Abstract
A Gram-stain positive, rod-shaped, endospore-forming and aerobic bacterium, designated BZ1T, was isolated from a soil sample collected from a sunflower field in Wuyuan county, Inner Mongolia, China. On the basis of 16S rRNA gene sequence analysis, the isolate was found to be a member of the genus Bacillus and the close phylogenetic relatives to be Bacillus gottheilii WCC 4585T, Bacillus oceanisediminis H2T, Bacillus mesonae FJAT-13985T and Bacillus horneckiae DSM 23495T with 98.3, 98.1, 98.0 and 97.6 % sequence similarity, respectively. Strain BZ1T was found to grow at 6–40 °C (optimum 30–33 °C), pH 6.0-9.0 (optimum pH 7.0) and 0–5.5 % (w/v) NaCl (optimum 0.5 %). The cell wall diamino acid of the peptidoglycan of strain BZ1T was identified as meso-diaminopimelic acid and the predominant respiratory quinone as MK-7. The major cellular fatty acids were found to be iso-C15:0, anteiso-C15:0 and iso-C14:0, and the polar lipids to consist of diphosphatidylglycerol, phosphatidylglycerol and phosphatidylethanolamine. The novel strain was found to have a DNA G + C content 44.5 mol%. DNA–DNA hybridization with closely related strains was low. Based on phenotypic, phylogenetic and chemotaxonomic results, it is concluded that strain BZ1T represents a novel species within the genus Bacillus, for which we propose the name Bacillus depressus sp. nov. The type strain is BZ1T (= CGMCC 1.15124T = KCTC 33643T).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Bacillus was initially proposed by Cohn (1872) and since then the genus has been expanded with many further novel species. At the time of writing, the genus encompasses about several hundred species with validly published names (List of Prokaryotic Names with Standing in Nomenclature; http://www.bacterio.net/) and members of the genus have been found in various environments, such as Bacillus panacisoli CJ32T isolated from soil (Choi and Cha 2014), Bacillus andreesenii 8-4-E13T from a composit (Kosowski et al. 2014), Bacillus gottheilii WCC 4585T from a pharmaceutical manufacturing site (Seiler et al. 2013), Bacillus beringensis BR035T from the Bering Sea (Yu et al. 2011) and Bacillus alkalisediminis K1-25T from a pond (Borsodi et al. 2011). Members of the genus Bacillus are Gram-stain positive and endospore-forming rods. The major fatty acid component of the members of the genus Bacillus is iso-C15:0.
During the course of a study of bacterial diversity in a soil sample from a sunflower field in Wuyuan county of Inner Mongolia in China, a Gram-stain positive bacterium, designated BZ1T, was isolated and studied here by a polyphasic characterisation. It is concluded that strain BZ1T represents a novel species within the genus Bacillus, for which we propose the name Bacillus depressus sp. nov.
Materials and methods
Isolation and culture conditions
Strain BZ1T was isolated from a saline-alkaline soil sample [pH 8.5; salt content 0.4 % (w/w)] collected from a sunflower field in Wuyuan county in Inner Mongolia in China (GPS coordinates 40°46′30″N and 107°36′10″E). For isolation, the soil sample was suspended in sterilised water, serially diluted and then incubated at 35 °C for 3 days on nutrient agar with the following composition: beef extract 3 g L−1, peptone 5 g L−1, NaCl 5 g L−1 and agar 15 g L−1 (pH 7.6). Then representative colonies were transferred to the same medium. Pure cultures were obtained by streaking repeatedly and the purified strains stored on trypticase soy agar (TSA; Difco) at 4 °C and in 20 % (v/v) glycerol suspensions at −20 °C.
31 colonies were isolated, purified and identified. Among the isolates, only one isolate was considered to represent a novel species and designated as strain BZ1T. Routine cultivation of the strain was performed on TSA plates at 30 °C. B. gottheilii WCC 4585T and Bacillus horneckiae DSM 23495T were obtained from the German Collection of Microorganisms and Cell Cultures (DSMZ), Bacillus oceanisediminis H2T and Bacillus mesonae FJAT-13985T were obtained from the China General Microbiological Culture Collection Center (CGMCC). These strains were cultured under comparable conditions and used as reference strains in the phenotypic tests.
Morphological, physiological and biochemical characteristics
Cell morphology was examined by light microscopy (CX31; Olympus). The colony morphology, size and pigmentation were observed on TSA after 48 h of incubation at 30 °C. The Gram reaction was performed according to the methods described by Gerhardt et al. (1981). Endospores were observed using the Schaeffer-Fulton staining method (Murray et al. 1994). Oxidase activity was determined using 1 % (w/v) tetramethyl-p-phenylenediamine. Catalase activity was detected by the production of oxygen bubbles with 3 % (v/v) H2O2. Growth at different temperatures (0–50 °C) was tested in TSB for 3–5 days. The pH range for growth was determined at various pH values (pH 3.0-12.0, in increments of 0.5 pH units) at 30 °C by culturing the strain in TSB by using the appropriate biological buffers as described by Xu et al. (2005). NaCl tolerance tests were assessed at different NaCl concentrations [0–10 % (w/v) NaCl, at intervals of 0.5 % and 11–20 % (w/v), at intervals of 1 %] for up to 14 days at 30 °C in TSB prepared according to the formula of the Difco medium except that NaCl was omitted. Sensitivity to antibiotics was examined on TSA plates using antibiotic discs (Goodfellow and Orchard 1974). Nitrate reduction, hydrolysis of casein, gelatin, starch, Tween 20, 40, 60 and 80, urease activity and H2S production were studied as described by Smibert and Krieg (1994). Resistance to lysozyme, egg-yolk reaction, decomposition of adenine, xanthine, hypoxanthine, guanine or tyrosine were determined by the method of Logan and De Vos (2009). The tests of carbon source utilisation were determined using methods described by Gordon and Mihm (1957) and acid production from carbohydrates was determined according to Gordon et al. (1974). Nitrogen assimilation was assessed using TSB. Additional enzyme activities and other physiological properties were performed using the API 20E, API 20NE and API ZYM kits (bioMérieux) according to the manufacturer’s instructions.
Chemotaxonomic characterisation
For fatty acid analyses, cells of strain BZ1T were cultivated on TSA for 24 h at 28 °C. Fatty acid methyl esters were prepared and analysed according to the standard protocol of the Microbial Identification System (MIDI) tested by GC (Sasser 1990; Kämpfer and Kroppenstedt 1996); the database used was TSBA6. Isoprenoid quinones were isolated according to the method of Collins et al. (1987) and analysed using reversed-phase HPLC (Wu et al. 1989). For cell wall peptidoglycan analysis, the cells were hydrolysed (4 N HCl, 100 °C, 15 h) and the peptidoglycan composition was analysed by two-dimensional ascending TLC on cellulose plates using the solvent system of Schleifer (1985). The cellular polar lipids were extracted and then examined by two-dimensional TLC and identified by using procedures described by Minnikin et al. (1984).
Molecular analysis
For 16S rRNA gene sequencing and phylogenetic analysis, extraction of genomic DNA, PCR amplification of the 16S rRNA gene and sequencing of purified PCR products were performed as described previously (Rainey et al. 1996). The 16S rRNA gene was amplified by PCR using 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-ACGGCTACCTTGTTACGACTT-3′) (Weisburg et al. 1991). Sequencing reactions of the PCR products were carried out by the method of Lu et al. (2001). The resulting 16S rRNA gene sequence was compared with those sequences available from Genbank using the BLAST program and the EzTaxon server (http://eztaxon-e.ezbiocloud.net/, Kim et al. 2012). Multiple alignments with sequences of closely related species of the genus Bacillus was performed using the CLUSTAL_X 1.8 software (Thompson et al. 1997). Aligned sequences were analysed by using the MEGA 5.0 software (Tamura et al. 2011) to reconstruct unrooted trees with the neighbour-joining method of Saitou and Nei (1987), maximum-parsimony (Fitch 1971) and maximum-likelihood (Felsenstein 1981) methods. A distance matrix was generated using Kimura’ s two-parameter model (Kimura 1980). The stability of clusters was ascertained by performing a bootstrap analysis based on 1000 replications of the sequences (Felsenstein 1985).
Determination of the DNA G + C content of strain BZ1T was achieved by the thermal denaturation method (Marmur and Doty 1962) using the genomic DNA of Escherichia coli strain K-12 as the standard for calibration. DNA–DNA hybridization was carried out based on the liquid renaturation method (De Ley et al. 1970; Huss et al. 1983; Jahnke 1992). Every hybridization experiment was performed with five replications. The highest and lowest values obtained were excluded and the means of the remaining three values were quoted as DNA–DNA relatedness values.
Results and discussion
Strain BZ1T was observed to be Gram-stain positive rods (0.5 − 0.8 × 1.5 − 4.0 μm) and aerobic. Colonies that formed after 2 days incubation on TSA at 30 °C were observed to be circular, opaque, mud coloured and 2-5 mm in diameter. No diffusible pigment was observed. Ellipsoidal endospores were observed to be located centrally or paracentrally in swollen sporangia. Growth was found to occur at temperatures from 6 to 40 °C (optimum 30–33 °C) and pH 6.0–9.0 (optimum pH 7.0) and in the presence of up to 5.5 % (w/v) NaCl. The results for the presence of catalase, oxidase, urease and reduction of nitrate were positive. The results of hydrogen sulfide and indole production, Voges-Proskauer reaction and nitrite reduction were negative.
Strain BZ1T was found to hydrolyse gelatin, starch, hippurate and hypoxanthine but not casein, Tween 20, 40, 60, 80, adenine, xanthine, guanine or tyrosine. Strain BZ1T was found to be susceptible to ampicillin (10 μg), chloramphenicol (30 μg), clindamycin (2 μg), erythromycin (15 μg), gentamicin (10 μg), kanamycin (30 μg), neomycin (30 μg), novobiocin (30 μg), penicillin (10 μg), rifampicin (5 μg), streptomycin (10 μg), tetracycline (30 μg) and vancomycin (30 μg), but resistant to bacitracin (0.04 μg), lyncomycin (2 μg) and nystatin (100 μ g).
The detailed physiological and biochemical characteristics of strain BZ1T are presented in the species description and Table 1. Strain BZ1T produces phenylalanine deaminase, alkaline phosphatase, esterase (C4), esterase lipase (C8), leucine arylamidase, α-chymotrypsin, acid phosphatase and naphthol-AS-BI-phosphohydrolase but not arginine dihydrolase, lysine decarboxylase, ornithine decarboxylase, tryptophan deaminase, lipase (C14), trypsin, α-galactosidase, β-galactosidase, α-glucosidase, β-glucosidase, β-glucuronidase, N-acetyl-β-glucosaminidase, α-mannosidase or β-fucosidase; and is weak positive for valine arylamidase and cystine arylamidase. The following substrates are used as sole carbon sources: acetate, benzoate, fumarate, d-galactose, glycogen, inulin, lactic acid, oxalate, d-raffinose and succinate but not L-arabinose, cellobiose, citrate, d-fructose, d-glucose, glycerol, lactose, malate, malonate, d-mannitol, pyruvic acid, l-rhamnose, d-ribose, l-sorbose, sucrose, tartrate, d-trehalose, xylitol or d-xylose; weakly positive for erythritol. The following compounds are used as sole nitrogen sources: l-cysteine and l-tyrosine but not l-alanine, l-arginine, l-aspartate, glycine, l-hydroxyproline, l-leucine, l-lysine, l-ornithine, l-phenylalanine, l-proline, l-tryptophan or l-valine. Acid is produced from aesculin, arbutin, d-fructose, d-glucose, inulin, d-ribose, l-sorbose and d-xylose but not from l-arabinose, d-cellobiose, erythritol, d-galactose, glycerol, inositol, lactose, maltose, d-mannitol, d-raffinose, l-rhamnose, sorbitol, starch, sucrose, d-trehalose or xylitol.
The main cellular fatty acids of the novel strain were identified as iso-C15:0 (32.1 %), anteiso-C15:0 (28.8 %) and iso-C14:0 (17.0 %); these iso- and anteiso-branched fatty acids of the 14–17-carbon series are typical of those observed in profiles of the type strains of members of the genus Bacillus (Kämpfer 1994; Albert et al. 2005). However, strain BZ1T could be differentiated significantly from the closely related species B. gottheilii WCC 4585T in the amount of iso-C14:0 (17.0 % vs. 3.6 %) and anteiso-C17:0 (2.0 % vs. 9.5 %), as shown in Table 2. In particular, the amount of iso-C14:0 in strain BZ1T was higher than that of the closely related species. There were notable differences observed in the percentages of other fatty acids between strain BZ1T and B. gottheilii WCC 4585T, confirming that strain BZ1T could be distinguished from B. gottheilii WCC 4585T.
Analysis of the quinones showed that the novel strain contains MK-7 as the predominant respiratory quinone. The diagnostic diamino acid in the cell wall peptidoglycan was identified as meso-diaminopimelic acid, which is typical of the large majority of members of the genus Bacillus (Priest et al. 1988). The major polar lipids present in strain BZ1T were identified as diphosphatidylglycerol, phosphatidylglycerol and phosphatidylethanolamine (Supplementary Fig. S1). The DNA G + C content of the genomic DNA of strain BZ1T was determined to be 44.5 mol%. This value is in the range of the genomic G + C contents of the members of the genus Bacillus (Slepecky and Hemphill 2006).
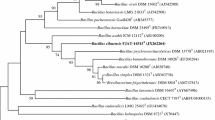
The 16S rRNA gene sequence of strain BZ1T was determined (1459 bp; GenBank/EMBL/DDBJ accession number KP259553). Phylogenetic analysis based on the neighbour-joining method revealed that strain BZ1T was included in the clusters of species of the genus Bacillus (Fig. 1). The relative position of the novel isolate was also confirmed in the trees constructed using maximum-parsimony and maximum-likelihood methods (data not shown). The close relatives in terms of pairwise 16S rRNA gene sequence similarity were B. gottheilii WCC 4585T (98.3 %), B. oceanisediminis H2T (98.1 %), B. mesonae FJAT-13985T (98.0 %), B. horneckiae DSM 23495T (97.6 %), Bacillus drentensis LMG 21831T (97.5 %), Bacillus firmus NCIMB 9366T (97.5 %), and Bacillus purgationiresistens DS22T (97.4 %), respectively. Low DNA–DNA relatedness was observed between strain BZ1T and its close phylogenetic neighbours: 33.1 ± 1.2 % with B. gottheilii WCC 4585T and 28.6 ± 0.7 % with B. oceanisediminis H2T. These values are below the threshold value of 70 % recommended by Wayne et al. (1987) for assignment of strains to the same species.
Therefore, on the basis of the above-mentioned phenotypic, phylogenetic and genetic data, strain BZ1T should be placed in the genus Bacillus. However, strain BZ1T could tolerate lower NaCl than all the reference strains except for B. mesonae FJAT-13985T. Strain BZ1T could be separated from its close relative B. gottheilii WCC 4585T in several phenotypic properties, such as colony colour, cell size, positive for activity of oxidase and urease, hydrolysis of complex substrates, as well as assimilation of carbon substrates. The strain differed from B. oceanisediminis H2T in its inability to produce arginine dihydrolase and, unlike B. mesonae FJAT-13985T, it is able to hydrolyse gelatin. Strain BZ1T is able to produce acid from d-glucose, inulin and d-xylose, in contrast to B. mesonae FJAT-13985T and B. horneckiae DSM 23495T. Characteristics useful for differentiating between strain BZ1T and the reference strains are shown in Tables 1 and 2. Thus, strain BZ1T should be classified as the type strain of a novel species of the genus Bacillus, for which the name Bacillus depressus sp. nov. is proposed.
Description of Bacillus depressus sp. nov
Bacillus depressus (de.pres′sus. L. masc. adj. depressus, depressed in shape, referring to the colonies).
Cells are Gram-stain positive, strictly aerobic rods, 0.5-0.8 μm in width and 1.5-4.0 μm long. Colonies are circular, opaque, mud coloured and 2-5 mm in diameter after 2 days incubation on TSA at 30 °C. Ellipsoidal endospores are observed centrally or paracentrally in swollen sporangia. Growth occurs at pH 6.0-9.0 and the optimal value for growth is pH 7.0; the temperature range for growth is 6–40 °C, optimum growth occurs at 30–33 °C; tolerant of up to 5.5 % (w/v) NaCl and optimum NaCl for growth is 0.5 % (w/v). Oxidase positive and catalase positive. Under aerobic conditions, nitrate is reduced, but nitrite not. Hydrogen sulfide and indole production, Voges-Proskauer and citrate utilisation are negative. Hydrolyses aesculin, starch, gelatin, urea, hippurate and hypoxanthine but not casein, Tween 20, 40, 60, 80, adenine, xanthine, guanine or tyrosine. The cell wall peptidoglycan contains meso-diaminopimalic acid and the predominant respiratory quinone is MK-7. The principal fatty acids are iso-C15:0, anteiso-C15:0 and iso-C14:0. The major polar lipids are diphosphatidylglycerol, phosphatidylglycerol and phosphatidylethanolamine. The DNA G + C content of the type strain is 44.5 mol%.
The type strain, BZ1T(= CGMCC 1.15124T = KCTC 33643T), was isolated from a soil sample in Wuyuan county, Inner Mongolia, China. The GenBank/EMBL/DDBJ accession number for the 16S rRNA gene sequence of strain BZ1T is KP259553.
References
Albert RA, Archambault J, Rosselló-Mora R, Tindall BJ, Matheny M (2005) Bacillus acidicola sp. nov., a novel mesophilic, acidophilic species isolated from acidic Sphagnum peat bogs in Wisconsin. Int J Syst Evol Microbiol 55:2125–2130
Borsodi AK, Pollák B, Kéki Z, Rusznyák A, Kovács AL, Spröer AL, Schumann P, Márialigeti K, Tóth EM (2011) Bacillus alkalisediminis sp. nov., an alkaliphilic and moderately halophilic bacterium isolated from sediment of extremely shallow soda ponds. Int J Syst Evol Microbiol 61:1880–1886
Choi JH, Cha CJ (2014) Bacillus panacisoli sp. nov., isolated from ginseng soil. Int J Syst Evol Microbiol 64:901–906
Cohn F (1872) Untersuchungen über Bakterien. Beitrage zur Biologie der Pflanzen 1:127–224
Collins MD, Howarth OW, Grund E, Kroppenstedt RM (1987) Isolation and structural determination of new members of the vitamin K2 series in Nocardia brasiliensis. FEMS Microbiol Lett 41:35–39
De Ley J, Cattoir H, Reynaerts A (1970) The quantitative measurement of DNA hybridization from renaturation rates. Eur J Biochem 12:133–142
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Fitch WM (1971) Toward defining the course of evolution: minimum change for a specific tree topology. Syst Biol 20:406–416
Gerhardt P, Murray RGE, Costilow RN, Nester EW, Wood WA, Krieg NR, Phillips GB (eds) (1981) Manual of methods for general bacteriology. American Society for Microbiology, Washington DC
Goodfellow M, Orchard VA (1974) Antibiotic sensitivity of some nocardioform bacteria and its value as a criterion for taxonomy. J Gen Microbiol 83:375–387
Gordon RE, Barnett DA, Handerhan JE, Pang CHN (1974) Nocardia coeliaca, Nocardia autotrophica, and the nocardin strain. Int J Syst Bacteriol 24:54–63
Gordon RE, Mihm JM (1957) A comparative study of some strains received as nocardiae. J Bacteriol 73:15–27
Huss VAR, Festl H, Schleifer KH (1983) Studies on the spectrophotometric determination of DNA hybridization from renaturation rates. Syst Appl Microbiol 4:184–192
Jahnke KD (1992) BASIC computer program for evaluation of spectroscopic DNA renaturation data from Gilford System 2600 spectrophotometer on a PC/XT/AT type personal computer. J Microbiol Methods 15:61–73
Kämpfer P (1994) Limits and possibilities of total fatty acid analysis for classification and identification of Bacillus species. Syst Appl Microbiol 17:86–98
Kämpfer P, Kroppenstedt RM (1996) Numerical analysis of fatty acid patterns of coryneform bacteria and related taxa. Can J Microbiol 42:989–1005
Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S, Chun J (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Kosowski K, Schmidt M, Pukall R, Hause G, Kämpfer P, Lechner U (2014) Bacillus pervagus sp. nov. and Bacillus andreesenii sp. nov., isolated from a composting reactor. Int J Syst Evol Microbiol 64:88–94
Liu B, Liu GH, Hu GH, Chen MH (2014) Bacillus mesonae sp. nov., isolated from the root of Mesona chinensis. Int J Syst Evol Microbiol 64:3346–3352
Logan NA, De Vos P (2009) Genus I. Bacillus. In: De Vos P, Garrity G, Jones D, Krieg NR, Ludwig W, Rainey FA, Schleifer KH, Whitman WB (eds) Bergey’s mannual of systematic bacteriology, vol 3. Springer, New York, pp 21–128
Lu Z, Liu Z, Wang L, Zhang Y, Qi W, Goodfellow M (2001) Saccharopolyspora flava sp. nov. and Saccharopolyspora thermophila sp. nov., novel actinomycetes from soil. Int J Syst Evol Microbiol 51:319–325
Marmur J, Doty P (1962) Determination of the base composition of deoxyribonucleic acid from its thermal denaturation temperature. J Mol Biol 5:109–118
Minnikin DE, O’Donnell AG, Goodfellow M, Alderson G, Athalye M, Schaal A, Parlett JH (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2:233–241
Murray RGE, Doetsch RN, Robinow CF (1994) Determinative and cytological light microscopy. In: Gerhardt P, Murray RGE, Wood WA, Krieg NR (eds) Methods for general and molecular bacteriology. American Society for Microbiology, Washington, pp 21–41
Priest FG, Goodfellow M, Todd C (1988) A numerical classification of the genus Bacillus. J Gen Microbiol 134:1847–1882
Rainey FA, Ward-Rainey N, Kroppenstedt RM, Stackebrandt E (1996) The genus Nocardiopsis represents a phylogenetically coherent taxon and a distinct actinomycete lineage: proposal of Nocardiopsiaceae fam. nov. Int J Syst Bacteriol 46:1088–1092
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids. USFCC Newsl 20:1–6
Schleifer KH (1985) Analysis of the chemical composition and primary structure of murein. Methods Microbiol 18:123–156
Seiler H, Wenning M, Schmidt V, Scherer S (2013) Bacillus gottheilii sp. nov., isolated from a pharmaceutical manufacturing site. Int J Syst Evol Microbiol 63:867–872
Slepecky RA, Hemphill HE (2006) The genus Bacillus-Nonmedica. Prokaryotes 4:530–562
Smibert RM, Krieg NR (1994) Phenotypic characterization. In: Gerhardt P, Murray RGE, Wood WA, Krieg NR (eds) Methods for general and molecular bacteriology. American Society for Microbiology, Washington DC, pp 607–654
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) Mega 5: molecular evolutionary genetic analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acid Res 25:4876–4882
Vaishamoayan P, Probst A, Krishnamurthi S, Ghosh S, Osman S, Mcdowall A, Ruckmani A, Mayilraj S, Venkateswaran K (2010) Bacillus horneckiae sp. nov., isolated from a spacecraft-assembly clean room. Int J Syst Evol Microbiol 60:1031–1037
Wayne LG, Brenner DJ, Colwell RR et al (1987) International Committee on Systematic Bacteriology. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol 37:463–464
Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703
Wu C, Lu X, Qin M, Wang Y, Ruan J (1989) Analysis of menaquinone compound in microbial cells by HPLC. Microbiology [English translation of Microbiology (Beijing)] 16:176–178
Xu P, Li WJ, Tang SK, Zhang YQ, Chen GZ, Chen HH, Xu LH, Jiang CL (2005) Naxibacter alkalitolerans gen. nov., sp. nov., a novel member of the family ‘Oxalobacteraceae’ isolated from China. Int J Syst Evol Microbiol 55:1149–1153
Yu Y, Li HR, Zeng YX, Chen B (2011) Bacillus beringensis sp. nov., a psychrotolerant bacterium isolated from the Bering Sea. Antonie Van Leeuwenhoek 99:551–557
Zhang JL, Wang JW, Fang CY, Song F, Xin YH, Qu L, Ding K (2010) Bacillus oceanisediminis sp. nov., isolated from marine sediment. Int J Syst Evol Microbiol 60:2924–2929
Acknowledgments
This research was supported by the Beijing Natural Science Foundation (Grant number 5152017) and by the National Natural Science Foundation of China (NSFC, Grant number 31070002).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wei, X., Xin, D., Xin, Y. et al. Bacillus depressus sp. nov., isolated from soil of a sunflower field. Antonie van Leeuwenhoek 109, 13–20 (2016). https://doi.org/10.1007/s10482-015-0605-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-015-0605-1