Abstract
Finite element (FE) models of the brain are crucial for investigating the mechanisms of traumatic brain injury (TBI). However, FE brain models are often limited to a single neuroanatomy because the manual development of subject-specific models is time consuming. The objective of this study was to develop a pipeline to automatically generate subject-specific FE brain models using previously developed nonlinear image registration techniques, preserving both external and internal neuroanatomical characteristics. To verify the morphing-induced mesh distortions did not influence the brain deformation response, strain distributions predicted using the morphed model were compared to those from manually created voxel models of the same subject. Morphed and voxel models were generated for 44 subjects ranging in age, and simulated using head kinematics from a football concussion case. For each subject, brain strain distributions predicted by each model type were consistent, and differences in strain prediction was less than 4% between model type. This automated technique, taking approximately 2 h to generate a subject-specific model, will facilitate interdisciplinary research between the biomechanics and neuroimaging fields and could enable future use of biomechanical models in the clinical setting as a tool for improving diagnosis.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Traumatic brain injuries (TBI) are a significant and costly public health issue. Recent epidemiological studies have estimated that TBI account for approximately one third of all injury-related deaths in the United States.10 However, despite major scientific pushes to reduce their societal cost, the incidence of TBI-related injuries and deaths continues to rise.16
Scientists and clinicians have studied TBI using modern imaging modalities to improve diagnosis and gain understanding of the short and long term consequences.36,51 Magnetic resonance imaging (MRI) is a technique used to diagnose macrostructural changes associated with severe TBI (e.g., hematoma, contusion, axonal injury), but the microstructural injuries associated with mild TBI (mTBI or concussion) present without visible change to the brain parenchyma,28,44 Consequently, mTBI imaging research has shifted towards examining patients with other quantitative MRI techniques, such as functional MRI (fMRI)26,36,49 and diffusion tensor imaging (DTI).9,37,59 Much of this neuroimaging research is based on statistical models that do not consider the spatial heterogeneity of mTBI,46 which is necessary to advance quantitative brain imaging research toward utility in individual patients.
In the biomechanics field, finite element (FE) models of the brain have been the gold standard tool for investigating TBI injury mechanisms.8,35,47,58 They have also been used to develop and evaluate injury criteria and exposure thresholds,19,48,53 which relate measured head kinematics to risk of brain injury. Furthermore, FE brain models have been used to assess the efficacy of safety gear and injury mitigation systems.12,13,50 Today’s state-of-the-art FE brain models generally include a detailed representation of the neuroanatomy obtained from medical imaging and sophisticated constitutive models derived from experimental material testing of the brain parenchyma.33
One of the major limitations of current brain FE models is that these models have generally been created to match the anthropometry of a single, representative person within the population, and historically this has been a 50th percentile male adult.29,34,53,60 This presents a significant limitation as structural differences between two brains under similar impact conditions may lead to different mechanical responses of the brain. As a result, little is known about the effects of anatomical changes to the brain that occur naturally with development and aging on brain biomechanics. Limited morphing methods have been developed and utilized to create subject-specific brain models that are derived by inner cranial skull shape,57,58 but these methods do not capture internal neuroanatomical differences. Alternatively, brain models could be developed on a subject-by-subject basis, but creating new models is very time-consuming, making this approach infeasible for studies using many subjects.
While the fields of neuroimaging and biomechanics have made significant contributions toward the understanding of TBI, both disciplines suffer from limitations the other could potentially address. Biomechanical analyses may improve the power of advanced neuroimaging techniques by predicting regions of interest (ROI) based on the impact conditions sustained by the patient. For instance, Wilde et al. defined the corpus callosum as a ROI in their investigation of microstructural changes to adolescent brains following mild TBI. This was largely informed by a prior study that utilized a FE brain model to investigate brain deformation patterns sustained during concussive impacts.55,56 Conversely, neuroimaging techniques may improve the accuracy of biomechanical models by providing the tools necessary to rapidly create patient-specific models for individual risk assessment.
Image registration is an industry-standard in neuroimaging research. Image registration is the process by which two anatomical images (typically T1-weighted MRI) are geometrically aligned through a combination of linear and nonlinear transformations.3 These registration pipelines are commonly used to map subject-specific results to a common brain anatomy (usually a representative template brain), which normalizes for individual anatomical differences and facilitates statistical analyses within a group. As such, image registration has become a fundamental step in most image processing pipelines.54 We hypothesize that these image registration transformations can be utilized to generate subject-specific brain models.
The objective of this study was to develop an automated process that leverages image registration algorithms to rapidly and accurately generate subject-specific FE models of the brain using the subject’s structural MRI data. The ability to generate subject-specific FE brain models with high throughput will enable direct comparisons between subject-specific biomechanical and imaging analyses and will lend insight into how individual neuroanatomical characteristics affect subject-specific brain deformation patterns.
Methods
Image Registration and Transformations
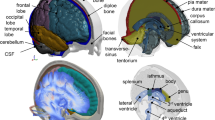
The foundation of the registration-based morphing (RBM) pipeline is image registration (or geometric normalization; Fig. 1). In short, image registration is the process by which a “moving” image I is geometrically aligned with a “fixed” image J.54 This is performed by determining the optimal transformation that transforms each voxel, \(\varvec{x}\), in the moving image, \(I\left( \varvec{x} \right)\), to the corresponding voxel, \(\varvec{y}\), in the fixed image, \(J\left( \varvec{y} \right)\), by minimizing some cost function that describes the differences between \(I\left( \varvec{x} \right)\) and \(J\left( \varvec{y} \right)\).5 Registration is crucial for creating brain templates and is also utilized to perform automated segmentation of brain tissues and quantify anatomical differences within and across subjects.3,6 This process is fundamental to many neuroimaging studies, with a common application being mapping subject imaging results to a common space, usually a brain template, to facilitate inter-subject comparisons.
Image registration operations. The affine transformation step globally translates, rotates, and shears the moving image. The elastic transformation step applies voxel-level deformations to geometrically align the moving image with the fixed image. The fixed image is usually a population template image.
Generally, image registration is a two-stage process. The first stage consists of an affine transformation, \(\Phi\), that operates to globally align the moving image \(I\left( \varvec{x} \right)\) with the fixed image \(J\left( \varvec{y} \right)\). This affine transformation operates to translate, rotate, scale, and shear the moving image.5 These global transformation operations are illustrated in Fig. 1.
where \(I^{\prime}\left( \varvec{x} \right)\) is the affine transformed image. Following initial alignment, the second stage is a non-linear transformation, \({\mathbf{\Psi }}\), that deforms each voxel \(\varvec{x}\) in image \(I^{\prime}\left( \varvec{x} \right)\) to the corresponding voxel \(\varvec{y}\) in \(J\left( \varvec{y} \right)\).
where \(I^{\prime\prime}\left( \varvec{x} \right)\) is the registered image and \(\delta \left( \varvec{y} \right)\) is the registration error. For this study, the Advanced Normalization Tools (ANTs) software package was used to compute all registrations.3,5 Specifically, the diffeomorphic SyN algorithm, which preserves image topology, was used to ensure that the transformations were symmetric.4,5
Registration-Based Morphing
Consider a FE model \(\hat{I}\left( {\hat{\varvec{x}}} \right)\), composed of nodes \(\hat{\varvec{x}}\), that corresponds directly to image \(I\left( \varvec{x} \right)\). In this case, the objective is to morph the model \(\hat{I}\left( {\hat{\varvec{x}}} \right)\) to the geometry depicted in the fixed image, \(J\left( \varvec{y} \right)\), by applying the registration transformations required to align images \(I\left( \varvec{x} \right)\) and \(J\left( \varvec{y} \right)\). The ANTs registration algorithm outputs 15 affine transformation parameters. The first 9 parameters, a–i, form a 3 × 3 matrix, \({\varvec{\Phi}}_{\varvec{M}}\) that defines the rotation, scaling, and shearing components of the affine transformation. The remaining 6 parameters, m–r, relate to the 3 × 1 translation vector \({\varvec{\Phi}}_{\varvec{t}}\). Matrices \({\varvec{\Phi}}_{\varvec{M}}\) and \({\varvec{\Phi}}_{\varvec{t}}\) can be combined into a 4 x 4 affine transformation matrix, \({\varvec{\Phi}}\), that operates to rotate, scale, shear, and translate the set of nodes \(\hat{\varvec{x}}\), resulting in an affine transformed model \(\hat{I}\left( {\varvec{\hat{x}^{\prime}}} \right)\).
However, when applying an arbitrary transformation Λ that registers image \(I\left( \varvec{x} \right)\) to \(J\left( \varvec{y} \right)\), the inverse transformation, Λ−1, must be used to transform nodes \(\hat{\varvec{x}}\) in FE model \(\hat{I}\left( {\hat{\varvec{x}}} \right)\).4
where, \(\varvec{\hat{x}^{\prime}}\) are the affine transformed nodes in model \(\hat{I}\left( {\varvec{\hat{x}^{\prime}}} \right)\).
To complete the RBM algorithm, the nonlinear transformation \({\varvec{\Psi}}\) is applied to the affine transformed model, \(\hat{I}\left( {\varvec{\hat{x}^{\prime}}} \right)\). The nonlinear transformation is a 4-dimensional matrix that is composed of 3D x-, y-, and z-deformation fields, where in each 3D image, the intensity of each voxel represents the applied x, y, or z deformation at the centroid of the voxel (Fig. 2). Since the transformation is being applied to nodes (i.e., points), the inverse nonlinear transformation must be applied to the affine transformed nodes.
(a) Depiction of the 3D deformation fields used in the non-linear transformation step. For the highlighted voxel, the non-linear deformation applied during registration is \(u\left( \varvec{x} \right)\) = [5.7, 0.1, 4.4] mm. (b) The nonlinear deformation field can be depicted as a 3D deformation vector field. (c) From the deformation vector field, the Jacobian determinant image can be computed. Dark values indicate contraction and bright values indicate expansion, relative to the fixed image. This nonlinear deformation field corresponds to the moving image depicted in Fig. 1. This subject was chosen for depiction purposes only and was not included in the analyses performed in this study.
where, \(u\) is the applied 3D deformation for each voxel \(\varvec{x}\). To apply the 3D deformation field, defined at the centroid of each voxel, a linear interpolation function was used to determine the corresponding deformation at each affine transformed node \(\varvec{\hat{x}^{\prime}}\).
where, \(\varvec{\hat{x}^{\prime\prime}}\) is the fully morphed model that corresponds to image \(J\left( \varvec{y} \right)\).
Template Image and Brain Model
A template brain image was constructed from T1-weighted anatomical scans (MPRAGE; 1.0 mm isotropic voxels, TR = 2300 ms, TE = 2.98 ms, TI = 900 ms, flip angle = 9°, GRAPPA factor = 2) obtained from 20 young, healthy male participants (age = 22 ± 3.0 years; height = 177.4 ± 3.5 cm; mass = 78.2 ± 9.5 kg) in a separate study.45 A total of 40 images were used to construct a symmetric template. A symmetric template was chosen to reduce, on average, the anatomical discrepancy between the template image and any subject it is morphed to.17 To force the template to be symmetric, the 40 images consisted of the 20 MRIs from each subject and the same 20 MRIs that were flipped along the left-right (i.e., ear-to-ear) axis. Since the center plane of the subject images was roughly located on the mid-sagittal plane, this flip was approximately a reflection about the mid-sagittal plane.17 The ANTs multivariate template construction tool was used in this study.7 This script uses an iterative nonlinear registration algorithm to determine the average brain anatomy from the 40 included T1 images. During this process, any misalignments induced by the left-to-right flip (i.e., not perfectly reflecting about the mid-sagittal plane) were automatically corrected, resulting in an accurate and symmetric template.17 Henceforth, this template will be referred to as the “CAB-20MSym” template (Center for Applied Biomechanics, N = 20, male, symmetric template). From the CAB-20MSym template, an intracranial volume (ICV) segmentation mask was created and used to extract the ICV from the whole-head MRI template. The ICV segmentation mask was obtained using a combination of automated brain extraction (“antsBrainExtraction.sh” in ANTs5) and manual correction. Finally, the ICV image was segmented to identify cerebrospinal fluid (CSF), grey matter, and white matter using a combination of automated (“antsAtroposN4.sh” in ANTs6) and manual segmentation.22 The resulting template maintained subject image resolution, with 1 mm3 isotropic voxels.
A custom Matlab (Mathworks, Natick, MA) script was used to generate a voxel model of the segmented ICV template image.21 In this process, each CSF, grey matter, and white matter voxel in the segmented image was converted to a corresponding hexahedral element and categorized using its segmented tissue type (Fig. 3). For example, a voxel identified as CSF is converted into a hexahedral element classified as CSF. This process was performed at the resolution of the template image, yielding 1 × 1 × 1 mm voxel elements. Quadrilateral shell elements were created on the external surface of the CSF layer to represent the dura/skull boundary and was made rigid.18,38 Material properties, adapted from Miller et al., were assigned to the corresponding parts. In this study, grey and white matter were modeled with the same linear viscoelastic constitutive model.38
Subject Images and Assessment
To verify the RBM algorithm, the CAB-20MSym template model was morphed to represent the anatomy of 44 subjects. These included the 20 subjects used to construct the template, as well as 24 obtained from the Enhanced Nathan Kline Institute—Rockland Sample (NKI-RS) database.40 The 24 NKI-RS subjects were randomly sampled from 391 scans obtained from male and female subjects between the ages of 21-80 years. All scans were grouped by sex and age (by decade) and two subjects were randomly selected from each group. For each subject, a voxel model, with 1 x 1 x 1 mm elements, was also generated using the same process used for the CAB-20MSym template model (Fig. 3). Subject segmentations were obtained by applying image registration transformations to the CAB-20MSym template segmentation image using a nearest neighbor interpolation. In this study, a total of 88 models were created.
All models were simulated under identical loading conditions using head kinematics from a reconstructed football impact (Case 71, Player 2; Sanchez et al.47). This case had peak linear acceleration and peak angular velocity of 123 g and 37.7 rad/s, respectively, and resulted in a concussion. Head kinematics were prescribed to the rigid shell element dura part that encased the brain model.18 All simulations were performed using the LS-Dyna explicit solver (mpp971R9.1.0 with double precision, LSTC, Livermore, CA, USA). Simulations were run with 20 CPUs.
For each subject, the difference between the morphed and voxel model was quantified. Model results included maximum resultant nodal displacements (MRD), sampled from 1000 nodes located in equivalent locations between the two models for each subject, and maximum principal strain (MPS) measured at voxel and global resolution. As metrics of global deformation, the 95th percentile MRD (across 1000 sampled nodes) and MPS (across all brain elements) were calculated.41 These metrics were chosen as nodal displacements are commonly used for model validation and element strains are used for investigating injury mechanisms.34,39,53,57
Analysis of Model Results
An advantage of the RBM technique is that the nonlinear deformation is applied at the voxel-level. In other words, each voxel is individually deformed to match the corresponding voxel in the fixed image. When applying these voxel-level deformations to a voxel mesh, the previously isotropic voxel elements are distorted. As characteristics of a brain model mesh have been shown to affect model results,21 it is possible that the strain response in a morphed model could deviate from the corresponding voxel model. This could be exacerbated in brains with anatomies that significantly differ from the CAB-20MSym template and require larger deformations during image registration (e.g., if the relative volume of the ventricles in the subject is much larger than the relative volume of the ventricles in the template model).
To assess the effect of RBM-induced mesh distortions, the difference in model response between the morphed and voxel models for each subject were investigated. For global metrics of brain deformation (e.g., 95th percentile MRD and MPS), a linear regression between the morphed and voxel results was performed. To assess differences in the spatial strain distributions between the voxel and morphed models for each subject, a strain difference map was computed. The MPS for each element was mapped back to the template image space and the difference map was generated by taking the absolute value of the difference in MPS between the voxel and morphed model (\({\text{abs}}\left( {{\text{MPS}}_{\text{vox}} - {\text{MPS}}_{\text{morphed}} } \right)\)). These difference maps were used to quantify the mean difference in MPS between the voxel and morphed models and identify regions where differences were greatest. Finally, to assess the effect of mesh distortion on MPS, the difference in MPS between the voxel and morphed model for each element was compared to its log Jacobian. The Jacobian of a voxel represents the volumetric expansion or contraction of that voxel in subject space relative to the corresponding voxel in the template space. For example, a Jacobian value of 2 signifies that during image registration, that voxel expanded by a factor of 2. In the imaging community, the log Jacobian is used to normalize the Jacobian distribution, and is commonly used to quantify brain growth and atrophy in various healthy and disease states.14,15,23,24,27,30,32
Differences in MPS between the voxel and morphed models were also compared to various mesh, imaging, and geometric metrics. Mesh metrics included assessments of mesh quality and characteristics (characteristic element length, scaled Jacobian, aspect ratio). The scaled Jacobian of an element indicates its deviation from the ideal shape (i.e., a hexahedron with 90° corners) and is not to be confused with the Jacobian (or log Jacobian) assessed in medical image analysis. The characteristic length is calculated as the ratio of the volume to the maximum area of an element and the aspect ratio is the ratio of maximum to minimum edge length of an element. For an ideal element, both scaled Jacobian and aspect ratio will be 1. Imaging metrics included assessments of image similarity between the subject and CAB-20MSym template and included cross-correlation (CC), mutual information (MI), mean squares (MS), and global correlations (GC). Details on these similarity metrics can be found in Avants et al.5
Results
Registration-Based Morphing
The CAB-20MSym template had 1,493,500 nodes and 1,543,976 elements. For all 44 subjects, the RBM algorithm was successful in generating a subject-specific model with no user intervention. On average, the time required to morph an individual model was approximately 2 h, of which 90% was consumed by the image registration algorithm. Across all subjects, intracranial volumes ranged from 1120 to 1750 cm3, with a mean volume of 1470 ± 160 cm3, which was similar to the CAB-20MSym template (1440 cm3). Element quality was preserved with average element Jacobian ratio of 0.96 ± 0.01 (range: 0.94–0.97), aspect ratio of 1.25 ± 0.05 (range: 1.14–1.34), and characteristic length of 0.89 ± 0.05 (range: 0.80–0.98) across all morphed models. As a demonstration, exemplary voxel and morphed models for two subjects are compared in Fig. 4. Subject HB-008 was a 20-year old male and subject sub-A00038519 was a 78-year old female. For both subjects, the morphed models accurately captured external and internal anatomical features while preserving mesh quality (Fig. 4).
Global Deformation Analysis
Using the head kinematics from the football concussion case, the 95th percentile values were calculated from the MPS and MRD distributions. Overall, 95th percentile MPS and MRD values ranged from approximately 0.3–0.4 and 9–16 mm. MPS and MRD exhibited a strong linear relationship across the voxel and morphed models, with R2 values of 0.98 and 0.95, respectively (Fig. 5). However, deformation in the morphed models was consistently lower than in the voxel models, with mean differences of 0.01 ± 0.005 strain and 0.36 ± 0.25 mm, respectively. These differences were not related to any measure of mesh quality or image similarity (Fig. 5).
Relationships between 95th percentile MPS (a) and 95th percentile MRD (b) in the voxel and morphed models. Differences in MPS (c) and MRD (d) were not related to the image similarity between the template and subject images, assessed using the mutual information image similarity metric. Similar results were found for the cross-correlation, mean squares, and global correlation image similarity metrics.
Spatial Distribution of Deformation
For each model, MPS measured at each voxel was mapped to the template image space. This facilitated a direct spatial comparison between the voxel and morphed model for each subject, by computing a spatial difference map (\({\text{abs}}\left( {{\text{MPS}}_{\text{vox}} - {\text{MPS}}_{\text{morphed}} } \right)\)). For each voxel, the MPS difference and log Jacobian value were assessed, and no relationship was observed between these metrics. For all subjects, the mean median MPS difference was 0.006 ± 0.004 strain and 95% of all voxels had an MPS difference of less than 0.04 ± 0.01 strain. The largest discrepancies were observed at the brain-CSF boundaries. These included the periventricular surface, sulci, and the regions where the falx and tentorium would be located (Fig. 6). The strains in these areas were consistently larger in the voxel models than in the morphed models, which were all derived from a common template mesh.
Discussion
In this study, a technique for developing subject-specific finite element models using image registration transformations was developed. Unlike traditional biomechanical morphing methods which are driven by external surface differences,42 registration-based morphing is a nonlinear technique that captures both external and internal anatomical features. Subject-specific model results can be mapped to either template or subject space, and these results can be correlated to neuroimaging diagnostics and/or be used to identify regions of interest for investigating neurological function following injury. In the biomechanics field, these models can be utilized to investigate the relationships between neuroanatomy and injury risk and facilitate research across a spectrum of populations.
An important concern when morphing FE models is that the mesh can potentially be heavily distorted, which can result in inaccurate mechanical calculation or model instability. This is especially a concern with RBM as the nonlinear deformations are applied at the voxel-level, which could induce large mesh distortions in regions of significant anatomical variation between the subject and template images. Given the sensitivity of a brain model’s response to the underlying mesh characteristics (element type, resolution, and quality), it was imperative to assess the effect of RBM on the simulated brain deformations.52,61 For the 44 subject images used in this study, which covered a spectrum of neuroanatomies, the morphed and voxel models resulted in similar deformations. Globally, the 95th percentile MPS and MRD demonstrated a strong linear relationship between the voxel and morphed models, with R2 values of 0.98 and 0.95, respectively. Despite this strong trend, the 95th percentile MPS and MRD were marginally lower in the morphed models compared to the voxel models, with mean differences of 0.01 strain and 0.36 mm, respectively. Compared to the magnitude of strain observed in these simulations, which varied between 0.3–0.4, this difference in strain is inconsequential. For the boundary conditions investigated in this study, which represented a concussive American football impact,43,47 the average 95th percentile MPS and MRD in the voxel models were 0.36 ± 0.03 strain and 12.7 ± 1.8 mm, respectively.
Spatially, the largest differences in MPS distributions between the voxel and morphed models were at the CSF-brain boundaries such as the periventricular surface, sulci, and regions proximal to the falx and tentorium (Fig. 6). Strains in these regions were generally larger in the voxel models. During the development of the CAB-20MSym template image set, the brain was segmented using an automated segmentation algorithm.6 However, to ensure that the CSF-brain boundaries were accurately defined, each slice was manually edited to correct for any segmentation inaccuracies and to ensure that the CSF-brain boundaries were accurately represented. Since the CAB-20MSym template was the baseline model, these carefully segmented CSF-brain boundaries were preserved in the RBM-morphed models. To generate the segmentation images that defined the structure of the voxel models, the registration transformations were similarly applied to the CAB-20MSym template segmentation image using a nearest neighbor interpolation, which differed slightly from the linear interpolation used to generate the morphed models. In doing so, the definition of various CSF-brain boundaries was lost, especially in the sulci and areas proximal to the tentorium. Since the CSF was modeled as an elastic fluid material,38 CSF elements served as shear-decoupling structures, resulting in slightly higher deformations in these regions for the voxel models, which resulted in slightly larger MPS in the voxel models. However, since the MPS differences between the morphed and voxel models were small relative to the overall magnitude of deformation in these simulations, these slight discrepancies are negligible. While this could have also contributed to the differences in MRD between models, it is more likely that the differences in MRD were attributed to slight discrepancies between the nodes sampled in the voxel and morphed models.
The most significant advantage of the RBM technique is that the morphing operations are nonlinear and applied at the voxel level, unlike traditional morphing techniques in which all elements are morphed using transformations that are defined by external surface differences alone.42 As such, in RBM both external and internal anatomical features are preserved in the morphed model. As a comparison to traditional surface-based morphing (SBM), the CAB-20MSym template was morphed to sub-A00038519 using SBM. As expected, the external shape of the brain was accurately represented, however, neuroanatomical structures, such as the cortex and ventricles, were not preserved (Fig. 7). Nonetheless, SBM has been used in the literature57,58 and is appropriate in many cases. For instance, SBM is effective in minimizing shape and size discrepancies between a model and post-mortem human surrogate specimen used to obtain experimental data in the model validation process.1,2 This reduces the effect of geometrical error in the model response, providing a more accurate assessment of model biofidelity. Nonetheless, if subject-specific strain distributions are required, or if ROI assessment is needed, SBM is insufficient and a nonlinear morphing technique is required.
Comparison between RBM and traditional surface-based morphing for subject sub-A00038519.42 The largest differences are observed in the ventricles and cortex.
Another advantage is that RBM preserves the mesh topology of the template model. In fact, the RBM method is completely model- and mesh-independent. Although the baseline model used in this study was simplified (e.g., did not include falx or tentorium), complex features included in a template model that would be difficult to implement on a per-subject basis using voxel models, can be morphed. These complex features could include anatomical interfaces, such as the falx and tentorium, meso-scale anatomical structures, such as axonal bundles or cerebrovasculature, or non-anatomical features required for contacts defined in the brain model (e.g., null shells or segment sets needed for implementing the brain-skull boundary condition using contacts). Furthermore, the development of a voxel model is dependent on the availability of a segmented brain image and the anatomical biofidelity of the model is dependent on the accuracy of the segmentation image. While there is ongoing work to automatically segment the falx, tentorium, meninges, and other brain structures, user intervention can often be required to some extent.22 The RBM pipeline streamlines the generation of subject-specific models as image segmentation (automated, manual, or a combination of both) is only required for the baseline template image. Morphing a singular template model also simplifies the model validation process as only a singular template model would require validation. Furthermore, although voxel models were utilized in this study, the RBM pipeline can be used to morph models constructed with other 3D and 2D element types (e.g., hexahedral, tetrahedral, quadrilaterals, and triangles), provided the FE mesh corresponds to the template image. Finally, the entire RBM process, including image registration, took approximately 2 h. This is a vast improvement over the weeks or months required to manually develop, assess, validate, and possibly calibrate a subject-specific brain FE model.
Given these results, RBM is applicable for the variation in brain anatomies across the subjects used in this study. Although several of the registration-induced mesh distortions were moderate, these did not drastically influence the RBM model results as 95% of all voxels had a mean discrepancy less than 0.04 strain, compared to the equivalent voxel model under the loading condition simulated in this study. This impact case was selected as it represented an omnidirectional, real-world, concussive impact with substantial brain deformation. Although magnitude, direction, and duration of head kinematics are important aspects of brain biomechanics, the mesh configuration that arises from morphing the model to a specific subject is not dependent on these factors.
The differences in strain between the voxel and morphed models used in this study were substantially less than those commonly observed between brain models used in the literature. Model discrepancy can arise due to differences in numerical implementation (e.g., mesh size), material properties, and boundary conditions within the brain (e.g., brain-skull boundary).21 For instance, Ji et al. observed MPS values between approximately 0.3–1.5 using three similarly validated models under identical loading conditions, similar in rotational acceleration magnitude to those used in this study.25 Similarly, reducing voxel mesh size from 4 mm isotropic voxels to 1 mm isotropic voxels has been reported to increase the 95th percentile MPS from 0.33 to 0.77.21
It is important to note that applicability of this method from an anatomical perspective was not considered in this study. For instance, the anatomical fidelity between the RBM-morphed model and original MRI was not assessed. These discrepancies are related to image registration error, which has been discussed at length in the literature.11,20,31 Since RBM morphs a baseline template model to the anatomy of a subject, the mesh topology of the template model will be preserved in the morphed model. This may result in an inaccurate morphed model if the subject anatomy differs substantially from the original template model. For instance, the template model developed in this study cannot be used to investigate patients with structural lesions, such as tumors, atrophy, or injury. However, the anatomical applicability of RBM can be expanded by developing additional template images and models that include alternative neuroanatomical representations. Although RBM was originally developed for modeling the human brain, this technique can be easily applied to other body regions, image modalities, and image resolutions, provided image registration transformations can be obtained.
In this study, simplified models were utilized to demonstrate and investigate the applicability of RBM. Across all 44 subjects, 95th percentile MPS and MRD varied from approximately 0.3–0.4 and 9–16 mm, respectively, and were generally correlated to overall brain volume. More importantly, strain distributions varied across subjects, which suggests that strain distribution may be dependent on internal neuroanatomical characteristics. These results suggest that when investigating an individual’s tolerance to brain injury, or comparing biomechanical and imaging results, subject-specific models are necessary. Nonetheless, it must be noted that the deformation results obtained in this study should not be interpreted for injury as the models were simplified and not validated to experimental brain deformation data. Although the material properties were obtained from a previously validated FE brain model,38 this does not ensure model validation.21
Subject-specific brain models have several important applications in the research and clinical spaces. In the research sector, subject-specific brain deformation patterns are easily mapped to either subject or template image space and can be assessed in conjunction with other brain imaging diagnostics for injury. They could also be used to identify regions of high deformation that could be of interest for investigating the neurological effects of TBI. In the biomechanics field, subject-specific models are being used to investigate the relationships between neuroanatomy and brain injury risk and allow for previously underrepresented populations (e.g., pediatric or elderly) to be included in studies. Furthermore, RBM is not limited to human brains. This pipeline can be easily extended to animal models for TBI, which would allow the integration of biomechanics, imaging, and physiology in the pursuit of characterizing TBI using multidisciplinary research tools. Finally, the automated and rapid nature of this technique could enable future use in the clinical setting as a tool for improving diagnosis, especially when paired with wearable sensor data, and determining optimal surgical interventions.
References
Alshareef, A., J. S. Giudice, J. Forman, R. S. Salzar, and M. B. Panzer. A novel method for quantifying human in situ whole brain deformation under rotational loading using sonomicrometry. J. Neurotrauma 35:780–789, 2018.
Alshareef, A., J. S. Giudice, J. Forman, D. F. Shedd, K. A. Reynier, T. Wu, S. Sochor, M. R. Sochor, R. S. Salzar, and M. B. Panzer. Biomechanics of the human brain during dynamic rotation of the head. J. Neurotrauma 2020. https://doi.org/10.1089/neu.2019.6847.
Avants, B., C. Epstein, M. Grossman, and J. Gee. Symmetric diffeomorphic image registration with cross-correlation: Evaluating automated labeling of elderly and neurodegenerative brain. Med. Image Anal. 12:26–41, 2008.
Avants, B. B., P. T. Schoenemann, and J. C. Gee. Lagrangian frame diffeomorphic image registration: morphometric comparison of human and chimpanzee cortex. Med. Image Anal. 10:397–412, 2006.
Avants, B. B., N. J. Tustison, G. Song, P. A. Cook, A. Klein, and J. C. Gee. A reproducible evaluation of ANTs similarity metric performance in brain image registration. NeuroImage 54:2033–2044, 2011.
Avants, B. B., N. J. Tustison, J. Wu, P. A. Cook, and J. C. Gee. An open source multivariate framework for n-tissue segmentation with evaluation on public data. Neuroinformatics 9:381–400, 2011.
Avants, B. B., P. Yushkevich, J. Pluta, D. Minkoff, M. Korczykowski, J. Detre, and J. C. Gee. The optimal template effect in hippocampus studies of diseased populations. NeuroImage 49:2457–2466, 2010.
Beckwith, J. G., W. Zhao, S. Ji, A. G. Ajamil, R. P. Bolander, J. J. Chu, T. W. McAllister, J. J. Crisco, S. M. Duma, S. Rowson, S. P. Broglio, K. M. Guskiewicz, J. P. Mihalik, S. Anderson, B. Schnebel, P. Gunnar Brolinson, M. W. Collins, and R. M. Greenwald. Estimated brain tissue response following impacts associated with and without diagnosed concussion. Ann. Biomed. Eng. 46:819–830, 2018.
Bendlin, B. B., M. L. Ries, M. Lazar, A. L. Alexander, R. J. Dempsey, H. A. Rowley, J. E. Sherman, and S. C. Johnson. Longitudinal changes in patients with traumatic brain injury assessed with diffusion-tensor and volumetric imaging. NeuroImage 42:503–514, 2008.
Centers for Disease Control and Prevention. Report to Congress on Traumatic Brain Injury in the United States: Epidemiology and Rehabilitation., 2015.
Crum, W. R., L. D. Griffin, D. L. G. Hill, and D. J. Hawkes. Zen and the art of medical image registration: correspondence, homology, and quality. NeuroImage 20:1425–1437, 2003.
Darling, T., J. Muthuswamy, and S. D. Rajan. Finite element modeling of human brain response to football helmet impacts. Comput. Methods Biomech. Biomed. Eng. 19:1432–1442, 2016.
de Grau, S., A. Post, T. B. Hoshizaki, and M. D. Gilchrist. Effects of surface compliance on the dynamic response and strains sustained by a player’s helmeted head during ice hockey impacts. Proc. Inst. Mech. Eng. Part P J. Sports Eng. Technol. 1754337119871866, 2019.
Dennis, E. L., X. Hua, J. Villalon-Reina, L. M. Moran, C. Kernan, T. Babikian, R. Mink, C. Babbitt, J. Johnson, C. C. Giza, P. M. Thompson, and R. F. Asarnow. Tensor-based morphometry reveals volumetric deficits in moderate/severe pediatric traumatic brain injury. J. Neurotrauma 33:840–852, 2016.
Farbota, K. D. M., A. Sodhi, B. B. Bendlin, D. G. McLaren, G. Xu, H. A. Rowley, and S. C. Johnson. Longitudinal volumetric changes following traumatic brain injury: a tensor-based morphometry study. J. Int. Neuropsychol. Soc. 18:1006–1018, 2012.
Faul, M., M. M. Wald, L. Xu, and V. G. Coronado. Traumatic brain injury in the United States; emergency department visits, hospitalizations, and deaths, 2002-2006., 2010.
Fonov, V., A. C. Evans, K. Botteron, C. R. Almli, R. C. McKinstry, D. L. Collins, and Brain Development Cooperative Group. Unbiased average age-appropriate atlases for pediatric studies. Neuroimage 54(1):313–327, 2011.
Gabler, L. F., J. R. Crandall, and M. B. Panzer. Investigating brain injury tolerance in the Sagittal Plane using a finite element model of the human head. Int. J. Automot. Eng. 7:37–43, 2016.
Gabler, L. F., J. R. Crandall, and M. B. Panzer. Development of a metric for predicting brain strain responses using head kinematics. Ann. Biomed. Eng. 46:972–985, 2018.
Ghosh, S. S., S. Kakunoori, J. Augustinack, A. Nieto-Castanon, I. Kovelman, N. Gaab, J. A. Christodoulou, C. Triantafyllou, J. D. E. Gabrieli, and B. Fischl. Evaluating the validity of volume-based and surface-based brain image registration for developmental cognitive neuroscience studies in children 4 to 11years of age. NeuroImage 53:85–93, 2010.
Giudice, J. S., W. Zeng, T. Wu, A. Alshareef, D. F. Shedd, and M. B. Panzer. An analytical review of the numerical methods used for finite element modeling of traumatic brain injury. Ann. Biomed. Eng. 47:1855–1872, 2019.
Glaister, J., A. Carass, D. L. Pham, J. A. Butman, and J. L. Prince. Automatic falx cerebri and tentorium cerebelli segmentation from magnetic resonance images., 2017.
Hanson, J. L., M. K. Chung, B. B. Avants, E. A. Shirtcliff, J. C. Gee, R. J. Davidson, and S. D. Pollak. Early stress is associated with alterations in the orbitofrontal cortex: a tensor-based morphometry investigation of brain structure and behavioral risk. J. Neurosci. 30:7466–7472, 2010.
Hua, X., A. D. Leow, N. Parikshak, S. Lee, M.-C. Chiang, A. W. Toga, C. R. Jack, M. W. Weiner, and P. M. Thompson. Tensor-based morphometry as a neuroimaging biomarker for Alzheimer’s disease: an MRI study of 676 AD, MCI, and normal subjects. NeuroImage 43:458–469, 2008.
Ji, S., H. Ghadyani, R. P. Bolander, J. G. Beckwith, J. C. Ford, T. W. McAllister, L. A. Flashman, K. D. Paulsen, K. Ernstrom, S. Jain, R. Raman, L. Zhang, and R. M. Greenwald. Parametric comparisons of intracranial mechanical responses from three validated finite element models of the human Head. Ann. Biomed. Eng. 42:11–24, 2014.
Johnson, B., K. Zhang, M. Gay, S. Horovitz, M. Hallett, W. Sebastianelli, and S. Slobounov. Alteration of brain default network in subacute phase of injury in concussed individuals: resting-state fMRI study. Neuroimage 59:511–518, 2012.
Kim, J., B. Avants, S. Patel, J. Whyte, B. H. Coslett, J. Pluta, J. A. Detre, and J. C. Gee. Structural consequences of diffuse traumatic brain injury: a large deformation tensor-based morphometry study. NeuroImage 39:1014–1026, 2008.
Kim, J. J., and A. D. Gean. Imaging for the diagnosis and management of traumatic brain injury. Neurotherapeutics 8:39–53, 2011.
Kimpara, H., Y. Nakahira, M. Iwamoto, K. Miki, et al. Investigation of anteroposterior head-neck responses during severe frontal impacts using a brain-spinal cord complex FE model. Stapp Car Crash J. 50:509, 2006.
Kipps, C. M., A. J. Duggins, N. Mahant, L. Gomes, J. Ashburner, and E. A. McCusker. Progression of structural neuropathology in preclinical Huntington’s disease: a tensor based morphometry study. J. Neurol. Neurosurg. Psychiatry 76:650–655, 2005.
Klein, A., J. Andersson, B. A. Ardekani, J. Ashburner, B. Avants, M.-C. Chiang, G. E. Christensen, D. L. Collins, J. Gee, P. Hellier, J. H. Song, M. Jenkinson, C. Lepage, D. Rueckert, P. Thompson, T. Vercauteren, R. P. Woods, J. J. Mann, and R. V. Parsey. Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. NeuroImage 46:786–802, 2009.
Lepore, N., C. Brun, Y.-Y. Chou, M.-C. Chiang, R. A. Dutton, K. M. Hayashi, E. Luders, O. L. Lopez, H. J. Aizenstein, and A. W. Toga. Generalized tensor-based morphometry of HIV/AIDS using multivariate statistics on deformation tensors. IEEE Trans. Med. Imaging 27:129–141, 2008.
Madhukar, A., and M. Ostoja-Starzewski. Finite element methods in human head impact simulations: a review. Ann. Biomed. Eng. 47:1832–1854, 2019.
Mao, H., L. Zhang, B. Jiang, V. V. Genthikatti, X. Jin, F. Zhu, R. Makwana, A. Gill, G. Jandir, A. Singh, et al. Development of a finite element human head model partially validated with thirty five experimental cases. J. Biomech. Eng. 135:111002, 2013.
McAllister, T. W., J. C. Ford, S. Ji, J. G. Beckwith, L. A. Flashman, K. Paulsen, and R. M. Greenwald. Maximum principal strain and strain rate associated with concussion diagnosis correlates with changes in corpus callosum white matter indices. Ann. Biomed. Eng. 40:127–140, 2012.
McDonald, B. C., A. J. Saykin, and T. W. McAllister. Functional MRI of mild traumatic brain injury (mTBI): progress and perspectives from the first decade of studies. Brain Imaging Behav. 6:193–207, 2012.
Miles, L., R. I. Grossman, G. Johnson, J. S. Babb, L. Diller, and M. Inglese. Short-term DTI predictors of cognitive dysfunction in mild traumatic brain injury. Brain Inj. 22:115–122, 2008.
Miller, L. E., J. E. Urban, and J. D. Stitzel. Development and validation of an atlas-based finite element brain model. Biomech. Model. Mechanobiol. 15:1201–1214, 2016.
Miller, L. E., J. E. Urban, and J. D. Stitzel. Validation performance comparison for finite element models of the human brain. Comput. Methods Biomech. Biomed. Eng. 2017. https://doi.org/10.1080/10255842.2017.1340462.
Nooner, K. B., et al. The NKI-rockland sample: a model for accelerating the Pace of discovery science in psychiatry. Front. Neurosci. 6:152, 2012.
Panzer, M. B., B. S. Myers, B. P. Capehart, and C. R. Bass. Development of a finite element model for blast brain injury and the effects of CSF cavitation. Ann. Biomed. Eng. 40:1530–1544, 2012.
Park, G., T. Kim, J. Forman, M. B. Panzer, and J. R. Crandall. Prediction of the structural response of the femoral shaft under dynamic loading using subject-specific finite element models. Comput. Methods Biomech. Biomed. Engin. 20:1151–1166, 2017.
Pellman, E. J., D. C. Viano, A. M. Tucker, I. R. Casson, and J. F. Waeckerle. Concussion in professional football: reconstruction of game impacts and injuries. Neurosurgery 53:799–814, 2003.
Pulsipher, D. T., R. A. Campbell, R. Thoma, and J. H. King. A critical review of neuroimaging applications in sports concussion. Curr. Sports Med. Rep. 10:14–20, 2011.
Reynier, K., A. Alshareef, E. J. Sanchez, D. F. Shedd, S. R. Walton, N. K. Erdman, B. T. Newman, J. S. Giudice, M. J. Higgins, J. R. Funk, D. K. Broshek, T. J. Druzgal, J. E. Resch, and M. B. Panzer. The effect of muscle activation on head kinematics during non-injurious head impacts in human subjects. Ann. Biomed. Eng (in review). 2020.
Rosenbaum, S. B., and M. L. Lipton. Embracing chaos: the scope and importance of clinical and pathological heterogeneity in mTBI. Brain Imaging Behav. 6:255–282, 2012.
Sanchez, E. J., L. F. Gabler, A. B. Good, J. R. Funk, J. R. Crandall, and M. B. Panzer. A reanalysis of football impact reconstructions for head kinematics and finite element modeling. Clin. Biomech. 2018. https://doi.org/10.1016/j.clinbiomech.2018.02.019.
Sanchez, E. J., L. F. Gabler, J. S. McGhee, A. V. Olszko, V. C. Chancey, J. Crandall, and M. B. Panzer. Evaluation of head and brain injury risk functions using sub-injurious human volunteer data. J. Neurotrauma 2017. https://doi.org/10.1089/neu.2016.4681.
Scheibel, R. S., D. A. Pearson, L. P. Faria, K. J. Kotrla, E. Aylward, J. Bachevalier, and H. S. Levin. An fMRI study of executive functioning after severe diffuse TBI. Brain Inj. 17:919–930, 2003.
Siegkas, P., D. J. Sharp, and M. Ghajari. The traumatic brain injury mitigation effects of a new viscoelastic add-on liner. Sci. Rep. 9:1–10, 2019.
Slobounov, S., M. Gay, B. Johnson, and K. Zhang. Concussion in athletics: ongoing clinical and brain imaging research controversies. Brain Imaging Behav. 6:224–243, 2012.
Takhounts, E. G., R. H. Eppinger, J. Q. Campbell, R. E. Tannous, et al. On the development of the SIMon finite element head model. Stapp Car Crash J. 47:107, 2003.
Takhounts, E. G., S. A. Ridella, V. Hasija, R. E. Tannous, J. Q. Campbell, D. Malone, K. Danelson, J. Stitzel, S. Rowson, and S. Duma. Investigation of traumatic brain injuries using the next generation of simulated injury monitor (SIMon) finite element head model. Stapp Car Crash J. 52:1, 2008.
Toga, A. W., and P. M. Thompson. The role of image registration in brain mapping. Image Vis. Comput. 19:3–24, 2001.
Viano, D. C., I. R. Casson, E. J. Pellman, L. Zhang, A. I. King, and K. H. Yang. Concussion in Professional Football: Brain Responses By Finite Element Analysis: Part 9. Neurosurgery 57:891–916, 2005.
Wilde, E. A., S. R. McCauley, J. V. Hunter, E. D. Bigler, Z. Chu, Z. J. Wang, G. R. Hanten, M. Troyanskaya, R. Yallampalli, X. Li, J. Chia, and H. S. Levin. Diffusion tensor imaging of acute mild traumatic brain injury in adolescents. Neurology 70:948–955, 2008.
Wu, T., A. Alshareef, J. S. Giudice, and M. B. Panzer. Explicit modeling of white matter axonal fiber tracts in a finite element brain model. Ann. Biomed. Eng. 47:1908–1922, 2019.
Wu, T., J. Antona-Makoshi, A. Alshareef, J. S. Giudice, and M. B. Panzer. Investigation of cross-species scaling methods for traumatic brain injury using finite element analysis. J. Neurotrauma. 2019. https://doi.org/10.1089/neu.2019.6576.
Zhang, K., B. Johnson, D. Pennell, W. Ray, W. Sebastianelli, and S. Slobounov. Are functional deficits in concussed individuals consistent with white matter structural alterations: combined FMRI & DTI study. Exp. Brain Res. 204:57–70, 20
Zhang, L., K. H. Yang, R. Dwarampudi, K. Omori, T. Li, K. Chang, W. N. Hardy, T. B. Khalil, and A. I. King. Recent advances in brain injury research: a new human head model development and validation. Stapp Car Crash J 45:369–394, 2001.
Zhao, W., and S. Ji. Mesh convergence behavior and the effect of element integration of a human head injury model. Ann. Biomed. Eng. 47:475–486, 2019.
Acknowledgments
The authors gratefully acknowledge the UVA Brain Institute and Achievement Rewards for College Scientists Foundation (ARCS) for supporting this research. We would also like to thank Grant Kim for his assistance. The authors are thankful to Dr. Alayna Panzer for her thorough revisions to the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Associate Editor Joel D. Stitzel oversaw the review of this article.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Giudice, J.S., Alshareef, A., Wu, T. et al. An Image Registration-Based Morphing Technique for Generating Subject-Specific Brain Finite Element Models. Ann Biomed Eng 48, 2412–2424 (2020). https://doi.org/10.1007/s10439-020-02584-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10439-020-02584-z