–189
Acknowledgments
We thank Professor Iain Sutcliffe for valuable comments and suggestions for improvement of this manuscript. This work was supported by a research grant from the Natural Science and Engineering Research Council of Canada to RSG.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Naushad, S., Adeolu, M., Wong, S. et al. A phylogenomic and molecular marker based taxonomic framework for the order Xanthomonadales: proposal to transfer the families Algiphilaceae and Solimonadaceae to the order Nevskiales ord. nov. and to create a new family within the order Xanthomonadales, the family Rhodanobacteraceae fam. nov., containing the genus Rhodanobacter and its closest relatives. Antonie van Leeuwenhoek 107, 467–485 (2015). https://doi.org/10.1007/s10482-014-0344-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-014-0344-8