Abstract
On-field monitoring of head impacts, combined with finite element (FE) biomechanical simulation, allow for predictions of regional strain associated with a diagnosed concussion. However, attempts to correlate these predictions with in vivo measures of brain injury have not been published. This article reports an approach to and preliminary results from the correlation of subject-specific FE model-predicted regions of high strain associated with diagnosed concussion and diffusion tensor imaging to assess changes in white matter integrity in the corpus callosum (CC). Ten football and ice hockey players who wore instrumented helmets to record head impacts sustained during play completed high field magnetic resonance imaging preseason and within 10 days of a diagnosed concussion. The Dartmouth Subject-Specific FE Head model was used to generate regional predictions of strain and strain rate following each impact associated with concussion. Maps of change in fractional anisotropy (FA) and median diffusivity (MD) were generated for the CC of each athlete to correlate strain with change in FA and MD. Mean and maximum strain rate correlated with change in FA (Spearman ρ = 0.77, p = 0.01; 0.70, p = 0.031), and there was a similar trend for mean and maximum strain (0.56, p = 0.10; 0.6, p = 0.07), as well as for maximum strain with change in MD (−0.63, p = 0.07). Change in MD correlated with injury-to-imaging interval (ρ = −0.80, p = 0.006) but change in FA did not (ρ = 0.18, p = 0.62). These results provide preliminary confirmation that model-predicted strain and strain rate in the CC correlate with changes in indices of white matter integrity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The incidence of sports-related mild traumatic brain injury (MTBI), also referred to as concussion, ranges from 1.6 to 3.8 million individuals each year in the USA, including those who do not seek medical care.34 These figures likely underestimate the magnitude of the problem, reflecting lack of appreciation of the medical implications of the symptoms and reluctance to report symptoms for fear of losing playing time.49 Concussions are particularly common in football and ice hockey.1,11 Concussed athletes typically have signs and symptoms initially but are free of symptoms within 7 days, although recovery may take longer in a small percentage of individuals.37,38 Concern has been raised about the effects of multiple concussions and the role they may play in the long-term sequelae (termed chronic traumatic encephalopathy) observed in some professional football players and other athletes,10,39,41,47 and recently observed in a young college football player without an identified history of concussion.53
Despite its importance, there are some fundamental gaps in our knowledge of concussion—including which specific brain regions are associated with the alteration in mental status that defines the injury and which components of biomechanical forces most affect those brain regions. These gaps reflect several important limitations in our ability to study MTBI. The first relates to the lack of accurate measures of head kinematics associated with human MTBI. Historically, small animal surrogates have been used to study brain tissue response to impact; however, these models often translate poorly to the human condition,12 and large animal models are limited due to a variety of concerns including ethical constraints for using primates and a lack of suitable cognitive and functional outcome measures most pertinent to human MTBI. In addition, anthropometric test devices (ATDs) have been utilized to simulate on-field impacts in sports; however, these devices are limited in their ability to replicate individual characteristics that influence impact response (e.g., directional neck strength, state of awareness at time of impact, player body differences, etc.), restricting their utility to primarily parametric evaluations. In the last 7 years, these limitations have been addressed by directly measuring head kinematics following impact of athletes during play with the Head Impact Telemetry (HIT) System.13
The second gap in our knowledge of human MTBI relates to attempts to visualize and quantify the neural effects of injury with neuroimaging. Conventional neuroimaging methods are unrevealing in the large majority of individuals with MTBI, particularly sports concussion.3 More recently. however, techniques, such as diffusion-weighted imaging [in particular diffusion tensor imaging (DTI)], which capitalize on the ability to detect the diffusion of water molecules in the brain have shown great promise in probing white matter integrity,44 particularly in mild and moderate TBI where white matter injury is believed to underlie the neuropathology of the disorder.14,33,36 Nevertheless, it is rarely feasible to study individuals both before and after their injury.
Finite element (FE) modeling has been widely used as a numerical surrogate to simulate head mechanical responses under a variety of impact and inertial rotational conditions. A number of studies have utilized FE-modeling techniques to investigate brain mechanical responses during impact.30,31,58,66,67 For example, using an experimentally validated FE brain model, Kleiven30 reported that strain in the bridging veins, which correlates with occurrence and severity of subdural hematoma, varied with impact direction for purely translational and purely rotational impulses. Zhang et al.66,67 used biomechanical measures obtained from ATD simulations as inputs into another FE brain model to predict tissue responses during impact, including intracranial pressure and brain shear stress, and concluded that the occurrence of concussion was best predicted by shear stress at the midbrain. Viano et al.61 reported intracranial pressure, stress, strain, and strain rate data at various regions in the brain based on FE modeling of laboratory reconstructions from on-field impacts associated with concussion in professional football players and correlated these results with clinical symptoms.
It is difficult to know how accurately these simulations reflect actual strains and strain rates within a live individual’s brain. Although numerous head FE models exist with varying complexity, most do not represent the true anatomic and physiologic details of the head. Each model is constructed with varying simplifications and/or approximations regarding anatomic geometry and structures, boundary and loading conditions, constitutive properties, and element formulations. For example, even though model's head size significantly influences intracranial mechanical responses, FE computations are often based on a 50th percentile male head geometry. Further, mechanical loading data (translational and rotational accelerations of the head center of gravity) in these simulations are typically generated with ATD headforms during accident reconstructions of injuries captured on video. One approach to enhance accuracy of brain region-specific strain predictions is to utilize subject-specific FE models based on neuroanatomic data obtained by high field magnetic resonance imaging (MRI). In addition, the accuracy of the input to the model can be enhanced by means of actual head kinematics measured during an impact associated with a diagnosed concussion for that subject. To better understand regional effects of tissue mechanical responses on structural and functional alterations in the brain, we have developed an automatic meshing technique to generate subject-specific FE head models based on each individual’s own MR image scans.27 We hypothesized that regions of FE model predicted high strain would show disrupted white matter integrity when assessed with DTI.
Methods
Overview
For the last 4 years, our group has studied the biomechanical basis of MTBI in collegiate and high school contact sport athletes. The collegiate athletes are varsity athletes on the Dartmouth College football and men’s and women’s ice hockey teams, and the high school athletes compete on the Hanover High School varsity football team. Participating athletes undergo cognitive assessment and neuroimaging both before and after a season of play. Any participants diagnosed by the team medical staff as sustaining a concussion are also studied as soon as possible post-injury. The protocol was approved by the Dartmouth College Committee for the Protection of Human Subjects, and the college participants gave written informed consent. The high school athletes gave their assent, and consent forms were signed by their parents/guardians. In this article, we report on 10 male athletes who were diagnosed as having a concussion during their sport season.
Head Impact Measurement
During all practices and games, players wore either football (Riddell Inc., Rosemont, IL) or hockey (Easton S9, Easton Sports, Scotts Valley, CA; CCM Vector, Reebok, Saint-Laurent, QC) helmets instrumented with the HIT System (Simbex, Lebanon, NH). The HIT System is designed to record in vivo acceleration of the head following impact by integrating six single-axis linear accelerometers, a single electronics board combining data acquisition (10 bit, 1000 Hz), radiofrequency (RF), and telemetry (903–927 MHz) components, and a rechargeable battery into each player’s helmet. Description of both HIT System function and validation has been previously described in the literature.8,13,21–24,35 In brief, each accelerometer is positioned against the head to differentiate head acceleration from helmet acceleration, and when any single accelerometer channel exceeds a pre-set threshold of 14.4 g, 40 ms of data (8 ms pre-trigger and 32 ms post-trigger) were transmitted to a sideline receiver connected to a laptop computer. Accelerometer data recorded from each impact was then post-processed using a simulated annealing optimization algorithm to solve for the entire 40-ms three-dimensional linear and rotational accelerations at the head center of gravity.6 The Cartesian coordinate system was defined by right-hand rule with the X direction toward the front of the head, the Y direction toward the left ear, and the Z direction toward the top of the head. Accuracy of this approach was confirmed with laboratory validation experiments similar to those previously reported.23,50
Dartmouth Subject-Specific Head FE Model (SSM)
We have developed an automatic meshing technique to generate subject-specific FE head models based on the individual’s own MR images.27 High-resolution T1-weighted MR images from a single template-defining subject were used to create an atlas mesh via a dedicated meshing script. Specifically, the brain images of the template subject were automatically segmented (FreeSurfer, Version 4.5.0) to generate a triangular outer-boundary surface with an in-house MATLAB (version 7.11/R2010b, The MathWorks Inc., Natick, MA) routine. The dedicated meshing script was developed using a commercial software package (TrueGrid, version 2.3; XYZ Scientific Applications Inc., Livermore, CA) that created a multi-block with its size and position determined by the triangular brain surface. The multi-block was subsequently subdivided and corner/edge nodes of each sub-block were projected onto the triangulated brain surface to generate whole-brain hexahedral elements.
Using the meshing script, a subject-specific FE mesh was generated for each of the remaining subjects via rigid body image registration. For each subject, the brain was similarly segmented and the corresponding MR images were rigidly registered with those of the template. Using the same meshing script but with the subject’s triangular brain surface transformed into the template space as geometrical input, whole-brain hexahedral meshes were subsequently produced and transformed back into the subject’s MR image space using the inverse of the rigid body transformation.
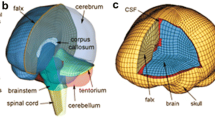
The skull inner-surface was represented as a rigid body shell (1 mm thick), with the nodes produced by displacing the brain boundary nodes outward in the average nodal normal direction by 1 mm. The falx was manually segmented for each subject by tracing the boundary on the mid-sagittal MR image. A purpose-written MATLAB routine was utilized to identify brain nodes corresponding to the falx on the mid-sagittal MR image, and they were then duplicated and displaced by 0.5 mm along both directions perpendicular to the mid-sagittal plane to generate two hemispheres. The same set of brain nodes were subsequently used to represent the falx as a membrane (1 mm thick). A representative subject-specific brain mesh (subject 3863) is shown in Fig. 1.
Illustration of a subject-specific brain mesh showing the falx and head CG (a and b), and a coronal (c) and a sagittal (d) view of the mesh corresponding to Figs. 3, 4, 5, 6. The skull inner-surface (seen in (c) and (d)) was generated based on the brain outer boundary elements (see text for details)
Material Properties and Boundary Conditions
A hyperelastic material model of a second-order Odgen form recently employed by Kleiven31 was chosen for all subject-specific models:
where λ i are principal stretches, and μ i and α i are the material constants (Table 1). The density and the bulk modulus of the brain were set to 1040 kg/m3 and 2.19 GPa, respectively.
In addition, a Prony series of six terms was employed to characterize viscoelasticity for a dimensionless relaxation modulus of the form:
where g i and τ i are the material constants, which are listed in Table 2. These material property values were derived in Kleiven31 by fitting the corresponding parameters using discrete spectrum approximation to include the non-linear elasticity described by Franceschini20 and Franceschini et al.19 as well as the high-frequency relaxation moduli determined by Nicolle et al.46 The falx was assumed to be linearly elastic in keeping with previous investigations in the literature (e.g., Kleiven31; Zhang et al.67; Takhounts et al.57), and the corresponding material constants are listed in Table 3.
A frictional contact boundary condition (frictional coefficient of 0.243) was employed to simulate the interface between the brain and skull, and between the brain and the falx using a general contact formulation. In addition, the falx was tied to the skull using a node-based approach at the interface. For all simulations in this study, the Abaqus/Explicit (Version 6.8; Dassault Systemes Simulia Corp., Providence, RI) simulation package was employed.
Model Validation
The most recent head impact tests performed on inverted and re-pressurized cadavers by Hardy et al.26 provide valuable experimental data for dynamic response of the human brain under impact loading conditions. High-speed X-ray was employed to measure relative brain–skull motion by placing a cluster of neutral density targets at representative locations within the cadaveric brain. In order to validate simulations of the Dartmouth SSM, two representative cadaver impact tests were selected (288-T1: occipital impact and C380-T5: temporal impact) against which the model-simulated relative brain–skull motion over the course of impact was compared with experimental measurements. For both simulations, the atlas mesh was used and scaled to match the reported head dimension of the cadaver used in the test. Biomechanical head acceleration traces were applied to the rigid body skull, and relative brain–skull trajectories at two neutral density target locations (corresponding to NDT 4 and 11 in Hardy et al.26, respectively) were obtained and compared with those measured (Fig. 2). SSM-computed brain–skull relative motion for both cases was correlated in all directions of motion with measures obtained through high-speed X-ray, and was also found to be in agreement for measures of maximum displacement (difference in peak values <2 mm relative to the measurements, which was similar to that found in Kleiven and Hardy32). Interestingly, the agreement between the measured and simulated displacement was noticeably better for the temporal impact (i.e., C380-T5 in Fig. 2, bottom), while the discrepancy in displacement phase was more evident for the occipital impact (i.e., C288-T1 in Fig. 2, top). This outcome was likely because the SSM explicitly included the falx (expected to be more important in a temporal impact) but not the tentorium (expected to have more influence in an occipital impact). In this study, we did not include the tentorium because techniques for automatic and accurate segmentation of this structure from T1-weighted MRI scans are not presently available.48 Nonetheless, these validation results demonstrate that the Dartmouth SSM is able to reproduce intracranial displacement responses that agree with measurements from representative cadaver tests. Enhancing the anatomical sophistication of the SSM to include important structures (such as the tentorium) is, no doubt, important for future development and is expected to improve both the accuracy and completeness of the validation results presented here (e.g., against the relative brain–skull displacement data as well as intracranial pressure data from Nahum et al.45 and Trosseille et al.60).
Comparison of simulated and measured brain–skull relative motion over the course of impact for two representative cadaver impact tests (top: C288-T1; bottom: C380-T5; The left and right columns correspond to location of NDT-4 and NDT-11 in Hardy et al.,26 respectively)
Clinical Diagnosis
For this analysis, concussion was defined as an alteration in mental status that was reported or observed by the player or team’s medical staff and associated with a blow to the head. A certified athletic trainer or team physician diagnosed and treated all instances of concussion at their professional discretion. As is typical of sports-related concussion,42 none of the 10 cases of diagnosed concussion was associated with loss of consciousness, and so to identify the impact most likely associated with the injury, the medical staff provided the date of injury, the suspected time of injury, and the approximate time of diagnosis. In addition, anecdotal observations of the events surrounding injury (e.g., description of the impact, method of identifying the injury, and comments from coaches, trainers, teammates, or parents) were provided. An impact recorded by the HIT System was retrospectively associated with each case of diagnosed injury by cross-referencing the information provided by the team with the time stamp of each impact sustained by the player on the day of injury. Processed time series data (X, Y, and Z linear and rotational acceleration) for each impact associated with concussion was used as input for the subject-specific FE model.
Neuroimaging
All scans were acquired in the Dartmouth Advanced Imaging Center, which houses a research-dedicated 3T Philips Achieva magnet. This 3.0T system has a Quasar Dual gradient set with strengths up to 80 mT/m and slew rates up to 200 T/m/s and an anatomic landmark based longitudinal repositioning system (SameScan/SmartScan). A Philips 8-channel SENSE Head Coil was used.
Diffusion Imaging
Diffusion imaging was carried out using 46 diffusion directions (collected with b = 1000 s/mm2, NEX = 1) plus one volume without diffusion gradients (b = 0). The scan data were used to estimate a second order symmetric diffusion tensor at each voxel, from which scalar diffusion parameters (FA, etc.) were then computed. Calculation of tensors was carried out using FDT 2.0 (FMRIB Software Library (FSL) package,56 version 4.1.6) which corrected the raw diffusion images for eddy currents and motion prior to fitting a tensor model, and subsequently brain-masked the results using BET.54 The DTI ToolKit (DTI-TK)65 was employed to generate DTI scalar diffusion parameters.
Diffusion tensor imaging processing also included a sequence of custom MATLAB scripts for preprocessing, quality assessment, and configuration of FSL programs. Preprocessing included gradient vector correction, which adjusts the vectors used in analysis to account for any offset in the imaging data relative to the magnet bore.15 Quality assessments consisted of manual verification of the correct appearance of all images plus tests using ExploreDTI59 and additional tests based on Tournier et al.59 Scans with significant corruption (affected b = 0 image or 3+ affected diffusion images), all ascribed to motion, were discarded (N = 1), while scans with 1–2 affected diffusion images were run without those images (N = 4).
Anatomic Reference
An MPRAGE (Magnetization Prepared Rapid Gradient Echo) T1-weighted sequence was used in each session with the following parameters: 140 contiguous 1.2 mm sagittal slices, TR: 6.8 ms, TE: 3.3 ms, TI: 852.9 ms, TFE prepulse delay: shortest, flip angle: 8°, NEX: 1, BW/Pixel: 241, FOV: 256 mm, matrix 256, and 1.0 × 1.0 mm in-plane resolution. This series balances scan time, signal-to-noise ratio, high gray/white tissue contrast, and high spatial resolution, and the near-isotropic acquisition allows re-slicing in any plane without significant loss of volumetric information.
Individual Anatomic Labeling
FreeSurfer was employed to generate automatic subcortical segmentations of one anatomic reference scan for each individual17,18,25 and to carry out bias correction and skull stripping. Each FreeSurfer processed brain was then coregistered with an unweighted diffusion image from each of the subject’s DTIs after it had been skull-stripped with BET (v2.1)54; this step used the Statistical Parametric Mapping package (SPM5) with the nearest-neighbor interpolation (other options at defaults) to propagate the FreeSurfer anatomic labels to each DTI image.
Template based Analysis
Individual FA images were non-linearly registered onto the FSL MNI template (2-mm version) using the FSL FNIRT tool (build 416), and the individually calculated alignment transforms were applied to all other DTI maps. Since strain and other SSM-derived maps are generated in the subject’s MPRAGE space, we also generated and applied an MPRAGE to DTI transform (using the b = 0 image as a reference for DTI) for each scan and combined that with the FA to MNI transform to warp SSM results to the same space. After alignment, all SSM and DTI maps were smoothed with a 3D Gaussian filter with σ = 2 (FWHM = 4.7 mm) to allow for minor alignment inaccuracies.
Statistical Analysis
We used Spearman’s ρ (rho), a nonparametric (rank-based) correlation measure suitable for small sample sizes, non-normal distributions, and nonlinear associations to assess correlations between FA and MD, and strain and strain rate in the corpus callosum (CC).5 Our assumption is that the collected preseason DTI scans represent a personal baseline for each subject, and that the effects of a concussion will result in changes detectable on these maps. We hypothesized that injury effects would correlate with the strain experienced in each brain region (i.e., effects are assumed for simplicity not be variable within a given region), and thus used FA-strain and MD-strain correlation as our fundamental measures. To assess whether DTI would provide sufficient precision and reliability to track individual concussion-induced changes, we acquired two DTI scans per subject back-to-back on a separate group of 27 mild TBI subjects and used a one-way ANOVA model to estimate within and between subject variance. We found that the CC mean FA had a within-subject standard deviation of 0.0152 and a between-subject SD of 0.0624 (mean MD: 0.00041 within and 0.101 between).
Results
Participants
Demographics for the ten athletes diagnosed with a concussion and with completed pre- and post-injury neuroimaging are summarized in Table 4. Five participants were college football players, four were high school football players, and one was a college ice hockey player. All participants were right handed and male. The mean peak linear and rotational resultant acceleration associated with the ten cases of concussion were 73.6 ± 21.3 g and 5,025 ± 1,226 rad/s2. Contributions of linear and rotational accelerations at the head CG in the X, Y, and Z directions of the head coordinate system at the time of peak resultant acceleration are summarized in Table 5 along with the peak maximum principal strain (mean, 0.280 ± 0.089) and peak maximum principal strain rate (mean, 54.3 ± 33.9) in the corpus callosum associated with each of the impacts. Corresponding mean FA and MD values for each subject are also summarized for each subject.
Subject-Specific FEM Strain Maps
The Dartmouth SSM27 was used to generate strain maps for each impact associated with a concussion. Figure 3 shows a representative strain map from Subject 3863. In addition, the strain maps from all 10 concussive impacts were used to generate a group concussive strain map. Figure 4 shows the pattern of regional strain displayed in a normalized brain. Of note is the significant overlap between the concussion associated high strain regions and the CC.
FA and MD Maps
Preseason and post-concussion DTI scans were utilized to generate FA and MD change maps for each subject. Figure 5 shows a representative FA change map, also from Subject 3863. In addition, FA change maps were generated for the group. Figure 6 shows the pattern of FA change associated with all 10 concussions displayed in a normalized brain.
Relationship of Strain Parameters to DTI Change in Corpus Callosum
FreeSurfer segments the CC into five subregions, which we used together to create a CC region. We also tested the anterior and posterior regions separately. Table 6 shows that either the mean or maximum strain rate within the CC ROI correlates significantly with FA changes, and to a lesser extent strain does as well.
In addition, we used the MNI normalized mean maps similar to those shown in Figs. 4 (strain) and 6 (FA) to calculate the correlation of strain or strain rate with FA or MD on the Wake Forest University MNI Brodmann atlas CC region of interest. The results (Table 7) show that strain rate is a significant predictor of FA changes, while strain is a significant predictor of MD changes, but a weaker predictor of ΔFA.
Relationship of Injury-to-Imaging Interval and DTI
Recent evidence suggests that changes in DTI parameters may be sensitive to the time interval between injury and post-injury imaging.7,33,36 Therefore, we also assessed the effect of this variable on the correlation of strain parameters and DTI parameters. Figure 7 shows that there is a significant correlation between change in MD and injury-to-imaging interval, but this did not hold for change in FA. Table 7 provides a revised version of CC ROI correlations of note when data are adjusted for the injury-scan interval.
Discussion
Results from this pilot study suggest that integration of directly measured head impact data with a corresponding subject-specific FE head model and advanced white matter imaging techniques may provide additional insights into the biomechanical basis of MTBI. Our study provides a first attempt to correlate FE model-predicted regional strains and strain rates associated with concussion with indicators of white matter change as measured by diffusion imaging. The finding that model-predicted strain rate correlated with change in FA in the CC using two different approaches for segmenting this structure is encouraging. The relationship between strain and MD in the MNI CC is also reassuring. FA and MD are the most widely used DTI measures in the literature44 and so were selected here as the most useful values for comparison to existing work.
The overall relationship between FA and MD is typically negative (i.e., as one increases, the other decreases); MD is maximized when diffusion is unrestricted and FA is maximized when it is narrowly restricted to a single orientation, and so they are in that sense opposites. However, they are not coupled tightly, since the eigenvalues from which both are derived can vary up or down based on local diffusivity and the tensor fit, potentially resulting in decrease or increase in both measures in some situations.
The differential relationship of injury-to-imaging interval and changes in MD and FA may account for some of the differences found between correlations of strain measures and FA and MD in this relatively small sample. It is now recognized that, after injury, FA may initially increase substantially (up to 1 week) but subsequently decrease over time, and FA as well as other DTI parameters (e.g., axial diffusivity and radial diffusivity) may also be affected differently at different stages because of a variety of biological processes acting over different time intervals.7,33,36 The explanation of these differences is not immediately apparent, though it has been hypothesized that FA reductions result from misalignment of fibers, cytotoxic edema, fiber disruption, or axonal degeneration (see e.g., Kumar33 and Mayer36 for a useful discussion). With respect to our cohort, it may be that these subjects did not experience injuries severe enough to trigger all of these contributory mechanisms. MD changes, on the other hand, may result from loss of microstructural integrity in early stages or expansion of extracellular space in later stages (both resulting in a net increase) and from decreased water content in extracellular space due to axonal swelling (a net decrease),7 and it has been suggested that MD may be more sensitive to minor diffuse axonal injury than FA.9 Further study is needed to refine the definition of these temporal relationships as this may be an important variable when looking for small signal change in white matter integrity. For example if confirmed, our results would suggest that changes in MD will become more apparent over a 10–14-day interval, whereas changes in FA appear somewhat less sensitive to timing over this window.
The fact that these athletes’ structural scans were read as normal despite the noted changes in white matter associated with measures of strain is consistent with what is known about the neuropathology of brain injury including MTBI.14 Furthermore, the CC is a common site for axonal injury of varying severities including mild injury,4,55 and thus the observed correlations of change in white matter indices with strain measures in this structure are consistent with the known neurobiology of MTBI. It is important to note, however, that the clinical descriptions of these ten concussions suggested that they were quite mild injuries (symptom resolution 11.4 ± 17 days) and the biomechanical parameters associated with these injuries were in the lower percentile of all impacts associated with concussion when compared with larger samples of on-field injury data previously reported in the literature. For example, from a sample of 32 collegiate football players clinically diagnosed with concussion, Rowson et al.51 reported a mean linear acceleration of 105 ± 27 g for impacts associated with those injuries compared to 74 ± 21 g for the impacts modeled in this study. While head kinematics after impact have yet to be correlated with injury severity, lower head acceleration has been previously correlated with a reduction in brain tissue strain, which could affect the outcome of our results. The finding of a relationship between strain and white matter change is noteworthy in individuals with injuries of this mild degree.
An important issue is the specificity of our findings. Figures 3 and 4 suggest brain regions other than the CC might also be subject to relatively high strains and contribute to the injury process. In this pilot study, we have chosen to focus on the CC in that our primary hypothesis was that we would find correlations between indices of white matter change and model-predicted strain in this region. From a biological perspective, the CC is known to be vulnerable to diffuse axonal injury in more severe forms of TBI, thus it seemed to be a compelling region to focus on. Second, (and related to the first), the CC is a large white matter structure. This allows us to be confident that the segmentation and normalization techniques used are reasonably reliable and valid, and that we are sampling white matter indices from a large volume of predominantly white matter tissue. This is particularly important in our relatively small sample of concussed individuals, where a single outlier could have undue influence on results of smaller regions or regions with a smaller ratio of white to gray matter. Preliminary secondary analyses in other brain regions with higher model predicted strain, showed some interesting trends that might reach significance with a larger sample; however, none achieved statistical significance with the current sample under this study. There are several potential reasons for this including the modest sample size of our study and/or regional inaccuracies in model's predicted strain. Data collection is ongoing to increase our sample size. In addition, correlation of DTI output with strain and strain rate maps from additional FEM head models is planned as part of a newly funded project to assess variability in FEM model outputs and relationship to white matter changes.
It is also possible that high impacts not associated with diagnosed concussion might result in changes in white matter measures, and that those changes could correlate with model-predicted strain. This was not tested in this study but is an important next step.
This article presents preliminary findings, and there are important limitations that we would like to acknowledge. In any study involving concussion, the recognition or diagnosis of “concussion” is not foolproof. Although there are generally agreed criteria,40 the limitations of on-field diagnosis and self-reporting mean that they are unevenly applied. Furthermore, while our study is predicated on the assumption that we can match each concussion to a single impact that causes it, based on on-field recordings and player debriefings, this relationship is sometimes challenging. For example, in many cases, a player may sustain multiple impacts before an identified concussion (the majority being of relatively low magnitude) leading to a suspicion that earlier harder impacts may have played a contributory role. We are evaluating methods for modeling multiple impacts and combining the resulting strains to address this. There are other factors involved in concussion identification. Recent attention to the issue has heightened the desire of some athletes to hide or not report symptoms suggestive of concussion, out of concern for loss of playing time. Conversely, other athletes, as well as parents and athletic training staff may reports symptoms leading to the diagnosis of concussion that would have been ignored several years ago.
The reproducibility of FA and MD in DTI is sufficient to track relatively small changes longitudinally in a single subject,15 but there are of course limits to the size of changes that can be detected, and physiological motion, image registration, and inherent noise combine to make assessing the precision with which comparisons can be made in any individual case difficult. Furthermore, a limitation that must be recognized in any study involving conventional DTI is its inability to properly measure diffusion in “crossing fiber” areas,62 which occur in about 5–15% of the human brain.2 New techniques, such as HYDI (Hybrid Diffusion Imaging), a q-space imaging technique, which carries out diffusion imaging in multiple HARDI shells63 have been developed to address these issues and will be incorporated in further studies in this area.
FE model validation is important to ensure its simulation accuracy and relevance. Our current Dartmouth SSMs are generated automatically based on the individual’s own MR images (with the exception of the falx that requires manual segmentation) with high mesh quality,27 which is especially important when studying a pool of subjects and provides a direct link between model simulation outputs and subject-specific neuroimaging data. However, the automation of SSM as applied here comes at the cost of having relatively few internal structures compared to other anatomically more sophisticated models (e.g., Zhang et al.67; Kleiven31; Takhounts et al.57). Although certain model simplifications are not expected to influence the simulation results (e.g., exclusion of the facial and neck components, rigid skull assumption in mild accelerations where no skull deformation was observed, etc.) and are, perhaps, desirable for model simulation efficiency, the tentorium (which is structurally similar to the falx) may play an important role in intracranial response that our SSM did not capture. Unfortunately, segmentation of the tentorium from T1-weighted MRI scans remains a challenging task (3–4 hours manual segmentation for an expert, or at least 20 min for a trained rater using a semi-automatic segmentation approach according to a recent study48) and we opted to exclude it from these studies in favor of the efficiencies gained from automation which are essential when evaluating a multiplicity of individual subjects. We expect that exclusion of the tentorium will influence model responses in the inferior region of the brain, but may not significantly alter the results in the CC region, which is structurally closer to the falx but farther away from the tentorium. Other structures such as the ventricles or differentiation of the gray/white matter may also influence the modeled brain response to some degree. However, their effects on the computations in the CC region may be negligible compared to those from the falx/tentorium because much larger strain/stress concentrations are expected in this vicinity due to the much higher rigidity of these structures.
Nonetheless, we have validated the model by comparing relative brain–skull displacements with measurements from the latest cadaver head impact tests.26 A new study is currently underway to further improve model sophistication, and as these improvements occur, additional steps to validate the model will be taken (e.g., against Nahum et al.45 and Trosseille et al.60 pressure data). However, it is important to note that whether an FE model validated against these cadaver tests is sufficiently representative of brain biomechanics in live subjects remains an open question because experimental data derived from cadavers are not entirely suitable for head model validation64 because of significant tissue property deterioration and loss of perfusion and vasculature pressure; nonetheless, at present measurements from cadaver tests are essential for validation of any head FE model subject to high rate impact. Although experimental in vivo brain mechanical data are emerging with the use of high-resolution MR techniques, it warrants further investigation whether an FE model validated against these in vivo data under quasi-static28,29 and/or low-magnitude impact (2–3 g and ~100 to 200 rad/s2)16,52 settings is suitable for application under much higher magnitude impact conditions (e.g., ~100 g and ~10 k rad/s2 typical for injured subjects).
Nevertheless, in this proof-of-concept study using data from 10 subjects diagnosed with concussion, we have found that mean and maximum strain rate correlated with change in FA. The correlation trend for mean and maximum strain vs. change in FA as well as for maximum strain vs. change in MD was similar. Change in MD was correlated with injury-to-imaging interval but not for change in FA. These results provide promising evidence that our Dartmouth SSM, in conjunction with DTI, can provide important information regarding the mechanism of sports-related concussion.
References
Agel, J., R. Dick, B. Nelson, S. W. Marshall, and T. P. Dompier. Descriptive epidemiology of collegiate women’s ice hockey injuries: National Collegiate Athletic Association Injury Surveillance System, 2000–2001 through 2003–2004. J. Athl. Train. Dev. J. 42:249–254, 2007.
Alexander, D. C., G. J. Barker, and S. R. Arridge. Detection and modeling of non-Gaussian apparent diffusion coefficient profiles in human brain data. Magn. Reson. Med. 48:331–340, 2002.
Belanger, H., R. Vanderploeg, G. Curtiss, and D. Warden. Recent neuroimaging techniques in mild traumatic brain injury. J. Neuropsychiatr. Clin. Neurosci. 19:5–20, 2007.
Blumbergs, P. C., G. Scott, J. Manavis, H. Wainwright, D. A. Simpson, and A. J. McLean. Staining of amyloid precursor protein to study axonal damage in mild head injury. Lancet 344:1055–1056, 1994.
Bonett, D. G., and T. A. Wright. Sample size requirements for estimating Pearson, Kenall, and Spearman correlations. Psychometrika 64:23–28, 2000.
Chu J., J. G. Beckwith, J. J. Crisco, and R. M. Greenwald. A novel algorithm to measure linear and rotational acceleration using single-axis accelerometers. Presented at 5th World Congress of Biomechanics, Munich, Germany, 2006.
Chu, Z., E. A. Wilde, J. V. Hunter, S. R. McCauley, E. D. Bigler, et al. Voxel-based analysis of diffusion tensor imaging in mild traumatic brain injury in adolescents. AJNR 31:340–346, 2010.
Crisco, J. J., R. Fiore, J. G. Beckwith, J. J. Chu, P. G. Brolinson, et al. Frequency and location of head impact exposures in individual collegiate football players. J. Athl. Train. 45:549–559, 2010.
Cubon, V. A., M. Putukian, C. Boyer, and A. Dettwiler. A diffusion tensor imaging study on the white matter skeleton in individuals with sports related concussion. J. Neurotrauma 28:189–201, 2011.
DeKosky, S. T., M. D. Ikonomovic, and S. Gandy. Traumatic brain injury—football, warfare, and long-term effects. N. Engl. J. Med. 363:1293–1296, 2010.
Dick, R., M. S. Ferrara, J. Agel, et al. Descriptive epidemiology of collegiate men’s football injuries: National Collegiate Athletic Association Injury Surveillance System, 1988–1989 through 2003–2004. J. Athl. Train. 42:221–233, 2007.
Duhaime, A.-C. Large animal models of traumatic injury to the immature brain. Dev. Neurosci. 28:380–387, 2006.
Duma, S., S. Manoogian, W. Bussone, P. Brolinson, M. Goforth, et al. Analysis of real-time head accelerations in collegiate football players. Clin. J. Sports Med. 15:3–8, 2005.
Farkas, O., and J. T. Povlishock. Cellular and subcellular change evoked by diffuse traumatic brain injury: a COMPLEX web of change extending far beyond focal damage. Prog. Brain Res. 161:43–59, 2007.
Farrell, J. A. D., B. A. Landman, C. K. Jones, S. A. Smith, J. L. Prince, et al. Effects of SNR on the accuracy and reproducibility of DTI-derived fractional anisotropy, mean diffusivity, and principal eigenvector measurements at 1.5T. J. Magn. Reson. Imaging 26:756–767, 2007.
Feng, Y., T. M. Abney, R. J. Okamoto, R. B. Pless, G. M. Genin, and P. V. Bayly. Relative brain displacement and deformation during constrained mild frontal head impact. J. R. Soc. Interface 7:1677–1688, 2010.
Fischl, B., D. H. Salat, E. Busa, M. Albert, M. Dieterich, et al. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. 33:341–355, 2002.
Fischl, B., D. H. Salat, A. J. W. van der Kouwe, N. Markris, F. Ségonne, and A. M. Dale. Sequence-independent segmentation of magnetic resonance images. NeuroImage 23:S69–S84, 2004.
Franceschini, G., D. Bigoni, P. Regitnig, and G. A. Holzapfel. Brain tissue deforms similarly to filled elastomers and follows consolidation theory. J. Mech. Phys. Solids. 54:2592–2620, 2006.
Franceschini, G. The Mechanics of Human Brain Tissue. PhD-Thesis. University of Trento, 2006.
Funk, J. R., S. M. Duma, S. J. Manoogian, and S. Rowson. Biomechanical risk estimates for mild traumatic brain injury. Annu. Proc. Assoc. Adv. Automot. Med. 51:343–361, 2007.
Greenwald, R., J. Gwin, J. Chu, and J. Crisco. Head impact severity measures for evaluating mild traumatic brain injury risk exposure. Neurosurgery. 62:789–798, 2008.
Gwin, J., J. Chu, and R. Greenwald. Head impact telemetry system for measurement of head acceleration in ice hockey. J. Biomech. 39:S153, 2006.
Gwin, J., J. Chu, T. McAllister, and R. Greenwald. In situ measures of head impact acceleration in NCAA division I Men’s Ice Hockey: implications for ASTM F1045 and other ice hockey helmet standards. J. ASTM Int. 6:1–10, 2009.
Han, X., and B. Fischl. Atlas renormalization for improved brain MR image segmentation across scanner platforms. IEEE Trans. Med. Imaging. 26:479–486, 2007.
Hardy, W. N., M. J. Mason, C. D. Foster, C. S. Shah, J. M. Kopacz, et al. A study of the response of the human cadaver head to impact. Stapp Car Crash J. 51:17–80, 2007.
Ji, S., J. Ford, R. Greenwald, et al. Automated subject-specific, hexahedral mesh generation via image registration. Finite Elem. Anal. Des. 47:1178–1185, 2011.
Ji, S., and S. S. Margulies. Brainstem motion within the skull: measurement of the pons displacement in vivo. J. Biomech. 40:92–99, 2007.
Ji, S., L. Zhu, L. Dougherty, and S. S. Margulies. In vivo measurements of human brain displacement. Stapp Car Crash J. 48:527–539, 2004.
Kleiven, S. Influence of impact direction on the human head in prediction of subdural hematoma. J. Neurotrauma 20:365–379, 2003.
Kleiven, S. Predictors for traumatic brain injuries evaluated through accident reconstructions. Stapp Car Crash J. 51:81–114, 2007.
Kleiven, S., and W. H. Hardy. Correlation of an FE model of the human head with local brain motion—consequences for injury prediction. Stapp Car Crash J. 46:123–144, 2002.
Kumar, R., R. Gupta, et al. Comparative evaluation of CC DTI metrics in acute mild and moderate TBI: it’s correlation with np tests. Brain Inj. 23:675–685, 2009.
Langlois, J., W. Rutland-Brown, and K. Thomas. Traumatic Brain Injury in the United States: Emergency Department Visits, Hospitalizations, and Deaths. Atlanta, GA: Centers for Disease Control and Prevention, National Center for Injury Prevention and Control, 2004.
Manoogian, S., D. McNeely, S. Duma, G. Brolinson, and R. Greenwald. Head acceleration is less than 10 percent of helmet acceleration in football impacts. Biomed. Sci. Instrum. 42:383–388, 2006.
Mayer, A., J. Lin, M. Mannell, C. Gasparovic, J. Phillips, et al. A prospective diffusion tensor imaging study in mild traumatic brain injury. Neurology. 74:643–650, 2010.
McCrea, M., W. Barr, K. Guskiewicz, C. Randolph, S. Marchall, et al. Standard regression-based methods for measuring recovery after sport-related concussion. J. Int. Neuropsychol. Soc. 11:58–69, 2005.
McCrea, M., K. M. Guskiewicz, S. W. Marshall, W. B. Barr, C. Randolph, et al. Acute effects and recovery time following concussion in collegiate football players. JAMA. 290:2556–2563, 2003.
McCrory, P. Sports concussion and the risk of chronic neurological impairment. Clin. J. Sport Med. 21:6–12, 2011.
McCrory, P., W. Meeuwisse, K. Johnston, J. Dvorak, M. Aubry, et al. Consensus statement on concussion in sport—presented at the 3rd International Conference on Concussion in Sport in Zurich, November 2008. Clin. J. Sport Med. 19:185–200, 2009.
McKee, A. C., R. C. Cantu, C. J. Nowinski, E. T. Hedley-Whyte, B. E. Gavett, et al. Chronic traumatic encephalopathy in athletes: progressive tauopathy after repetitive head injury. J. Neuropathol. Exp. Neurol. 68:709–735, 2009. 10.1097/NEN.0b013e3181a9d503.
Meehan, W. P., P. d’Hemecourt, and R. Comstock. High school concussions in the 2008–2009 academic year: mechanism, symptoms, and management. Am. J. Sports Med. 38:2405–2409, 2010.
Miller, R., S. Margulies, M. Leoni, M. Nonaka, X. Chen, et al. Finite element modeling approaches for predicting injury in an experimental model of severe diffuse axonal injury. Proceedings of the 42nd Stapp Car Crash Conference, pp. 155–166, 1998.
Mukherjee, P., S. W. Chung, J. I. Berman, C. P. Hess, and R. G. Henry. Diffusion tensor MR imaging and fiber tractography: technical considerations. AJNR Am. J. Neuroradiol. 29:843–852, 2008.
Nahum, A. M., R. Smith, and C. Ward. Intracranial pressure dynamics during head impact. In: Society of Automotive Engineers. Proceedings of 21st Stapp Car Crash Conference, SAE Paper, Warrendale, PA, pp. 337–366, 1977.
Nicolle, S., M. Lounis, R. Willinger, and J. F. Palierne. Shear linear behavior of brain tissue over a large frequency range. Biorheology. 42:209–223, 2005.
Omalu, B. I., S. T. DeKosky, R. L. Minster, M. I. Kamboh, R. L. Hamilton, and C. H. Wecht. Chronic traumatic encephalopathy in a national football league player. Neurosurg. Clin. N. Am. 57:128–134, 2005.
Penumetcha, N., S. Kabadi, B. Jedynak, C. Walcutt, M. H. Gado, et al. Feasibility of geometric-intensity-based semi-automated delineation of the tentorium cerebelli from MRI scans. J. Neuroimaging 21:148–155, 2011.
Powell, J. W., and K. D. Barber-Foss. Traumatic brain injury in high school athletes. JAMA. 182:958–963, 1999.
Rowson, S., J. G. Beckwith, J. J. Chu, D. S. Leonard, R. M. Greenwald, and S. M. Duma. A six degree of freedom head acceleration measurement device for use in football. J. Appl. Biomech. 27:8–14, 2011.
Rowson, S., and S. M. Duma. Development of the star evaluation system for football helmets: integrating player head impact exposure and risk of concussion. Ann. Biomed. Eng. 39:2130–2140, 2011.
Sabet, A. A., E. Christoforou, B. Zatlin, G. M. Genin, and P. V. Bayly. Deformation of the human brain induced by mild angular head acceleration. J. Biomech. 41:307–315, 2008.
Schwarz, A. Suicide reveals signs of a disease seen in the N.F.L. The New York Times, 2010.
Smith, S. M. Fast robust automated brain extraction. Hum. Brain Mapp. 17:143–155, 2002.
Smith, C. Neuropathology. In: Textbook of Traumatic Brain Injury, edited by J. Silver, T. McAllister, and S. Yudofsky. Washington, DC: American Psychiatric Publishing, 2011.
Smith, S. M., M. Jenkinson, M. W. Woolrich, C. F. Beckmann, T. E. J. Behrens, et al. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage. 23(Suppl 1):S208–S219, 2004.
Takhounts, E. G., S. A. Ridella, V. Hasija, R. E. Tannous, J. Q. Campbell, et al. Investigation of traumatic brain injuries using the next generation of stimulated injury monitor (Simon) finite element head model. Stapp Car Crash J. 52:1–31, 2008.
Thibault, L., T. Gennarelli, S. Margulies, J. Marcus, and R. Eppinger. The strain dependent pathophysiological consequences of inertial loading on central nervous system tissue. Presented at IRCOBI Conference, Bron, Lyon, France, 1990.
Tournier, J. D., S. Mori, and A. Leemans. Diffusion tensor imaging and beyond. Magn. Reson. Med. 65:1532–1556, 2011.
Trosseille, X., C. Tarriere, F. Lavaste, F. Guillon, and A. Domont. Development of a F.E.M. of the human head according to a specific test protocol. Proceedings of the 36th Stapp Car Crash Conference, Seatttle, Washington, USA, SAE 922527, 1992.
Viano, D., I. Casson, E. Pellman, L. Zhang, A. King, and K. Yang. Concussion in professional football: brain responses by finite element analysis: part 9. Neurosurgery. 57:891–916, 2005.
Wiegell, M. R., H. B. Larsson, and V. J. Wedeen. Fiber crossing in human brain depicted with diffusion tensor MR imaging. Radiology. 217:897–903, 2000.
Wu, Z., H. Guo, N. Chow, J. Sallstrom, R. D. Bell, et al. Role of the MEOX2 homeobox gene in neurovascular dysfunction in alzheimer disease [see comment]. Nat. Med. 11:959–965, 2005.
Yang, K. H., J. Hu, N. A. White, A. I. King, C. C. Chou, and P. Prasad. Development of numerical models for injury biomechanics research: a review of 50 years of publications in the Stapp Car Crash Conference. Stapp Car Crash J. 50:429–490, 2006.
Zhang, H., P. Yushkevich, and J. Gee. DTI toolkit: a spatial normalization and atlas construction toolkit optimized for examining white matter morphometry using DTI data. Poster Presented at the 17th Scientific Meeting & Exhibition of the International Society for Magnetic Resonance in Medicine, 2009.
Zhang, L., K. H. Yang, and A. I. King. Comparison of brain responses between frontal and lateral impacts by finite element modeling. J. Neurotrauma. 18:21–30, 2001.
Zhang, L., K. H. Yang, and A. I. King. A proposed injury threshold for mild traumatic brain injury. J. Biomech. Eng. 126:226–236, 2004.
Acknowledgments
This study was supported through NIH RO1NS055020, R01HD048638, CDC R01/CE001254, the National Operating Committee on Standards for Athletic Equipment (NOCSAE 04-07 & SAC-1), and the William H. Neukom 1964 Institute for Computational Science at Dartmouth College.
Conflict of interest
Richard M. Greenwald, and Simbex have a financial interest in the instruments (HIT System, Sideline Response System (Riddell, Inc)) that were used to collect the biomechanical data reported in this study.
Author information
Authors and Affiliations
Corresponding author
Additional information
Associate Editor Stefan M. Duma oversaw the review of this article.
Rights and permissions
About this article
Cite this article
McAllister, T.W., Ford, J.C., Ji, S. et al. Maximum Principal Strain and Strain Rate Associated with Concussion Diagnosis Correlates with Changes in Corpus Callosum White Matter Indices. Ann Biomed Eng 40, 127–140 (2012). https://doi.org/10.1007/s10439-011-0402-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10439-011-0402-6