Abstract
A Gram-stain-positive, rod-shaped, facultatively anaerobic, motile and spore-forming bacterium with multiple flagella designated GXH0341T was isolated from the soil associated with decayed pine tree samples collected from Weizhou Island, Beihai, Guangxi, China. Growth occurred at 4–37 °C (optimum 30 °C), at pH 5.0–11.0 (optimum 8.0) and in the presence of 0–7% (w/v) NaCl (optimum 2%). Phylogenetic analysis based on 16S rRNA gene sequences indicated that strain GXH0341T was most closely related to Bacillus mesophilus DSM 101000 T (98.9%), followed by Bacillus salitolerans KC1T (96.95%) and Margalitia shackletonii DSM 18435 T (96.67%). Phylogenetic analysis revealed that strain GXH0341T represented a separate lineage within the genus Bacillus. Peroxidase is positive. The predominant quinone was MK-7 and the cell-wall diagnostic diamino acid was meso-diaminopimelic acid. The predominant polar lipids are diphosphatidylglycerol, phosphatidylethanolamine, phosphatidylglycerol, phosphatidylcholine, and two unidentified phospholipids. The major fatty acids are iso-C14:0, iso-C15:0, anteiso-C15:0 and iso-C16:0. The genome of GXH0341T comprises the biosynthetic gene cluster for T3PKS, terpene, lassopeptide and RRE-containing element as secondary metabolites. The average nucleotide identity values and the digital DNA-DNA hybridization values between GXH0341T and B. mesophilus DSM 101000 T were 78.22% and 21.00%, respectively, which were in the range of the recommended level for interspecies identity. The results of phenotypic, chemotaxonomic and genotypic analyses clearly indicated strain GXH0341T represents a novel species of the genus Bacillus, for which the name Bacillus pinisoli sp. nov. is proposed. The type strain is GXH0341T (= MCCC 1K07157T = JCM 35212 T).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Bacillus, first described by Cohn in 1872 [1], are composed of Gram-positive, rod shaped, aerobic or facultatively anaerobic and endospore-forming bacteria of the family Bacillaceae within the phylum Firmicutes. As the time went by, more and more bacteria were found and classified into this family/genus. Castellani and Chalmers assigned the tribe Bacteroideae, Haemophileae, Pasteurelleae and Proteeae to this family [2]. Alves et al. assigned the genus Nosocomiicoccus [3]. Krishnamurthi et al. assigned the genus Psychrobacillus [4]. Zhao et al. assigned the genus Lysinibacillus [5] and so on. Among the many criteria of classification, one of the standards for the description of new taxa in this group were published in 2009 according to Logan et al. [6] which proposed minimal standards for describing this genus based on their ability to form spores in the presence of oxygen. As a result, the member of the genus Bacillus became large and chaotic, and had been recognized taxonomically inoperable and in need for reclassification. Therefore, in order to clarify the evolutionary relationships and classification of Bacillus species, comprehensive phylogenomic and comparative analyses were performed on Bacillus/Bacillaceae genomes according to Gupta et al. [7], and they arranged most of the Bacillus clades, in addition to the Subtilis and Cereus clades, in 17 novel distinct clades. As a consequence, the genus Bacillus was restricted to only the members of the Subtilis and Cereus clades. However, despite the detailed classification, a number of strains are still classified as ambiguous genera, and are not recommended as new genera. In the course of investigating the bacteria diversity of Weizhou Island, Beihai, Guangxi, China, a novel strain GXH0341T, which was isolated from the soil associated with decayed pine tree and assessed by the analysis of phenotype and genotype.
Materials and Methods
Strain Isolation and Culture Conditions
Strain GXH0341T was isolated from the soil associated with decayed pine tree samples which were collected on Weizhou Island, Beihai, Guangxi, China (21°05′ 31.71″ N, 109°14′05.33″ E), and the samples were placed in a sterile sampling bag, stored at 4 °C and transported back to the laboratory. 2 g of soil samples were added to 20 mL of 0.85% NaCl (w/v) solution and incubated in 30 °C shaking for 1 h, and used it as a reserve solution and diluted to 10–2, 10–3, 10–4 and 10–5 with sterile distilled water. Various dilutions were coated on 2216E agar medium (MA) and incubated at 30 °C for 72 h. The GXH0341T was isolated by the method described above and stored in a glycerol suspension (35%, v/v) and stored in − 20 °C. Bacillus mesophilus (= CCTCC AB 2015209 T) was obtained from China General Microbiological Culture Collection Center (CGMCC), and the strain was incubated on MA medium and used as reference strain for all experiments.
16S rRNA Gene Sequencing and Phylogenetic Analysis
DNA was extracted using the chelex-100 method [8] and PCR amplification of the 16S rRNA gene was performed as described by Chun et al. [9]. The 16S rRNA gene sequence was determined by Sangon Biotech (Guangzhou, PR China). The 16S rRNA gene sequence of GXH0341T was identified through the EzBioCloud database (https://www.ezbiocloud.net) [10]. The Clustal W algorithm was used in MEGA 11 to perform multiple alignments of sequences. The neighbor-joining [11], maximum-likelihood [12] and maximum-parsimony [13] methods were used to reconstruct phylogenetic trees by MEGA software [14]. The evolutionary distances were computed by the Kimura two-parameter method [15]. The stability of the phylogenetic tree topology was evaluated using the bootstrap method, with a total of 1000 replicates [16].
Genomic Analysis
In order to further genotypic analysis, the whole genomic DNA of strain GXH0341T was sequenced using the Illumina Hiseq 4000 (Wekemo Tech Co., Ltd. Shenzhen China). The quality control of the raw data was carried out by Fastp v.0.20.1 software [14], and finally the clean data was assembled into genome sequence by Unicycler v.0.4.8 [15]. The estimated digital DNA-DNA hybridization (dDDH) values were calculated using the genome-to-genome distance calculator GGDC 3.0 (http://ggdc.dsmz.de/) [17]. The average nucleotide identity (ANI) was analyzed by JSpeciesWS online service (http://jspecies.ribohost.com/jspeciesws/) [18]. The genome sequence was annotated by the NCBI Prokaryote Genome Annotation Pipeline (PGAP). The draft genome of GXH0341T was examined using the antiSMASH 6.0 (https://antismash.secondarymetabolites.org/) server to detect putative biosynthetic gene clusters [19]. The phylogenomic tree was constructed through the PATRIC website (https://www.patricbrc.org/) [20].
Physiology and Chemotaxonomy
After culturing on MA for 72 h at 30 °C, the cell morphology of the strain GXH0341T was observed using a scanning electron microscopy (SUPRA 55 Sapphire, carl AEISS) and transmission electron microscope (HT-7700, Hitachi). Spores were observed with carbol-fuchsin solution. Oxidase activity was measured by 1% (w/v) N, N, N', N'-Tetramethyl-p-phenylenediamine dihydrochloride and oxidized by 3% (v/v) hydroperoxide solution [21]. Based on MA medium, growth was observed at different temperatures (0, 4, 15, 25, 30, 37, 45 °C) and NaCl (0–10% w/v, at intervals of 1% NaCl). To determine the range of growth pH (4.0–12.0, at intervals of 1.0 pH unit), we adjusted the pH value using HCl or NaOH and then determined the optimal pH growth value by measuring the value of optical density in 600 nm (OD600) such as Xu et al. [22]. According to Gonzalez et al. [23], we hydrolysed gelatin, starch, and Tweens (20, 60, and 80). GXH0341T was inoculated to MB and cultured on a shaker at 30 °C for 12 h, 75 µL of the bacterial solution was spread on MA, placed in an anaerobic generating bag produced by Merck (Anaerocult A mini) and cultured at 30 °C for 7 days for anaerobic growth test. The motility of the strain was determined using a motility medium (4 g/L agar). Gram staining was carried out according to the method of Allen [24]. Biolog’s Gen III ecological board was used to test the carbon utilization of strains. The test panels were performed strictly according to their respective instructions. Susceptibility test discs were used to determine the sensitivity of the strains to antibiotics containing the following amounts (μg per tablet): ampicillin (10), kanamycin (30), amoxicillin (10), cefoxitin (30), carbenicillin (100), bacitracin (0.04), gentamicin (10), tetracycline (30), chloramphenicol (30), streptomycin (10), erythromycin (15), rifampin (5), neomycin (30), streptomycin (300). Other tests were performed using API ZYM, API 50 CH, and API 20NE test strips (bioMérieux), according to the manufacturer's instructions.
Chemotaxonomic Characteristics
For determination of chemotaxonomic characteristics, strain GXH0341T and reference strain Bacillus mesophilus DSM 101000 T were grown in MB medium for 3 days at 30 °C, harvested by centrifugation, washed with distilled water and freeze-dried. The quinone component was analyzed by menaquinone system using high performance liquid chromatography (HPLC) as described by Collins et al. [25]. The cell-wall amino acids compositions were analyzed as described by Schleifer et al. [26]. The polar lipids were detected by solvent systems using thin-layer chromatography (TLC) [27]. The fatty acid composition was measured according to the instructions of the Sherlock Microbial Identification System (MIDI, version 6.0) [28].
Results and Discussion
Morphological, Physiological and Biochemical Characteristics
The colonies of strain GXH0341T were pink, rounded, with a convex and smooth surface. The cell of strain GXH0341T was Gram-stain-positive, rod-shaped approximately 0.5–0.7 μm wide and 2.2–2.6 μm long (Fig. S1) with multiple flagella (Fig. S2) and could produce oval spores outside the cells (Fig. S3). The strain was motile, facultatively anaerobic and its peroxidase was positive, but the oxidase activity was negative. The strain GXH0341T could grow at 4–37 °C (optimum 30 °C), at pH 5.0–11.0 (optimum 8.0) and in the presence of 0–7% (w/v) NaCl (optimum 2%). The Voges-Proskauer test was negative.
The experimental results indicated that strain GXH0341T could hydrolyse Tween20, Tween60, and Tween 80. In API 50CH strips, inositol, fructose, potassium 5-ketogluconate were positive. In API ZYM strips, alkaline phosphatase, esterase (C4), esterase lipase (C8), leucine arylamidase, valine arylamidase, trypsin, α-chymotrypsin, acid phosphatase, naphthol-AS-BI-phosphohydrolase, α-galactosidase, ß-galactosidase, α-glucosidase, ß-glucosidase were positive. Biochemical assays with an API 20NE kits revealed that d-glucose, esculin, 4-nitrophenyl-β-d-galactopyranoside were utilized. GEN III stripes indicated that l-alanine, glucuronamide, acetoacetic acid, l-malic acid, 1% sodium lactate, fusidic acid, d-serine, troleandomycin, rifamycin SV, minocycline, lincomycin, guanidine HCl, niaproof 4, vancomycin, nalidixic acid, lithium chloride, potassium tellurite, aztreonam, sodium butyrate, sodium bromate could be used as carbon sources. And susceptibility test discs experiments indicated GXH0341T were sensitive to antibiotics containing the following (μg per tablet): gentamicin (10), tetracycline (30), chloramphenicol (30), streptomycin (10), erythromycin (15), rifampin (5), neomycin (30), streptomycin (300), ampicillin (10), kanamycin (30), amoxicillin (10), carbenicillin (100), bacitracin (0.04) but not sensitive to cefoxitin (30). The comparison of the physiological characteristics of its closely members strains in different experiments are shown in Table 1, besides, all the negative results of test strips are shown in supplementary material Table S5.
16S rRNA Gene Sequencing and Phylogenetic Analysis
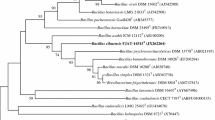
The complete 16S rRNA gene sequence of GXH0341T was determined (1506 bp, GenBank/EMBL/DDBJ accession number OL336472). Based on 16S rRNA gene sequence analyzed of GXH0341T showed the highest sequence similarity with Bacillus mesophilus DSM 101000 T (98.90%), followed by Bacillus salitolerans KC1T (96.95%) and Margalitia shackletonii DSM 18435 T (96.67%). Phylogenetic analysis using the neighbor-joining algorithm revealed that strain GXH0341T represented a separate lineage within the genus Bacillus (Fig. 1), and the phylogenetic position was also confirmed by the trees generated using the maximum-likelihood (Fig. S4) and maximum-parsimony (Fig. S5) methods. For further comparative observation, the phylogenetic tree with the addition of the core Bacillus species is shown in (Fig. S8). As the strain which we report a novel bacterium in this study and the most similar strains B. mesophilus DSM 101000 T, Bacillus salitolerans KC1T are all defined as members of ambiguous genera and do not belong to any of the genera as defined by Gupta et al. [5]. Therefore, this novel bacterium GXH0341T was temporarily classified as a member of Bacillus.
The neighbor-joining tree based on the 16S rRNA gene sequences showing the phylogenetic relationship of strain GXH0341T with related taxa. The evolutionary distances were computed using the Kimura 2-parameter method. The sequence of Brevibacillus brevis NBRC 15304 T (AB271756) was used as an outgroup. Bootstrap support values were calculated from 1000 replicates and only values above 50% were shown. Bar, 0.01 substitutions per nucleotide position
Genomic Analysis
The genome sequence of GXH0341T, which was deposited in the GenBank/EMBL/DDBJ database under accession number JAJQI000000000, was used for further analysis. The genome size of GXH0341T was about 4,004,187 bp, it contained 38 number of contigs, 302 number of subsystems, 4100 number of coding sequences and 111 number of RNAs. The N50 value was 242,785 bp, N75 value was 141,909 bp, and the average genome coverage was 149.72 × . The longest and the shorted contig size of the whole genome were 446,877 bp and 1311 bp, respectively. The results annotated by the antiSMASH 6.0 database showed that GXH0341T had one Type III Polyketide synthase (T3PKS) which involved in the biosynthesis of alkyl-O-dihydrogeranyl-methoxyhydroquinones. The genome of GXH0341T contains one terpene type of secondary metabolite biosynthesis gene cluster, with a similarity of 33% to synthesize carotenoid. It is also consistent with the phenotypic feature that the colonies color of GXH0341T is pink. In addition, GXH0341T also had two secondary metabolite biosynthesis gene clusters which were RRE-element containing cluster (RRE-containing), and Lasso peptide cluster (lassopeptide), which may involve in biosynthesis and regulatory responses, while no biosynthetic gene cluster was found in Bacillus mesophilus DSM 101000 T. Some genomic information was compared between GXH0341T and B. mesophilus DSM 101000 T (Table 2). The DNA G + C content was 37.0 mol%, which was slightly higher than those of strain B. mesophilus DSM 101000 T which of DNA G + C content was 36.8 mol% [29]. Based on JSpeciesWS online service, the ANI value between GXH0341T and B. mesophilus DSM 101000 T was 78.22%, which were less than the suggested cut-of 95–96% ANI value for species delineation [30]. The dDDH value between GXH0341T and B. mesophilus DSM 101000 T was 21.00%, which was clearly below the 70% cut-off for species delineation. The ANI and dDDH values between GXH0341T and the related type strain were seen in Table 3. The phylogenomic tree (Fig. S6) confirmed the relationship between GXH0341T and B. mesophilus DSM 101000 T. Based on above results we proposed the view temporarily that strain GXH0341T represents a novel species of the genus Bacillus.
Chemotaxonomic Characteristics
The predominant quinone of strain GXH0341T was found to be MK-7 and the peptidoglycan contained meso-diaminopimelic acid as the diagnostic diamino acid. The polar lipids of GXH0341T were found to be included diphosphatidylglycerol (DPG), phosphatidylethanolamine (PE), phosphatidylglycerol (PG), phosphatidylcholine (PC), and two unidentified phospholipids (Fig. S7) while Bacillus mesophilus DSM 101000T contained diphosphatidylglycerol, phosphatidylglycerol, phosphatidylethanolamine, phosphatidylcholine, two aminophospholipids (APL) and seven unidentified phospholipids [29]. The major fatty acid of strain GXH0341T were iso-C14:0 (23.8%), iso-C15:0 (18.5%), anteiso-C15:0 (18.2%) and iso-C16:0 (21.4%), which were similar to the reference strain B. mesophilus DSM 101000T. Besides, the content of iso-C14:0 in strain GXH0341T was much higher than in the reference strain B. mesophilus DSM 101000T while the other strains showed small differences (shown in supplement material Table 4).
Taxonomic Conclusion
Based on 16S rRNA gene sequencing, the strain GXH0341T formed a cluster with Bacillus mesophilus DSM 101000 T of the genus Bacillus. ANI and DDH values between strain GXH0341T and its closely relative Bacillus species were both much lower than the species definition threshold value (95–96% for ANI and 70% for DDH). The phenotypic, physiological, chemotaxonomic, and genetic comparative analysis of strain GXH0341T supported that it is a novel species of the genus Bacillus. Therefore, strain GXH0341T is considered to be a novel Bacillus species, and proposed as B. pinisoli sp. nov.
Description of Bacillus pinisoli sp. nov.
Bacillus pinisoli (pi.ni.so'li. L. fem. n. pinus pine; L. neut. n. solum soil; N.L. gen. n. pinisoli of pine soil).
Cells are Gram-stain-positive, rod-shaped approximately 0.5–0.7 μm wide and 2.2–2.6 μm long, with multiple flagella, facultatively anaerobic, motile and spore-forming. On MA, colonies are pink, rounded, with a convex and smooth surface. Growth occurs at 4–37 °C (optimum 30 °C), at pH 5.0–11.0 (optimum 8.0) and in the presence of 0–7% (w/v) NaCl (optimum 2%). Peroxidase was positive, but the oxidase activity and the Voges–Proskauer test was negative. Hydrolysis of Tween20, Tween 60, and Tween 80 are all positive. In API 50CH strips, inositol, fructose, potassium 5-ketogluconate are positive. In API ZYM strips, alkaline phosphatase, esterase (C4), esterase lipase (C8), leucine arylamidase, valine arylamidase, trypsin, α-chymotrypsin, acid phosphatase, naphthol-AS-BI-phosphohydrolase, α-galactosidase, ß-galactosidase, α-glucosidase, ß-glucosidase are positive. Biochemical assays with API 20NE kits reveal that d-glucose, esculin, 4-nitrophenyl-β-d-galactopyranoside are utilized. GEN III stripes indicate that l-alanine, glucuronamide, acetoacetic acid, l-malic acid, 1% sodium lactate, fusidic acid, d-serine, troleandomycin, rifamycin SV, minocycline, lincomycin, guanidine HCl, niaproof 4, vancomycin, nalidixic acid, lithium chloride, potassium tellurite, aztreonam, sodium butyrate, sodium bromate can be used as carbon sources. Sensitive to antibiotics containing the following (μg per tablet): gentamicin (10), tetracycline (30), chloramphenicol (30), streptomycin (10), erythromycin (15), rifampin (5), neomycin (30), streptomycin (300), ampicillin (10), kanamycin (30), amoxicillin (10), carbenicillin (100), bacitracin (0.04) but not sensitive to cefoxitin (30). The predominant quinone was MK-7 and the cell-wall diagnostic diamino acid was meso-diaminopimelic acid. The predominant polar lipids are diphosphatidylglycerol (DPG), phosphatidylethanolamine (PE), phosphatidylglycerol (PG), phosphatidylcholine (PC), and two unidentified phospholipids. The major fatty acid (> 5%) are iso-C14:0, iso-C15:0, anteiso-C15:0 and iso-C16:0. The DNA G + C content of the type strain is 37.0 mol%.
The type strain GXH0341T (= MCCC 1K07157T = JCM 35212 T), was isolated from the soil of a decayed pine tree samples in Weizhou Island, Beihai, Guangxi, China.
References
Skerman VBD, McGowan V, Sneath PHA (1980) Approved lists of bacterial names. Int J Syst Bacteriol 30:225–420. https://doi.org/10.1099/00207713-30-1-225
Castellani A, Chalmers AJ (1919) Manual of tropical medicine, 3rd ed. Williams, Wood and Co., New York. https://doi.org/10.1097/00007611-191010000-00018
Alves M, Nogueira C, de Magalhaes-Sant’ana A, Chung AP, Morais PV, da Costa MS (2008) Nosocomiicoccus ampullae gen. nov., sp. nov., isolated from the surface of bottles of saline solution used in wound cleansing. Int J Syst Evol Microbiol 58:2939–2944. https://doi.org/10.1099/ijs.0.65753-0
Krishnamurthi S, Ruckmani A, Pukall R, Chakrabarti T (2010) Psychrobacillus gen. nov. and proposal for reclassification of Bacillus insolitus Larkin & Stokes, 1967, B. psychrotolerans Abd-El Rahman et al., 2002 and B. psychrodurans Abd-El Rahman et al., 2002 as Psychrobacillus insolitus comb. nov., Psychrobacillus psychrotolerans comb. nov. and Psychrobacillus psychrodurans comb. nov. Syst Appl Microbiol 33:367–373. https://doi.org/10.1016/j.syapm.2010.06.003
Zhao F, Feng Y, Chen R, Zhang J, Lin X (2015) Lysinibacillus alkaliphilus sp. nov., an extremely alkaliphilic bacterium, and emended description of genus Lysinibacillus. Int J Syst Evol Microbiol 65:2426–2431. https://doi.org/10.1099/ijs.0.000280
Logan NA, Berge O, Bishop AH, Busse H-J, De Vos P et al (2009) Proposed minimal standards for describing new taxa of aerobic, endospore-forming bacteria. Int J Syst Evol Microbiol 59:2114–2121. https://doi.org/10.1099/ijs.0.013649-0
Gupta Radhey S, Sudip P, Navneet S, Shu C (2020) Robust demarcation of 17 distinct Bacillus species clades, proposed as novel Bacillaceae genera, by phylogenomics and comparative genomic analyses: description of Robertmurraya kyonggiensis sp. nov. and proposal for an emended genus Bacillus limiting it only to the members of the Subtilis and Cereus clades of species. Int J Syst Evolut Microbiol 70(11):5753–5798. https://doi.org/10.1099/ijsem.0.004475
de Lamballerie, Zandotti C, Vignoli C, Bollet C, de Micco P (1992) A one-step microbial DNA extraction method using Chelex 100 suitable for gene amplification. Res Microbiol 143:785–790. https://doi.org/10.1016/0923-2508(92)90107-Y
Chun J, Goodfellow M (1995) A phylogenetic analysis of the genus Nocardia with 16S rRNA gene sequences. Int J Syst Bacteriol 45(2):240–245. https://doi.org/10.1099/00207713-45-2-240
Yoon SH, Ha SM, Kwon S, Lim J, Kim Y (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol 67:1613–1617. https://doi.org/10.1099/ijsem.0.001755
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. https://doi.org/10.1093/oxfordjournals.molbev.a040454
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376. https://doi.org/10.1007/BF01734359
Fitch WM (1971) Toward defining the course of evolution: minimum change for a specific tree topology. Syst Biol 20:406–416. https://doi.org/10.1093/sysbio/20.4.406
Tamura K, Stecher G, Kumar S (2021) MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol 38(7):3022–3027. https://doi.org/10.1093/molbev/msab120
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16(2):111–120. https://doi.org/10.1007/BF01731581
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791. https://doi.org/10.1007/BF01734359
Meier-Kolthoff JP, Sardà Carbasse J, Peinado-Olarte RL, Göker M (2022) TYGS and LPSN: a database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acid Res 50:D801–D807. https://doi.org/10.1093/nar/gkab902
Richter M, Rosselló-Móra R, Oliver Glöckner F, Peplies J (2016) JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32(6):929–931. https://doi.org/10.1093/bioinformatics/btv681
Blin K, Shaw S, Kloosterman AM, Charlop-Powers Z, van Wezel GP et al (2021) AntiSMASH 6.0: improving cluster detection and comparison capabilities. Nucleic Acids Res 49(W1):W29–W35. https://doi.org/10.1093/nar/gkab335
Wattam AR, Davis JJ, Assaf R, Boisvert S, Brettin T et al (2017) Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res 45:D535–D542. https://doi.org/10.1093/nar/gkw1017
Brown AE (2007) Benson’s microbiological application laboratory manual in general microbiology, 10th edn. McGraw- Hill, New York. https://doi.org/10.1890/0012-9623(2004)85[173b:EA]2.0.CO;2
Kovacs N (1956) Identification of Pseudomonas pyocyaneaby the oxidase reaction. Nature 178:703–704. https://doi.org/10.1038/178703a0
Gonzalez C, Gutierrez C, Ramirez C (1978) Halobacterium vallismortis sp. nov. an amylolytic and carbohydrate-metabolizing, extremely halophilic bacterium. Can J Microbiol 24:710–715. https://doi.org/10.1139/m78-119
Allen JL (1992) A modified Ziehl-Neelsen stain for mycobacteria. Med Lab Sci 49(2):99–102
Collins MD, Pirouz T, Goodfellow M, Minnikin DE (1977) Distribution of menaquinones in actinomycetes and corynebacteria. J Gen Microbiol 100:221–230. https://doi.org/10.1099/00221287-100-2-221
Schleifer KH (1985) Analysis of the chemical composition and primary structure of murein. Methods Microbiol 18(08):123–156. https://doi.org/10.1016/S0580-9517(08)70474-4
Minnikin DE, O’Donnell DA, Goodfellow M, Alderson G, Athalye M, Schaal A, Parlett J (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2(5):233–241. https://doi.org/10.1016/0167-7012(84)90018-6
Sasser M, Kunitsky C, Jackoway G, Ezzell JW, Teska JD (2005) Identification of Bacillus anthracis from culture using gas chromatographic analysis of fatty acid methyl esters. J AOAC Int 88:178–181. https://doi.org/10.1093/jaoac/88.1.178
Zhou Y, Liu G, Liu B, Chen G, Du Z (2016) Bacillus mesophilus sp. nov., an alginate-degrading bacterium isolated from a soil sample collected from an abandoned marine solar saltern. Antonie Van Leeuwenhoek 109:937–943. https://doi.org/10.1007/s10482-016-0692-7
Chun J, Oren A, Ventosa A, Christensen H, Arahal DR, da Costa MS, Rooney AP (2018) Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol 68:461. https://doi.org/10.1099/ijsem.0.002516
Zhang WY, Hu J, Zhang XQ, Zhu XF, Wu M (2015) Bacillus salitolerans sp. nov., a novel bacterium isolated from a salt mine in Xinjiang province, China. Antonie Van Leeuwenhoek 108(2):443–51. https://doi.org/10.1007/s10482-015-0497-0
Acknowledgements
We thank Marine Culture Collection of China (MCCC) and Japan Collection of Microorganisms (JCM) for strain preservation, as well as members of the Guangxi Key Laboratory for Polysaccharide Materials and Modifications and Yunnan Institute of Microbiology of Yunnan University for helpful discussions.
Funding
This research was funded by the Science and Technology Major Project of Guangxi (AA18242026), the Science and Technology Major Project of Guangxi (AB21196020) and the National Natural Science Foundation of China (81960614).
Author information
Authors and Affiliations
Contributions
MJ and YJ designed the study; YH, HC and SQ performed research; YZ, JW, XC and YNI analyzed data. Ying Huang wrote the manuscript. All of the authors contributed to the manuscript revision process and read and approved the submitted version.
Corresponding authors
Ethics declarations
Conflict of interest
All authors declare that they have no conflicts of interest.
Ethical Approval
This article does not contain any studies with human participants performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The GenBank/EMBL/DDBJ accession numbers for the 16S rRNA gene and the genome sequences of strain GXH0341T are OL336472 and JAJQKI000000000, respectively. Strain GXH0341T has been deposited in Marine Culture Collection of China (MCCC) and Japan Collection of Microorganisms (JCM), the preservation number are MCCC 1K07157 and JCM 35212, respectively.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Huang, Y., Cai, H., Qin, S. et al. Bacillus pinisoli sp. nov., Isolated from Soil of a Decayed Pine Tree. Curr Microbiol 80, 55 (2023). https://doi.org/10.1007/s00284-022-03130-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-022-03130-x