Abstract
A novel Gram-stain-positive, slightly halophilic, catalase- and oxidase-positive, endospore-forming, motile, aerobic, rod-shaped bacterium, designated strain JSM 081003T, was isolated from non-saline forest soil in Hunan Province, China. Growth occurred with 0.5–15% (w/v) NaCl (optimum 2–4%) at pH 6.5–10.5 (optimum pH 7.5–8.5) and at 5–40°C (optimum 30°C). meso-Diaminopimelic acid was present in the cell-wall peptidoglycan. The major cellular fatty acids were iso-C15:0, anteiso-C15:0 and iso-C14:0. Strain JSM 081003T contained MK-7 as the predominant respiratory quinone, and diphosphatidylglycerol, phosphatidylethanolamine and phosphatidylglycerol as the major polar lipids. The genomic DNA G + C content of strain JSM 081003T was 40.9 mol%. A phylogenetic analysis based on 16S rRNA gene sequence comparisons revealed that strain JSM 081003T should be assigned to the genus Bacillus, and was related most closely to the type strains of Bacillus lehensis (sequence similarity 99.6%), Bacillus oshimensis (99.4%) and Bacillus patagoniensis (96.6%); lower than 96.0% sequence similarity was observed with other Bacillus species. The combination of phylogenetic analysis, DNA–DNA relatedness values, phenotypic characteristics and chemotaxonomic data supports the view that strain JSM 081003T represents a new species of the genus Bacillus, for which the name Bacillus hunanensis sp. nov. is proposed. The type strain is JSM 081003T (= DSM 23008T = KCTC 13711T).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Halophilic, halotolerant, alkaliphilic and/or alkalitolerant bacilli species are not only widely distributed throughout various types of saline environments (Ash et al. 1991; Nielsen et al. 1994; Ventosa et al. 1998; Arahal and Ventosa 2002; Romano et al. 2005; Lim et al. 2006a, b; Yumoto 2007; Chen et al. 2009a, b) but also were isolated from non-saline environments (Nielsen et al. 1995; Echigo et al. 2005, 2007; Usami et al. 2007). Those bacteria attracted increasing interest, attributable to their ability to grow under extreme conditions as well as to the potential use of their enzymes in biotechnological applications (Horikoshi 1999; Margesin and Schinner 2001; Nogi et al. 2005; Krulwich et al. 2007). During an investigation of the diversity of halophilic and halotolerant bacteria in Xiaoxi National Natural Reserve (28°42′15′′–28°53′15′′N 110°6′50′′–110°21′35′′E), China, a slightly halophilic, endospore-forming, Gram-stain-positive bacterium, designated strain JSM 081003T, was isolated from a non-saline forest soil sample. Based on the results of a polyphasic taxonomic study, this strain is considered to represent a novel species of the genus Bacillus.
Materials and methods
Strains and culture conditions
Strain JSM 081003T was isolated from a non-saline forest soil sample using the dilution plating technique on marine agar 2216 (MA; Difco) supplemented with 10% (w/v) NaCl and cultivated at 28°C for 2 weeks. After primary isolation, the strain was purified by repeated streaking and subculturing on MA plates (4–5 times) and examining by light microscopy. The isolate was maintained as serial transfers on MA slants, otherwise lyophilized cultures at 4°C and also deep-frozen at −80°C in 20% (v/v) glycerol. For comparison, two type strains, Bacillus lehensis DSM 19099T and Bacillus oshimensis DSM 18940T, were obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ, Braunschweig, Germany). Unless indicated otherwise, morphological, physiological, molecular and chemotaxonomic studies were performed with cells grown on MA (pH 8.0) at 30°C.
Phenotypic characterization
In order to compare strain JSM 081003T and strains B. lehensis DSM 19099T and B. oshimensis DSM 18940T under the same laboratory conditions, a phenotypic characterization of these three strains was carried out following the recommendations of the proposed minimal standards for describing new taxa of aerobic, endospore-forming bacteria (Logan et al. 2009). Cell morphology was examined by using phase-contrast microscopy (DM3000, 100×HCX PL Fluotar oil immersion objective, Ph3; Leica) with cells grown on MA plus 10 mg MnSO4 for 1–3 days at 30°C and 4–7 days at room temperature (18–20°C). The Gram staining and the KOH lysis test were carried out according to Smibert and Krieg (1994) and Gregersen (1978), respectively. Flagella were examined according to the methods described by Smibert and Krieg (1994). Growth in the absence of NaCl was investigated on nutrient agar (NA) and in nutrient broth (NB) prepared according to the formula of Atlas (1993) except the addition of NaCl. Tolerance of NaCl was tested on NA as well as in NB at different NaCl concentrations [0.1 and 0.5% (w/v), and 1–30% (w/v) in increments of 1%]. Growth was tested at various temperatures (4, 5–55°C, in increments of 5°C) and at different pH (5.0–11.0, in increments of 0.5 pH units) on MA as well as in NB supplemented with 2.5% (w/v) NaCl. The buffer solutions described by Chen et al. (2007) were used for pH experiments. Methyl red and Voges–Proskauer tests and determination of H2S production from l-cysteine, hydrolysis of aesculin, indole production, nitrate and nitrite reduction and activities of arginine dihydrolase, lysine and ornithine decarboxylase, phenylalanine deaminase and urease were performed as described by Smibert and Krieg (1994). Hydrolysis of casein, cellulose, DNA, gelatin, starch, Tween 20, 40, 60 and 80 was determined as described by Cowan and Steel (1965). Growth under anaerobic conditions was determined on MA supplemented with 0.5% (w/v) glucose and with or without 0.1% (w/v) nitrate using the GasPak Anaerobic Systems (BBL) according to the manufacturer’s instructions. Determination of acid production from carbohydrates and utilization of carbon and nitrogen sources was performed as described by Ventosa et al. (1982). Observation of motility and tests of catalase and oxidase activities were detected as described previously (Chen et al. 2007). Other enzymic activities were assayed using API ZYM strips (bioMérieux) according to the manufacturer’s instructions with 3% (w/v) NaCl. All the physiological and biochemical tests were repeated for three times.
Determination of 16S rRNA gene sequence, phylogenetic analysis and DNA–DNA hybridization
The 16S rRNA gene sequence was amplified by PCR and sequenced as described by Cui et al. (2001). Pairwise sequence similarities were calculated using a global alignment algorithm, implemented at the EzTaxon server (Chun et al. 2007). Phylogenetic analysis was performed using the software package MEGA version 4.1 (Tamura et al. 2007) after multiple alignment of sequence data by CLUSTAL_X (Thompson et al. 1997). Distances were calculated using distance options according to Kimura’s two-parameter model (Kimura 1980) and clustering was performed with the neighbour-joining method (Saitou and Nei 1987). Maximum-likelihood (Felsenstein 1981) and maximum-parsimony (Kluge and Farris 1969) trees were generated using the tree making algorithms contained in the PHYLIP package (Felsenstein 2002). Bootstrap analysis was used to evaluate the tree topology by means of 1000 resamplings (Felsenstein 1985). DNA–DNA hybridization experiments were performed using the optical renaturation method (De Ley et al. 1970; Huß et al. 1983; Jahnke 1992). Every hybridization experiment was performed with three replications and the relatedness value was expressed as the mean of the three values.
Chemotaxonomic characterization
Diamino acid in cell wall was analysed as described by Hasegawa et al. (1983). Isoprenoid quinones were analysed by HPLC as described by Groth et al. (1996). Polar lipids were extracted according to the method of Minnikin et al. (1979) and were identified by two-dimensional TLC and spraying with appropriate detection reagents (Collins and Jones 1980). Fatty acids were determined according to Sasser (1990) using the Microbial Identification System (Microbial ID) with cells grown in marine broth 2216 (Difco) in flasks on a rotary shaker (with shaking at 200 rpm) at 30°C for 2 days. Genomic DNA was isolated according to Hopwood et al. (1985) and the G + C content was determined using the HPLC method (Mesbah et al. 1989).
Results and discussion
Phenotypic characteristics
Strain JSM 081003T was slightly halophilic and strictly aerobic and the cells were motile, Gram-stain-positive rods, forming ellipsoidal endospores that lay in central unswollen sporangia. Colonies were yellow-pigmented, somewhat convex and opaque with smooth, glistening surfaces and circular margins and 2–3 mm diameter after incubation for 3–5 days at 30°C on MA. The strain grew optimally in the presence of 2–4% (w/v) NaCl, at pH 7.5–8.5 and at 30°C. Detailed phenotypic properties that differentiate strain JSM 081003T from related Bacillus species are summarized in Table 1 and also mentioned in the species description below.
Phylogenetic analysis based on 16S rRNA gene sequence comparison and DNA–DNA relatedness
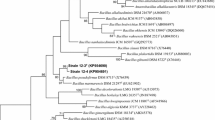
The almost-complete 16S rRNA gene sequence (1427 bp) of the organism was determined (GenBank/EMBL/DDBJ accession number HM054473). Phylogenetic analysis based on 16S rRNA gene sequences revealed that strain JSM 081003T should be assigned to the genus Bacillus, and was related most closely to the type strains of B. lehensis (16S rRNA gene sequence similarity 99.6%; Ghosh et al. 2007), B. oshimensis (99.4%; Yumoto et al. 2005) and B. patagoniensis (96.6%; Olivera et al. 2005); lower than 96.0% sequence similarity was observed with other Bacillus species. In the neighbour-joining phylogenetic tree, strain JSM 081003T formed a robust lineage with the type strains of B. lehensis and B. oshimensis supported by a significant bootstrap resampling value (100%) (Fig. 1). However, JSM 081003T occupied a distinct lineage in the phylogenetic trees constructed by using maximum-likelihood and maximum-parsimony methods (Supplementary Fig. S1). Levels of DNA–DNA relatedness between strain JSM 081003T and strains B. lehensis DSM 19099T and B. oshimensis DSM 18940T were 41% (SD, 2.5%) and 43% (SD, 2.6%), respectively, values that are well below the threshold value (70%) recommended by Wayne et al. (1987) for the definition of members of a species. Therefore, it would appear that, on the basis of the phylogenetic and DNA–DNA hybridization data, strain JSM 081003T represents a new species of the genus Bacillus according to accepted criteria (Wayne et al. 1987; Stackebrandt and Goebel 1994).
Phylogenetic tree showing the phylogenetic positions of strain JSM 081003T and related taxa based on 16S rRNA gene sequence analysis constructed using the neighbour-joining method. Numbers at nodes are bootstrap percentages (>50%) based on a neighbour-joining analysis of 1000 resampled datasets. Bar 1 substitutions per 100 nucleotides
Chemotaxonomic characteristics and DNA base composition
Chemotaxonomic data for strain JSM 081003T were consistent with the assignment of the strain to the genus Bacillus. The strain possessed a cell-wall type based on meso-diaminopimelic acid as the diagnostic diamino acid. Strain JSM 081003 T contained MK-7 (93.5%) as the predominant menaquinone, with MK-6 (4.2%) and MK-8 (2.3%) present in minor amounts. The polar lipids of this strain consisted of diphosphatidylglycerol, phosphatidylethanolamine, phosphatidylglycerol and two unknown phospholipids. The fatty acid profile of strain JSM 081003T was similar to those of the type strains of the two phylogenetically related Bacillus species, although there were differences in the proportions of some components (Table 2). The fatty acid profile of JSM 081003T contained the major compounds iso-C15:0 (43.6%), anteiso-C15:0 (23.4%) and iso-C14:0 (10.2%), which are characteristic of numerous members within the genus Bacillus (Kämpfer 1994). The DNA G + C content of strain JSM 081003T was 40.9 mol%.
Taxonomic conclusion
The results of the phylogenetic analysis and of morphological and chemotaxonomic investigations supported the affiliation of strain JSM 081003T to the genus Bacillus. However, the strain could be clearly distinguished from its phylogenetic relatives by several phenotypic differences, such as the yellow-pigmentation, the positive results of acid production from amygdalin and d-mannitol and the negative results of urease activity and acid production from trehalose and d-xylose (Table 1). Together with some chemotaxonomic differences (Tables 1, 2) and low levels of DNA–DNA relatedness between strain JSM 081003T and the type strains of B. lehensis and B. oshimensis, the results of the polyphasic taxonamic study presented here allow us to assign the isolate to a novel species, for which we propose the name Bacillus hunanensis sp. nov.
Description of B. hunanensis
B. hunanensis (hu.nan.en’sis. N.L. masc. adj. hunanensis pertaining to Hunan Province, China, the source of the sample from which the type strain was isolated).
Cells are Gram-stain-positive, catalase- and oxidase-positive, motile, aerobic, slightly halophilic, straight rods, approximately 0.6–0.8 μm wide and 1.5–3.5 μm long, occurring singly, as pairs or as short chains, producing ellipsoidal endospores that lie in central unswollen sporangia. Colonies are yellow-pigmented, somewhat convex and opaque, have smooth, glistening surfaces and circular margins and are 2–3 mm in diameter on MA. No diffusible pigments are produced. Growth occurs with 0.5–15% (w/v) NaCl (optimum 2–4%) and at pH 6.5–10.5 (optimum pH 7.5–8.5) and at 5–40°C (optimum 30°C). No growth occurs in the absence of salts. Nitrate and nitrite is not reduced. Negative for tests of methyl red, Voges–Proskauer, H2S and indole production. Aesculin, casein, gelatin, starch, Tween 20, 40 and 60 are hydrolyzed, but cellulose, DNA and Tween 80 are not. Acids are produced from amygdalin, d-glucose, glycerol, maltose, d-mannitol, melibiose and sucrose, but not from N-acetylglucosamine, adonitol, l-arabinose, cellobiose, dulcitol, d-fructose, d-galactose, glycogen, myo-inositol, lactose, d-mannose, melezitose, raffinose, l-rhamnose, d-ribose, d-salicin, d-sorbitol, trehalose or d-xylose. The following compounds are utilized as sole sources of carbon and energy or sole sources of carbon, nitrogen and energy: N-acetylglucosamine, dextrin, d-fructose, d-glucose, maltose, d-mannose, sucrose, d-mannitol, citrate and l-asparagine; the following are not utilized: l-arabinose, cellobiose, d-galactose, lactose, melezitose, melibiose, raffinose, l-rhamnose, d-ribose, d-salicin, trehalose, d-xylose, adonitol, d-arabitol, glycerol, myo-inositol, d-sorbitol, acetate, butyrate, gluconate, propionate, succinate, l-alanine, l-arginine, l-glutamic acid, glycine, l-histidine, hydroxy-l-proline, l-isoleucine, l-leucine, l-methionine, l-phenylalanine, l-proline, l-serine and l-valine. Constitutive enzymes expressed are acid and alkaline phosphatase, α-chymotrypsin, esterase (C4), esterase lipase (C8), -glucosidase, lipase (C14) and α-mannosidase; arginine dihydrolase, cystine arylamidase, α-fucosidase, α- and β-galactosidase, α-glucosidase, N-acetyl-β-glucosaminidase, β-glucuronidase, leucine arylamidase, lysine decarboxylase, naphthol-AS-BI-phosphohydrolase, ornithine decarboxylase, phenylalanine deaminase, trypsin, urease and valine arylamidase are not observed. meso-Diaminopimelic acid is present in the cell-wall peptidoglycan as the diagnostic diamino acid. Possesses MK-7 as the predominant menaquinone, and diphosphatidylglycerol, phosphatidylethanolamine and phosphatidylglycerol as the major polar lipids. Major fatty acids are iso-C15:0, anteiso-C15:0 and iso-C14:0. The DNA G + C content of the type strain is 40.9 mol% (HPLC method).
The type strain is JSM 081003T (= DSM 23008T = KCTC 13711T), which is the isolate on which the species description is based. It was isolated from non-saline forest soil in Hunan Province, China.
References
Arahal DR, Ventosa A (2002) Moderately halophilic and halotolerant species of Bacillus and related genera. In: Berkeley RCW, Heyndrickx M, Logan N, De Vos P (eds) Applications and systematics of Bacillus and relatives. Blackwell, Oxford, pp 83–99
Ash C, Farrow JAE, Wallbanks S, Collins MD (1991) Phylogenetic heterogeneity of the genus Bacillus as revealed by comparative analysis of small-subunit ribosomal-RNA sequences. Lett Appl Microbiol 13:202–206
Atlas RM (1993) In: Parks LC (ed) Handbook of microbiological media. CRC Press, Boca Raton, pp 666–672
Chen YG, Cui XL, Pukall R, Li HM, Yang YL, Xu LH, Wen ML, Peng Q, Jiang CL (2007) Salinicoccus kunmingensis sp. nov., a moderately halophilic bacterium isolated from a salt mine in Yunnan, south-west China. Int J Syst Evol Microbiol 57:2327–2332
Chen YG, Zhang YQ, Xiao HD, Liu ZX, Yi LB, Shi JX, Zhi XY, Cui XL, Li WJ (2009a) Pontibacillus halophilus sp. nov., a moderately halophilic bacterium isolated from sea urchin. Int J Syst Evol Microbiol 59:1635–1639
Chen YG, Zhang YQ, Wang YX, Liu ZX, Klenk HP, Xiao HD, Tang SK, Cui XL, Li WJ (2009b) Bacillus neizhouensis sp. nov., a halophilic marine bacterium isolated from a sea anemone. Int J Syst Evol Microbiol 59:3035–3039
Chun J, Lee JH, Jung Y, Kim M, Kim S, Kim BK, Lim YW (2007) EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int J Syst Evol Microbiol 57:2259–2261
Collins MD, Jones D (1980) Lipids in the classification and identification of coryneform bacteria containing peptidoglycans based on 2, 4-diaminobutyric acid. J Appl Bacteriol 48:459–470
Cowan ST, Steel KJ (1965) Manual for the identification of medical bacteria. Cambridge University Press, London
Cui XL, Mao PH, Zeng M, Li WJ, Zhang LP, Xu LH, Jiang CL (2001) Streptomonospora salina gen. nov., sp. nov., a new member of the family Nocardiopsaceae. Int J Syst Evol Microbiol 51:357–363
De Ley J, Cattoir H, Reynaerts A (1970) The quantitative measurement of DNA hybridization from renaturation rates. Eur J Biochem 12:133–142
Echigo A, Hino M, Fukushima T, Mizuki T, Kamekura M, Usami R (2005) Endospores of halophilic bacteria of the family Bacillaceae isolated from non-saline Japanese soil may be transported by Kosa event (Asian dust storm). Saline Systems 1:8
Echigo A, Fukushima T, Mizuki T, Kamekura M, Usami R (2007) Halalkalibacillus halophilus gen. nov., sp. nov., a novel moderately halophilic and alkaliphilic bacterium isolated from a non-saline soil sample in Japan. Int J Syst Evol Microbiol 57:1081–1085
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Felsenstein J (2002) PHYLIP (phylogeny inference package), version 3.6a. Distributed by the author. Department of Genome Sciences, University of Washington, Seattle
Ghosh A, Bhardwaj M, Satyanarayana T, Khurana M, Mayilraj S, Jain RK (2007) Bacillus lehensis sp. nov., an alkalitolerant bacterium isolated from soil. Int J Syst Evol Microbiol 57:238–242
Gregersen T (1978) Rapid method for distinction of Gram-negative from Gram-positive bacteria. Eur J Appl Microbiol Biotechnol 5:123–127
Groth I, Schumann P, Weiss N, Martin K, Rainey FA (1996) Agrococcus jenensis gen. nov., sp. nov., a new genus of actinomycetes with diaminobutyric acid in the cell wall. Int J Syst Bacteriol 46:234–239
Hasegawa T, Takizawa M, Tanida S (1983) A rapid analysis for chemical grouping of aerobic actinomycetes. J Gen Appl Microbiol 29:319–322
Hopwood DA, Bibb MJ, Chater KF, Kieser T, Bruton CJ, Kieser HM, Lydiate DJ, Smith CP, Ward JM (1985) Preparation of chromosomal, plasmid and phage DNA. In: Hopwood DA, Bibb MJ, Chater KF, Kieser T, Bruton CJ, Kieser HM, Lydiate DJ, Smith CP, Ward JM, Schrempf H (eds) Genetic manipulation of Streptomyces: a laboratory manual. F. Crowe and Sons, Norwich, pp 79–80
Horikoshi K (1999) Alkaliphiles: some applications of their products for biotechnology. Microbiol Mol Biol Rev 63:735–750
Huß VAR, Festl H, Schleifer KH (1983) Studies on the spectrophotometric determination of DNA hybridization from renaturation rates. Syst Appl Microbiol 4:184–192
Jahnke KD (1992) BASIC computer program for evaluation of spectroscopic DNA renaturation data from Gilford System 2600 spectrophotometer on a PC/XT/AT type personal computer. J Microbiol Methods 15:61–73
Kämpfer P (1994) Limits and possibilities of total fatty acid analysis for classification and identification of Bacillus species. Syst Appl Microbiol 17:86–98
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Kluge AG, Farris FS (1969) Quantitative phyletics and the evolution of anurans. Syst Zool 18:1–32
Krulwich TA, Hicks DB, Swartz TH, Ito M (2007) Bioenergetic adaptations that support alkaliphily. In: Gerday C, Glansdorff N (eds) Physiology and biochemistry of extremophiles. American Society for Microbiology, Washington, pp 311–329
Lim JM, Jeon CO, Kim CJ (2006a) Bacillus taeanensis sp. nov., a halophilic Gram-positive bacterium from a solar saltern in Korea. Int J Syst Evol Microbiol 56:2903–2908
Lim JM, Jeon CO, Lee SM, Xu LH, Jiang CL, Kim CJ (2006b) Bacillus salarius sp. nov., a halophilic, spore-forming bacterium isolated from a salt lake in China. Int J Syst Evol Microbiol 56:373–377
Logan NA, Berge O, Bishop AH, Busse HJ, De Vos P, Fritze D, Heyndrickx M, Kämpfer P, Rabinovitch L, Salkinoja-Salonen MS, Seldin L, Ventosa A (2009) Proposed minimal standards for describing new taxa of aerobic, endospore-forming bacteria. Int J Syst Evol Microbiol 59:2114–2121
Margesin R, Schinner F (2001) Potential of halotolerant and halophilic microorganisms for biotechnology. Extremophiles 5:73–83
Mesbah M, Premachandran U, Whitman WB (1989) Precise measurement of the G + C content of deoxyribonucleic acid by high-performance liquid chromatography. Int J Syst Bacteriol 39:159–167
Minnikin DE, Collins MD, Goodfellow M (1979) Fatty acid and polar lipid composition in the classification of Cellulomonas, Oerskovia and related taxa. J Appl Bacteriol 47:87–95
Nielsen P, Rainey FA, Outtrup H, Priest FG, Fritze D (1994) Comparative 16S rDNA sequence analysis of some alkaliphilic bacilli and the establishment of a sixth rRNA group within the genus Bacillus. FEMS Microbiol Lett 117:61–66
Nielsen P, Fritze D, Priest FG (1995) Phenetic diversity of alkaliphilic Bacillus strains: proposal for nine new species. Microbiology 141:1745–1761
Nogi Y, Takami H, Horikoshi K (2005) Characterization of alkaliphilic Bacillus strains used in industry: proposal of five novel species. Int J Syst Evol Microbiol 55:2307–2315
Olivera N, Sineriz F, Breccia JD (2005) Bacillus patagoniensis sp. nov., a novel alkalitolerant bacterium from the rhizosphere of Atriplex lampa in Patagonia, Argentina. Int J Syst Evol Microbiol 55:443–447
Romano I, Lama L, Nicolaus B, Gambacorta A, Giordano A (2005) Bacillus saliphilus sp. nov., isolated from a mineral pool in Campania, Italy. Int J Syst Evol Microbiol 55:159–163
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids. MIDI Technical Note 101. MIDI Inc, Newark
Smibert RM, Krieg NR (1994) Phenotypic characterization. In: Gerhardt P, Murray RGE, Wood WA, Krieg NR (eds) Methods for general and molecular bacteriology. American Society for Microbiology, Washington, DC, pp 607–654
Stackebrandt E, Goebel BM (1994) Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol 44:846–849
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetic analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Usami R, Echigo A, Fukushima T, Mizuki T, Yoshida Y, Kamekura M (2007) Alkalibacillus silvisoli sp. nov., an alkaliphilic moderate halophile isolated from non-saline forest soil in Japan. Int J Syst Evol Microbiol 57:770–774
Ventosa A, Quesada E, Rodriguez-Valera F, Ruiz-Berraquero F, Ramos-Cormenzana A (1982) Numerical taxonomy of moderately halophilic Gram-negative rods. J Gen Microbiol 128:1959–1968
Ventosa A, Nieto JJ, Oren A (1998) Biology of moderately halophilic aerobic bacteria. Microbiol Mol Biol Rev 62:504–544
Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, Krichevsky MI, Moore LH, Moore WEC, Murray RGE et al (1987) International committee on systematic bacteriology. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol 37:463–464
Yumoto I (2007) Environmental and taxonomic biodiversities of Gram-positive alkaliphiles. In: Gerday C, Glansdorff N (eds) Physiology and Biochemistry of Extremophiles. American Society for Microbiology, Washington, pp 295–310
Yumoto I, Hirota K, Goto T, Nodasaka Y, Nakajima K (2005) Bacillus oshimensis sp. nov., a moderately halophilic, non-motile alkaliphile. Int J Syst Evol Microbiol 55:907–911
Acknowledgments
This work was supported by grants from the National Basic Research Program of China (2010CB833800), National Natural Science Foundation of China (NSFC) (30970007, 30970008), Key Laboratory of Ecotourism’s Application Technology of Hunan Province (SL2010Z01), Key Laboratory of Hunan Forest Products and Chemical Industry Engineering (JDZ200907) and State-level Public Welfare Scientific Research Institutes for Basic R&D Special Fund (IMBF200915). We are grateful to Mr Zhu-Xiang Liu and Mr Ke Huang for their excellent technical assistance.
Author information
Authors and Affiliations
Corresponding author
Additional information
The GenBank/EMBL/DDBJ accession number for the 16S rRNA gene sequence of strain JSM 081003T is HM054473.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chen, YG., Hao, DF., Chen, QH. et al. Bacillus hunanensis sp. nov., a slightly halophilic bacterium isolated from non-saline forest soil. Antonie van Leeuwenhoek 99, 481–488 (2011). https://doi.org/10.1007/s10482-010-9512-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-010-9512-7