Abstract
The Spanish populations of Juniperus oxycedrus subsp. macrocarpa are restricted to coastal dune habitats of the Mediterranean and Atlantic coasts. Here, we investigate the genetic structure in this taxon and the possible existence of detailed phylogeographic structure associated with the Strait of Gibraltar using data from amplified fragment length polymorphism (AFLP) and plastid markers for 14 populations in three regions: Andalusia, the Valencian Community and the Balearic Islands. Principal coordinate analysis (PCOA) data revealed a lack of clear genetic structure in J. oxycedrus subsp. macrocarpa based on AFLP. The highest AFLP genetic diversity was found in Andalusia. Plastid markers revealed 14 haplotypes, only two of them were shared among the regions. Andalusian populations had the highest number of exclusive haplotypes. Our results support Andalusia as a reservoir of genetic diversity for J. oxycedrus subsp. macrocarpa, and, as for other gymnosperms, this area could be considered as a genetic refugium. The frequent presence of common and exclusive haplotypes in Andalusia might indicate the colonisation of the Spanish Mediterranean coast from the Atlantic populations. Colonisation of these new areas could be promoted by biological factors, such as the existence of long-lived individuals or potential for seed dispersal. Finally, the Strait of Gibraltar did not constitute a natural barrier to the expansion of this species along the coast, as identical haplotypes are found on the Atlantic and Mediterranean coasts.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The Mediterranean basin has been considered as a refugium (Quézel 1985), and the maintenance of plant biodiversity in this region during glacial periods is well established (Magri et al. 2006). Plant species and their genetic lineages experienced complex histories during successive glacial and interglacial periods, involving extinction, persistence, local differentiation or migration, and the Pleistocene glaciations appear to have been the most important paleoclimatic events in shaping the genetic structure of Mediterranean plants (Thompson 2005). As a direct consequence, the Mediterranean area has become a hotspot for phylogeographical and evolutionary studies (e.g. Petit et al. 2005; Terrab et al. 2008; Magri et al. 2007; Médail and Diadema 2009; Escudero et al. 2010). Recently, Fady and Conord (2010) analysed the relationships between genetic diversity of plant populations and paleoclimate data for the Last Glacial Maximum (LGM) around the Mediterranean basin, demonstrating that the LGM climate had a significance effect on current geographical and genetic diversity patterns, especially in woody taxa. Moreover, their studies revealed a pattern in which there was remarkably low within-population genetic diversity in western populations compared with eastern populations in the Mediterranean region, leading them to stress the need for a focus on new genetic research of western populations for conservation purposes.
The spatial genetic structure of plant populations around the western Mediterranean area has been analysed to assess the phylogeographical patterns mostly related to the identification of glacial refugia and therefore to determinate migration routes of plants (e.g. Piñeiro et al. 2007; Terrab et al. 2008). The Iberian Peninsula is potentially important phylogeographically, because it is the western extreme of the Mediterranean basin and because the Strait of Gibraltar links the Atlantic Ocean and the Mediterranean Sea and also constitutes the closest link between Europe and Africa. The role of biogeographical barriers such as the Strait of Gibraltar would be expected to influence phylogeographic patterns, though different views have been put forward. Kadereit et al. (2005) stated that the Strait of Gibraltar could act as an effective barrier for Eryngium maritimum L., Cakile maritima Scop. and Salsola kali L. along the coast of the Iberian Peninsula. In addition, the Strait of Gibraltar has been also reported to be a barrier between the Iberian Peninsula and northern Morocco (Escudero et al. 2008; Rodríguez-Sánchez et al. 2008; Terrab et al. 2008; Jaramillo-Correa et al. 2010). However, the ecological conditions of this Strait have not always been considered as an effective barrier as there is no obvious break in the genetic structure of populations of some other taxa along the European coast (Kadereit and Westberg 2007) or between the Iberian Peninsula and North Africa (Arroyo et al. 2008).
The Iberian Peninsula has also been regarded as a glacial refugium (Provan and Bennett 2008; Médail and Diadema 2009), and populations of some species there have high levels of genetic diversity (Comes and Kadereit 1998; Taberlet et al. 1998) and a high number of haplotypes (Hewitt 2001). No genetic data are currently available for any populations of maritime juniper (Juniperus oxycedrus L. subsp. macrocarpa (Sm.) Ball) relating to the identification of glacial refugia and subsequent recolonisation, but different migration routes have been postulated for other Juniperus species from southern Iberian territories to other Iberian or European areas as the climate warmed (cf. Terrab et al. 2008). Finally, Michalczyk et al. (2010) used genetic data to identify the geographical positions of genetic refugia and clarify recolonisation routes, in the absence of a fossil record. This allowed them to recognise several glacial microrefugia during the LGM in Central Europe for populations of common juniper (J. communis L).
Littoral sand dunes are characterised by a clear linear distribution of plant populations, and this might limit the spatial options for migration among populations on a large scale (Kadereit and Westberg 2007). Apart from the studies of Kadereit et al. (2005), a few studies of the genetic variability of sand dune plants are available (Stachys maritima L., López-Pujol et al. 2003; Pancratium maritimum L., Zahreddine et al. 2004; Grassi et al. 2005; Ammophila arenaria (Link.) L., Rodríguez-Echeverría et al. 2008). However, little information is available about the typical shrubby species from coastal sand dunes. In this study, we selected Juniperus oxycedrus subsp. macrocarpa, a woody coniferous shrub that is naturally distributed along the coastal dunes of the Mediterranean basin.
In Spain, the populations of J. oxycedrus subsp. macrocarpa grow on stabilised littoral sand dunes on the Atlantic and Mediterranean coasts, and no previous molecular studies have focused in detail on these populations. On the contrary, the widespread and close-related species J. communis has been much studied in Europe (Van der Merwe et al. 2000; Oostermeijer and de Knegt 2004; Provan et al. 2008; Khantemirova and Semerikov 2010; Michalczyk et al. 2010). Data about the spatial genetic structure would provide an opportunity to determine the phylogeography of this coastal species related to the identification of refugia and colonisation processes. Moreover, the existence of potentially important barriers, such as the Strait of Gibraltar, may influence migration routes among populations, as shown for other plants. The main aims of the present study were (1) to infer the spatial genetic structure of Spanish populations of J. oxycedrus subsp. macrocarpa and to test whether the current population fragmentation represented a barrier to gene flow and promoted genetic differentiation; (2) to reconstruct colonisation history from possible refugia, reflecting historical patterns of genetic differentiation in comparison to other Mediterranean gymnosperms or coastal plant species; and (3) to compare our genetic structure data with the dispersal patterns for other western Mediterranean species characterised by similar breeding systems, ecological habitat or fragmented population structure. Finally, some conservation aspects related to management are discussed in relation to the genetic information obtained.
Materials and methods
Plant sampling and DNA extraction
Spanish maritime juniper can be considered to have three main centres of distribution: Andalusia, the Valencian Community and the Balearic islands (Fig. 1). A total of 195 samples were collected from 14 populations, covering the range of this plant in Spain (Table 1). Total genomic DNA was extracted from silica gel-dried leaves (Chase and Hills 1991) using a modified 2× CTAB method (Doyle and Doyle 1987), and DNA was resuspended in 1,000 μL 0.1× TE buffer (10 mM Tris–HCl, 1 mM EDTA, pH 8.0). DNA was purified using QIAGEN and MOBIO purification kits, according to the manufacturers’ protocols.
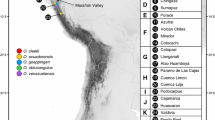
Map showing locations and geographical distribution of plastid haplotypes of Juniperus oxycedrus subsp. macrocarpa sampled in this study. For population codes, see Table 1
AFLP analysis
Among the collected individuals, an initial set of 100 individuals from the three regions was analysed to infer the genetic structure of J. oxycedrus subsp. macrocarpa. Restriction and ligation reactions were carried out in accordance with the AFLP™ Plant Mapping Protocol (Applied Biosystems). Juniperus oxycedrus subsp. macrocarpa is characterised by a large genome (Murray et al. 2004), and the preselective and selective primers were therefore modified as suggested by, e.g., Fay et al. (2005). The preselective primers had two extra 3′ bases (EcoR1-AC + MseI-CT), and three and four extra 3′ bases were added to EcoR1 and MseI selective primers. The rest of the reagents and amplification range of temperatures followed the original protocol. A primer trial was carried out with 12 different primers combinations, but only the primers EcoR1-ACT + MseI-CTAG (blue-labelled), EcoR1-ACG + MseI-CTAT (green-labelled) and EcoR1-ACC + MseI-CTAC (yellow-labelled) showed clearly distinguishable bands. Fluorescently labelled fragments from the selective amplifications were separated by electrophoresis on a 5.0% denaturing polyacrylamide gel using an ABI 3100 automated sequencer. Gel analysis was carried out using Genescan 3.1. (Applied Biosystems, Inc.). Subsequently, fragments of each primer combination were scored with Genotyper 2.0 (Applied Biosystems, Inc.). Only amplified fragments with sizes between 50 and 500 bases were scored because bands outside this size range cannot be accurately sized.
Plastid molecular markers
To assess the phylogeographical pattern of the Spanish populations, we added two new populations (PO, PC) from Andalusia (Table 1), since these populations represent the extremes of the Andalusian distribution area. Sixteen plastid regions were sequenced and aligned to screen for microsatellites or nucleotide polymorphisms, using a single accession from each population. The regions sequenced were trnT-trnL, trnL-trnF, trnS-trnG, rpl20-rps12, psbB-psbH, trnH-psbA, trnK–trnK, trnD-trnT, rps4, matK-trnK, rps16, petG-trnP, trnV, rpl16, rpl14 and accD-psaI (Taberlet et al. 1991; Johnson and Soltis 1994; Demesure et al. 1995; Jordan et al. 1996; Oxelman et al. 1997; Hamilton 1999; Saltonstall 2001; Wang et al. 2003). Samples were then run on an ABI 3100 automated sequencer, and complementary strands were assembled using Sequencer (Gene Codes Corp., USA). Sequences were then exported to PAUP v.4.0d64 (Swofford 2002), and the sequences were aligned by eye.
Most of the regions were amplified and sequenced successfully (except rpl16, rps4, rpl14, accD-psaI and rpl20-rps12), although several changes to the original protocol were made. Only three regions (petG-trnP, trnD-trnT and trnL-trnF) contained nucleotide polymorphisms (see Appendix S1): the petG-trnP intergenic spacer contained a polyA nucleotide microsatellite [(A)10–12], whereas the other two (trnL-trnF and trnD-trnT) contained one and two indels, respectively. These were used as molecular markers (GenBank accessions numbers HM594857-HM594865).
For petG-trnP, the whole region had to be sequenced according to Wang et al. (2003), because no amplification was obtained using several newly designed primers (data not shown). Sequences were aligned using PAUP v.4.0d64 (Swofford 2002), with adjustments by visual inspections. Length variable fragments of trnL-trnF and trnD-trnT were amplified using newly designed primers (Table 2). These primers were designed using the Primer3 program (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). The PCR conditions were an initial round of denaturing at 94°C for 5 min, followed by 30 cycles of denaturing at 94°C for 30 s, annealing at 48–50.0°C (depending on primer pairs) for 30 s and extension at 72°C for 30 s, followed by a final extension at 72°C for 10 min. PCR reactions were performed in a reaction volume of ca. 20 μL containing 17.5 μL ABGene 1.1 × Master mix, 0.5 μL labelled primer (1.5 pmol/μL), 0.5 μL unlabelled complementary primer (25 pmol/μL), 0.5 μL of 0.4% bovine serum albumin (BSA) and 1 μL template DNA. Sufficient differences in the size of the fragments lengths allowed all regions to be analysed in a single capillary. A 0.4-μL sample of each diluted PCR product was added to 0.2 μL Rox internal size standard (ABI) and 10 μL HiDi formaldehyde. Fragment lengths were analysed on a 3100 ABI automated sequencer. Analysis was carried out using Genescan 3.1. and Genotyper 2.0 (Applied Biosystems, Inc.).
Data analysis
AFLP data
Polymorphic bands were scored as present (1) or absent (0) and used to form a binary matrix. For each population, allele frequencies were estimated using the Bayesian approach proposed by Zhivotovsky (1999), based on non-uniform prior distribution of null-allele frequencies. Allele frequencies were then used to calculate gene diversity for each population (Hj), total gene diversity (Ht) and genetic differentiation between populations (Fst) (Lynch and Milligan, 1994). The percentage of polymorphic fragments (P) at the 5% level was calculated using AFLP-SURV (Vekemans 2002), and the number of private alleles was also estimated. As differences in sampling intensity between populations could bias our comparisons of genetic diversity, we computed the band richness (Br) standardised to the smallest sample size by means of a rarefaction method with the AFLP-DIV 1.0 software (Coart et al. 2005). An analysis of molecular variance (AMOVA) was also performed with the variance components for (1) within populations, without regional grouping, (2) among populations within regions and (3) among regions, using Arlequin v. 3.1 (Excoffier et al. 2005). In addition, SAMOVA (spatial analysis of molecular variance) was used to identify groups of populations that maximised the proportion of total genetic variance (Dupanloup et al. 2002). SAMOVA was undertaken assuming different groups (K, ranging from two to eleven) as simulated by the software SAMOVA v. 1.0 (Dupanloup et al. 2002). Population structuring was also determined using the STRUCTURE software v. 2.3.3. (Pritchard et al. 2000). This Bayesian method was used to assign individuals to genetic clusters (K) and to estimate admixture proportions (Q) for each individual. The proportion of membership for each cluster was calculated without the consideration of sampling localities. The analyses were performed under the admixture model assuming independent allele frequencies and using a burn-in period of 50,000 followed by 100,000 Markov Chain Monte Carlo, and the most likely value of K was determined according to Evanno et al. (2005). A principle coordinates analysis (PCOA) was performed in the R Package for Multivariate Analysis version 4.0 (Casgrain and Legrendre 1999) using Jaccard’s coefficient (Jaccard 1908). The table of eigenvalues produced by R was exported to Microsoft Excel to produce XY scatter plots.
To test for repeatability of AFLP results, we ran five samples from Andalusia region twice from DNA extraction through to selective PCR. In all cases, the banding patterns were similar and produced identical results for all selective primers, even though there was some variation in the quality of the PCR (as evidenced by differences in band intensity between replicates).
Plastid marker data
A matrix was prepared, using different numbers for alleles of different lengths of the size-variable fragments (see Appendix S1). In the case of petG-trnP region, the length of the microsatellite was used to identify alleles. Haplotypes numbered 1–15 were defined by the combination of the alleles for the four different loci. The haplotype definitions are given in Table 3.
The number of haplotypes (Nh) and number of private haplotypes (Hp) were computed for each population. Genetic diversity was calculated using Popgene 1.31 (Yeh et al. 1999) and haplotype richness following Petit et al. (1998) using CONTRIB program. Haplotype richness was corrected for differences in sample size using the rarefaction method. To provide robust estimates, the sample size of the smallest population sample (n = 7) was used for rarefaction. AMOVA was also conducted, as described previously for the AFLP data. The existence of a phylogeographic structure was tested following Pons and Petit (1996) by calculating two measures of genetic differentiation: Gst and Nst. Whereas Gst is a differentiation measure that is based on allele frequencies only, Nst takes into account the similarities between haplotypes (i.e. the number of mutations between haplotypes). These two parameters were compared using PERMUT software with 5,000 permutations. To identify hypothetical migration pathways of maritime juniper along the coast based on plastid markers, pairwise analyses along the cline based on correlations between genetic parameters (number of haplotypes and Nei’s gene diversity) and longitude were made using Kendall’s τ estimate according to Gómez et al. (2005). These correlations were calculated using the program PAST v. 1.97 (Hammer et al. 2001). Unrooted statistical parsimony networks were constructed using the program TCS version 1.06 (Clement et al. 2000). For that, the matrix was prepared using the nucleotide sequence of each size-variable fragment (see Appendix S1). Presence of loop structures was resolved following the systematic criteria of Pfenninger and Posada (2002).
Results
AFLP data
Results for AFLP were obtained for 85 samples since some samples showed weak electropherograms or did not amplify. The final matrix contained 508 characters generated from the combination of three selective primer pairs. No difference or pattern between female and male or monoecious and dioecious individuals was detected in any statistical analysis, and all individuals were therefore considered together for the statistical analysis. Genetic diversity estimators are given in Table 4. There was no obvious geographical pattern in relation to the distribution of genetic diversity. The Andalusian region was the most genetically diverse region, based on P, Hj, Br and Npal. The total gene diversity (Ht) and genetic distance (Fst) for the Spanish J. oxycedrus subsp. macrocarpa populations were 0.2097 and 0.1433, respectively.
The principal coordinate analysis (PCOA) did not reveal any clear geographical pattern among the three regions (Fig. 2) or within these regions (data not shown); most of the populations were intermixed though the samples from the Valencian Community and the Balearics showed a tendency to group. Plots of coordinates 2 versus 3, 1 versus 2 and 1 versus 4, and the graphs obtained showed similar results (not shown). As revealed by SAMOVA, the optimal number of groups of populations (K) was two, because F CT values decreased progressively as K was increased from two to 11. In this case, only the most northeastern Iberian population CS was differentiated from the others, but these results were not significantly statistic (P > 0.05). Bayesian clustering analysis using Structure showed no strong pattern of population differentiation. The admixture proportions (Q) for each individual plant are shown in Fig. 3. Heterogeneity or intermediacy among populations was reflected in the proportional assignment of their individuals. No geographical region clustered separately, indicating existence of gene flow or similar genetic structure among the current populations. The current geographical subdivision is not supported by the molecular analyses. The AMOVA indicated that the highest proportion of the molecular variation was within populations (70.84%, all P < 0.05), whereas only 6.56% corresponded to variation among regions (Table 5). This same analysis based on SAMOVA groups revealed a high percentage of molecular variation within populations (50.5%, F CT P > 0.05).
Bayesian admixture proportions (Q) of individual plants of J. oxycedrus subsp. macrocarpa for a K = 3 population model. Each individual is represented by a vertical line which is partitioned into K coloured segments, the length of each colour being proportional to the estimated membership coefficient
Plastid marker data
Plastid markers were scored for 171 samples. The four polymorphic markers revealed a total of 14 different haplotypes (Table 3, Appendix S2). The most common haplotype was H6 (Fig. 1). Four haplotypes (H5, H6, H11, H15) were shared among several populations, and two (H5, H6) were found in all three regions. Ten haplotypes were unique to a single region, mostly Andalusia (H1, H2, H3, H4, H8, H9, H14); only one (H12) and two haplotypes (H7, H10) were exclusive to the Valencian Community and the Balearics, respectively. Most of these exclusive haplotypes were only found in one or two individuals within each population. Populations generally possessed more than one haplotype (Table 4). The most diverse population in terms of haplotype richness was ROM, one of the more western Andalusia populations; seven haplotypes were detected among 11 individuals. However, haplotype H6 was the only one found in several other Andalusian populations (BB, PC and PP) (Pb = 0.000). Genetic divergence estimated with ordered haplotypes (Nst) and unordered haplotypes (Gst = 0.335) was 0.335 and 0.324, respectively. Values for Nst were significantly larger than Gst values (P < 0.05), indicating the existence of phylogeographical structure.
As with the AFLP data, SAMOVA showed that two groups (K = 2) yielded the highest F CT values (CS and the rest of populations), but it was not statistically significant (P > 0.05). Moreover, AMOVA always revealed the highest percentage of variation within populations based on original geographical fragmentation (47.42%) (P < 0.0001, Table 5) or on SAMOVA groups (50.11%). The pairwise analysis revealed a weak negative correlation between molecular parameters and longitude (number of haplotypes, Kendall’s Π = −0.0983 P > 0.05; Nei’s gene diversity, Kendall’s Π = −0.0915, P > 0.05). However, none of these analyses was statistically significant.
The plastid haplotype network indicated the most common haplotype H6 as the hypothetical centre of the network from which two areas could be delimited (Fig. 4). Firstly, H6 is directly connected to H5, which has been also obtained in the three studied regions. Most of the exclusive Andalusian haplotypes appear around the haplotype H6, without any clear geographical relationship within this region. Secondly, two short branches could be clearly distinguished from H5, which are characterised by Balearic (H7–H10) and Valencian (H15–H12) haplotypes, respectively.
Discussion
The lack of geographical structure based on AFLP data and the greater genetic variation within populations than among regions point to the existence of a single metapopulation without any clear geographical structure (Mediterranean vs. Atlantic or Iberian Peninsula vs. Balearic Islands). Our molecular data illustrate the general idea of high levels of gene flow and genetic diversity and low levels of differentiation in outcrossing wind-pollinated coniferous species (Hamrick et al. 1992), and the AMOVA and genetic diversity indices fall within the reported variability for wind-pollinated plants (Hamrick and Godt 1996). Lewandowski et al. (1996) found remarkably low levels of polymorphism (11%) for a single Greek island population of J. oxycedrus subsp. macrocarpa compared with the range obtained here (33.5–65.6%) for Spanish populations or values obtained for other Juniperus spp. (Huh and Huh 2000; Van der Merwe et al. 2000; Jiménez et al. 2003; Oostermeijer and de Knegt 2004; Meloni et al. 2006). We are not aware of any other molecular study of genetic variability in maritime juniper around the Mediterranean basin, but there are some based on comparative morphology. Klimko et al. (2004) reported low genetic differentiation of Italian populations based on the lack of significant differences for most morphological features (e.g. cone size). In contrast, some notable morphological variability was found in the SW Spanish populations by Juan et al. (2006). Our molecular data are more in agreement with these latter results. If there were any direct relationships between genetic variability and morphological differences, as Klimko et al. (2004) suggested, detailed genetic and morphological studies of Italian and other Spanish populations, respectively, are needed to draw final conclusions.
Spatial genetic structure
The high levels of genetic variability might indicate that J. oxycedrus subsp. macrocarpa in Spain has not lost much of its genetic variation despite the drastic population fragmentation in three well-separated areas. It seems that this species could currently reflect part of the original genetic population information, as reported for other Juniperus taxa (Huh and Huh 2000; Jiménez et al. 2003; Oostermijer and Knegt 2004; Meloni et al. 2006). Moreover, the reproductive biology should be effective in homogenising genetic structure and preventing clear molecular differentiation among populations or regions, as stated by Oostermeijer and de Knegt (2004). It is worth highlighting that the Balearic maritime juniper population is embedded within the continental populations, and no clear genetic isolation was detected with AFLP data. This spatial genetic pattern is not common, since many other Balearic populations of perennial plants are fairly genetic isolated from other island or continental populations (Rosselló et al. 2002; Juan et al. 2004; Rubio de Casas et al. 2006). The data obtained might indicate that this is an example in which the biological features (e.g. total life span, reproduction, seed dispersal) could have more influence on the current genetic variability than fragmentation and isolation of the populations. Such life-history traits and their relationships with patterns of genetic structure have attracted the interest of population geneticists (Hamrick and Godt 1996; Aguinagalde et al. 2005). These results could strengthen the suggestion that the original range of genetic variability of the maritime juniper has been preserved, despite the current natural fragmentation.
Phylogeography and migration routes
The existence of southern genetic refugia has been proposed commonly for forest trees in the different Mediterranean peninsulas (Hewitt 1999; Petit et al. 2003; Médail and Diadema 2009), as these populations are easily identified by higher haplotype richness (Hewitt 2001). Recently, Gómez et al. (2005) pointed out the presence of southern genetic refugia for other gymnosperms (Pinus spp.) characterised by their high genetic variability on the basis on rare and exclusive haplotypes. As shown in Fig. 4, the Andalusian region is characterised by high levels of plastid genetic diversity, combined with the presence of private and rare haplotypes. The genetic information presented here would support the existence of a natural diversity reservoir for J. oxycedrus subsp. macrocarpa in the SW of the Iberian Peninsula. In a similar way, Michalczyk et al. (2010) demonstrated the usefulness of genetic methods when fossil records are missing to identify German refugia for the close relative J. communis. Nevertheless, palynological fossil records would be also required to support and establish historical patterns of coastal plant distribution and to support possible migration routes.
Colonisation routes have been widely discussed for forest plants (Dumolin-Lapègue et al. 1997; Comes and Kadereit 1998; Hewitt 2001; Gómez et al. 2005), but few data are available concerning possible migration routes of coastal shrubby perennial plants. The current low level of genetic differentiation among the three Spanish regions and the pattern of plastid haplotype distribution support the existence of a possible route of migration from the southwestern (Andalusia) to the eastern populations (Valencian Community and Balearics). In addition, the SAMOVA for plastid data and the pairwise analyses between molecular parameters and longitude pointed out an easterly migration. Furthermore, the significant difference between Gst and Nst values indicates the existence of a phylogeographical structure among Spanish J. oxycedrus subsp. macrocarpa populations. The existence of migration routes from southern areas of Iberian Peninsula has also been postulated for other gymnosperms (Gómez et al. 2005; Benito Garzón et al. 2007). In this case, maritime juniper populations appear to have migrated from the Atlantic coast to the Mediterranean coast, as shown by Kadereit and Westberg (2007) for other coastal plant species. Therefore, the Strait of Gibraltar does not appear to represent a natural geographical barrier along the European coast for J. oxycedrus subsp. macrocarpa, as it is for other plants such as Eryngium maritimum and Halimione portulacoides (L.) Aellen (Kadereit et al. 2005). A detailed phylogeographical survey would be needed to corroborate the Strait of Gibraltar as a biogeographical barrier for Mediterranean conifers between North Africa and the Iberian Peninsula (cf. Jaramillo-Correa et al. 2010).
The lack of a clear spatial genetic structure based on AFLP and the presence of the most common plastid haplotypes (H6 and H5) or shared haplotypes (H11, H15) between Andalusia and the Balearic and Valencian regions together with the biological features of J. oxycedrus subsp. macrocarpa could explain the hypothesis of Austerlitz et al. (2000) about colonisation. Austerlitz et al. (2000) stated that life cycle characteristics of trees or long-lived plants are necessary for explaining the observed structure of genetic diversity. Biological factors such as the longevity of shrubby plants could be directly influencing the genetic pattern. In fact, long-lived woody plants usually maintain relatively higher within-population levels of variation than annuals and short-lived perennials (Hamrick et al. 1992; Hamrick and Godt 1996; Austerlitz et al. 2000). No direct data are currently available concerning the age of individuals of maritime juniper, but some data for its close relative J. communis could be used as a guideline. Most individuals sampled of the common juniper were over 85 years old (usual range 70–190 years), though some particular specimens could be over 200 years old (Ward 1982). Spanish populations of maritime juniper are mainly composed of old long-lived shrubs and juvenile plants are almost absent (Muñoz-Reinoso 2003). Some old individuals will persist even in the absence of regeneration, though a low percentage of seeds from older bushes are still potentially viable (Ward 1982). Our results support the hypothesis of a one-dimensional colonisation process postulated by Austerlitz et al. (2000). According to those authors, after a new founder population (e.g. any Balearic or Valencian Community populations), the growth during the first year is explained by the presence of new juvenile migrants resulting from seed dispersal by birds (Piotto et al. 2003) or by rabbits (Soriguer 1986) and not because of plant reproduction. An individual of Juniperus could be considered reproductive after at least nine years in male individuals and longer in females (Ward 1981), and the potential production of viable seeds can only occur a long time after establishment and development of seedlings. The growth rates for these gymnosperms are quite low, and hence, when long-lived plants reach their reproductive age, a non-negligible part of space is already occupied by individuals that arrived years before as seeds. Moreover, recruitment within these maritime juniper communities is quite rare, and many seedlings fail (Ortiz et al. 1998; McKinley and Van Auken 2005; Juan et al. 2003, 2006). Therefore, although active gene flow via seeds could happen among populations, no current replacement or new genetic combinations seem to exist. Similar results were also found for other wind-pollinated long-lived plants such as Fraxinus mandshurica Rupr. by Hu et al. (2008).
Conservation implications
Spanish maritime juniper populations have been in decline for decades, and in the last few years, local Governments (e.g. Junta de Andalucía, Generalitat Valenciana) have focused on maintaining and re-establishing natural populations. Nonetheless, no general conservation action is planned for the whole Spanish distribution area of J. oxycedrus subsp. macrocarpa. Describing units of conservation is one of the most important contributions of genetics to conservation (Frankham 2010). Therefore, our current results could have direct implications for the management and development of conservation strategies, since the maritime juniper is included on the national Red List as Vulnerable (Moreno 2008) and on local Red Lists as Endangered (Laguna 1998; Pastor and Juan 1999) or Vulnerable (Sáez and Rosselló 2001). Moreover, its threatened habitat has also been included in the European Union Habitat Directive as a priority plant community. One of the practical aspects of this work is that the results gained could be used to identify the genetic refugia for Mediterranean species that are crucial in delimitating zones for conservation of genetic resources. The identification of population units is necessary so that management and monitoring programmes can be efficiently targeted. According to our plastid results, it should be stressed that Andalusia would be considered as a natural genetic diversity reservoir for the Spanish populations and, as a consequence, these populations could be considered as the main management unit.
Consequently, this geographical area deserves special attention in conservation management programmes. Finally, the existence of several plastid haplotypes in populations such as ROM, PO and DO would also allow these populations to be designated as genetic conservation units, and thus, management programmes could be efficiently designed. In addition to a global conservation proposal, some local actions could be also suggested since each local Government has already undertaken its own conservation guidelines. For example, in the Valencian and Andalusian regions, the main conservation actions are mainly based on the reintroductions of new individuals from sometimes unspecified origin to reinforce certain current populations in combination with seed conservation in germplasm banks and the habitat protection under the conservation figures of National Park, Natural Park or Microrreserves. In the case of the Balearics, the mere protection of the natural environment is basically reported (Sáez and Rosselló 2001). The genetic data presented here should be also used to promote new conservation actions within each geographical region, e.g., to consider new protected populations or to keep seed separate according to their origin. Moreover, our data could support the proper management of those reintroduction actions using plant material from the same geographical origin or also from the same population as here suggested for CS, ALI, MLL, DO or ROM populations according to our genetic data.
References
Aguinagalde I, Hampe A, Mohanty A, Martín JP, Duminil J, Petit RJ (2005) Effects of life-history traits and species distribution on genetic structure at maternally inherited markers in European trees and shrubs. J Biogeogr 32:329–339
Arroyo J, Aparicio A, Albaladejo RG, Muñoz J, Braza R (2008) Genetic structure and population differentiation of the Mediterranean pioneer spiny broom Calicotome villosa across the Strait of Gibraltar. Biol J Linn Soc 93:39–51
Austerlitz F, Mariette S, Machon N, Gouyon PH, Godelle B (2000) Effects of colonization processes on genetic diversity: differences between annual plants and tree species. Genetics 154:1309–1321
Benito Garzón M, Sánchez de Dios R, Sainz Ollero H (2007) Predictive modelling of tree species distributions on the Iberian Peninsula during the Last Glacial Maximum and Mid-Holocene. Ecography 30:120–134
Casgrain P, Legrendre P (1999) The R PACKAGE for multivariate and spatial analysis./http://alize.ere.umontreal.ca/_casgrain/R/S
Chase MW, Hills HG (1991) Silica gel: an ideal material or field preservation of leaf samples for DNA studies. Taxon 40:215–220
Clement M, Posada D, Crandall K (2000) TCS: a computer program to estimate gene genealogies. Mol Ecol 9:1657–1660
Coart E, Van Glabeke S, Petit RJ, Van Bockstaele E, Roldán-Ruiz I (2005) Range wide versus local patterns of genetic diversity in hornbeam (Carpinus betulus L.). Conserv Genet 6:259–273
Comes PH, Kadereit JW (1998) The effect of Quaternary climatic changes on plant distribution and evolution. Trends Plant Sci 3:432–438
Demesure B, Sodzi N, Petit RJ (1995) A set of universal primers for amplification of polymorphic non-coding regions of mitochondrial and chloroplast DNA in plants. Mol Ecol 4:129–131
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure from small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Dumolin-Lapègue S, Demesure B, Fineschi S, Le Come V, Petit RJ (1997) Phylogeographic structure of white oaks throughout the European continent. Genetics 146:1475–1487
Dupanloup I, Schneider S, Excoffier L (2002) A simulated annealing approach to define the genetic structure of populations. Mol Ecol 11:2571–2581
Escudero M, Vargas P, Valcárcel V, Luceño M (2008) Strait of Gibraltar: an effective gene-flow barrier for wind-pollinated Carex helodes (Cyperaceae) as revealed by DNA sequences, AFLP, and cytogenetic variation. Am J Bot 95:745–755
Escudero M, Vargas P, Arens P, Ouborg NJ, Luceño M (2010) The east-west-north colonization history of the Mediterranean and Europe by the coastal plant Carex extensa (Cyperaceae). Mol Ecol 19:352–370
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Laval G, Schneider S (2005) Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol Bioinform Online 1:47–50
Fady B, Conord C (2010) Macroecological patterns of species and genetic diversity in vascular plants of the Mediterranean basin. Diversity Distrib 16:53–64
Fay MF, Cowan RS, Leitch IJ (2005) The effects of nuclear DNA content (C-value) on the quality and utility of AFLP fingerprints. Ann Bot 95:237–246
Frankham R (2010) Where are we in conservation genetics and where do we need to go? Conserv Genet 11:661–663
Gómez A, Vendramin GG, González-Martínez SC, Alía R (2005) Genetic diversity and differentiation of two Mediterranean pines (Pinus halepensis Mill. and Pinus pinaster Ait.) along a latitudinal cline using chloroplast microsatellite markers. Divers Distr 11:257–263
Grassi F, Cazzaniga E, Minuto L, Peccenini S, Barberis G, Basso B (2005) Evaluation of biodiversity and conservation strategies in Pancratium maritimum L. for the Northern Tyrrhenian Sea. Biodiv Conserv 14:2159–2169
Hamilton MB (1999) Four primer pairs for the amplification of chloroplast intergenic regions with intraspecific variation. Mol Ecol 8:521–523
Hammer Ø, Harper DAT, Ryan PD (2001) PAST: paleontological statistics software package for education and data analysis. Palaeontol Electr 4:9
Hamrick JL, Godt MJW (1996) Effects of life history traits on genetic diversity in plant species. Phil Trans Roy Soc B 351:1291–1298
Hamrick JL, Godt MJW, Sherman-Broyle SL (1992) Factors influencing levels of genetic diversity in woody plant species. New Forest 6:95–124
Hewitt GM (1999) Post-glacial re-colonization of European biota. Biol J Linn Soc 68:87–112
Hewitt GM (2001) Speciation, hybrid zones and phylogeography—or seeing genes in space and time. Mol Ecol 10:537–549
Hu LJ, Uchiyama K, Shen HL, Saito Y, Tsuda Y, Ide Y (2008) Nuclear DNA microsatellites reveal genetic variation but a lack of phylogeographical structure in an endangered species, Fraxinus mandshurica, across North-east China. Ann Bot 102:195–205
Huh MK, Huh HW (2000) Genetic diversity and population structure of Juniperus rigida (Cupressaceae) and Juniperus coreana. Evol Ecol 14:87–98
Jaccard P (1908) Nouvelles recherches sur la distribution florale. B Soc Vaud Sci Nat 44:223–270
Jaramillo-Correa JP, Grivet D, Terrab A, Kurt Y, de-Lucas AI, Wahid N, Vendramin GG, González-Martínez SC (2010) The Strait of Gibraltar as a major biogeographic barrier in Mediterranean conifers: a comparative phylogeographic survey. Mol Ecol 19:5452–5468
Jiménez JF, Werner O, Sánchez-Gómez P, Fernández S, Guerra J (2003) Genetic variations and migration pathway of Juniperus thurifera L. (Cupressaceae) in the Western Mediterranean region. Israel J Plant Sci 51:11–22
Johnson LA, Soltis DE (1994) matK DNA sequence and phylogenetic reconstruction in Saxifragaceae s.s. Syst Bot 19:143–156
Jordan WC, Courtney MW, Neigel JE (1996) Low levels of intraspecific genetic variation at a rapidly evolving chloroplast DNA locus in North American duckweeds (Lemnaceae). Am J Bot 83:430–439
Juan R, Pastor J, Fernández I, Diosdado JC (2003) Relationships between mature cones traits and seed viability in Juniperus oxycedrus subsp. macrocarpa (Sm.) Ball (Cupressaceae). Acta Biol Cracov 45:69–78
Juan A, Crespo MB, Cowan RS, Lexer C, Fay MF (2004) Patterns of variability and gene flow in Medicago citrina an endangered endemic of islands in the Western Mediterranean, as revealed by amplified fragment length polymorphism (AFLP). Mol Ecol 13:2679–2690
Juan R, Pastor J, Fernández I, Diosdado JC (2006) Seedling emergence in the endangered Juniperus oxycedrus subsp. macrocarpa (Sm.) Ball in Southwest Spain. Acta Biol Cracov 48:49–58
Kadereit JW, Westberg E (2007) Determinants of phylogeographic structure: a comparative study of seven coastal flowering plant species across their European range. Watsonia 26:229–238
Kadereit JW, Arafeh R, Somogyi G, Westberg E (2005) Terrestrial growth and marine dispersal? Comparative phylogeography of five coastal plant species at a European scale. Taxon 54:861–876
Khantemirova EV, Semerikov VL (2010) Genetic variation of some varieties of common juniper Juniperus communis L. inferred from analysis of allozyme loci. Russian J Genet 46:546–554
Klimko M, Boratyńska K, Boratyński A, Marcysiak K (2004) Morphological variation of Juniperus oxycedrus subsp. macrocarpa (Cupressaceae) in three Italian localities. Acta Soc Bot Pol 73:113–119
Laguna E (coord) (1998) Flora endémica, rara o amenazada de la Comunidad Valenciana. Conselleria de Medio Ambiente, Valencia
Lewandowski A, Boratyński A, Mejnartowicz L (1996) Low level of isozyme variation in an island population of Juniperus oxycedrus subsp. macrocarpa (Sm. ex Sibth.) Ball. Acta Soc Bot Pol 65:335–338
López-Pujol J, Orellana MR, Bosch M, Simon J, Blanché C (2003) Effects of habitat fragmentation on allozyme diversity and conservation status of the coastal sand dune plant Stachys maritima (Lamiaceae) in the Iberian Peninsula. Plant Biol 5:504–512
Lynch M, Milligan BG (1994) Analysis of population genetic structure with RAPD markers. Mol. Ecol 3:91–99
Magri D, Vendramin GG, Comps B, Dupanloup I, Geburek T, Gömöry D, Latałowa M, Litt T, Paule L, Roure JM, Tantau I, Van Der Knaap WO, Petit RJ, De Beaulieu J-L (2006) A new scenario for the Quaternary history of European beech populations: palaeobotanical evidence and genetic consequences. New Phytol 171:199–221
Magri D, Fineschi S, Bellarosa R, Buonamici A, Sebastiani F, Schirone B, Simeone MC, Vendramin GG (2007) The distribution of Quercus suber chloroplast haplotypes matches the palaeogeographical history of the western Mediterranean. Mol Ecol 16:5259–5266
McKinley DC, Van Auken OW (2005) Influence of interacting factors on the growth and mortality of Juniperus seedlings. Am Mid Nat 154:320–330
Médail F, Diadema K (2009) Glacial refugia influence plant diversity patterns in the Mediterranean Basin. J Biogeogr 36:1333–1345
Meloni M, Perini D, Filigheddu R, Binelli G (2006) Genetic variation in five Mediterranean populations of Juniperus phoenicea as revealed by inter-simple sequence repeat (ISSR) markers. Ann Bot 97:299–304
Michalczyk IM, Opgenoorth L, Luecke Y, Huck S, Ziegenhagen B (2010) Genetic support for periglacial survival of Juniperus communis L. in Central Europe. The Holocene 20:887–894
Moreno JC (2008). (coord.) Lista roja 2008 de la flora vascular española. Dirección General de Medio Natural y Política Forestal (Ministerio de Medio Ambiente, y Medio Rural y Marino, y Sociedad Española de Biología de la Conservación de Plantas), Madrid
Muñoz-Reinoso JC (2003) Juniperus oxycedrus ssp. macrocarpa in SW Spain: ecology and conservation problems. J Coastal Conserv 9:113–122
Murray BG, Leitch IJ, Bennett MD (2004) Gymnosperm DNA C-Values Database (Release 3.0, Dec. 2004). http://www.rbgkew.org.uk/cval/homepage.html
Oostermeijer JGB, de Knegt B (2004) Genetic population structure of the wind-pollinated, dioecious shrub Juniperus communis in fragmented Dutch heathlands. Plant Species Biol 19:175–184
Ortiz PL, Arista M, Talavera S (1998) Low reproductive success in two subspecies of Juniperus oxycedrus L. Int J Plant Sci 159:843–847
Oxelman B, Lidén M, Berglund D (1997) Chloroplast rps16 intron phylogeny of the tribe Sileneae (Caryophyllaceae). Plant Syst Evol 206:393–410
Pastor J, Juan R (1999) Juniperus oxycedrus subsp. macrocarpa. In: Blanca G, Cabezudo B, Hernández-Bermejo JE, Herrera CM, Molero Mesa J, Muñoz J, Valdés B (eds) Libro rojo de la flora silvestre amenazada de Andalucía. Tomo I. Especies en peligro de extinción. Consejera de Medio Ambiente, Sevilla, pp 156–159
Petit RJ, El Mousadik A, Pons O (1998) Indentifying populations for conservation on the basis of genetic markers. Conserv Biol 12:844–855
Petit RJ, Aguinagalde I, de Beaulieu J-L, Bittkau C, Brewer S, Cheddadi R, Ennos R, Fineschi S, Grivet D, Lascoux M, Mohanty A, Müller-Starck G, Demesure-Mush B, Palmé A, Martín JP, Rendell S, Vendramin GG (2003) Glacial refugia: hotspots but not melting pots of genetic diversity. Science 300:1563–1565
Petit RJ, Hampe A, Cheddadi R (2005) Climate changes and tree phylogeography in the Mediterranean. Taxon 54:877–885
Pfenninger M, Posada D (2002) Phylogeographic history of the land snail Candidula unifasciata (Helicellinae, Stylommatophora): fragmentation, corridor migration, and secondary contact. Evolution 56:1776–1788
Piñeiro R, Fuertes Aguilar J, Draper Munt D, Nieto Feliner G (2007) Ecology matters: Atlantic–Mediterranean disjunction in the sand-dune shrub Armeria pungens (Plumbaginaceae). Mol Ecol 16:2155–2171
Piotto B, Bartolini G, Bussotti F, Calderón García AA, Chessa I, Ciccarese C, Ciccarese L, Crosti R, Cullum FJ, Di Noi A, García-Fayos P, Lambardi M, Lisci M, Lucci S, Melini S, Muñoz Reinoso JC, Murranca S, Nieddu G, Pacini E, Pagni G, Patumi M, Pérez García F, Piccini C, Rossetto M, Tranne G, Tylkowski T (2003) Fact sheets on the propagation of Mediterranean trees and shrubs from seed. In: Piotto B, Di Noi A (eds) Seed propagation of Mediterranean trees and shrubs. APAT, Italy, pp 11–52
Pons O, Petit RJ (1996) Measuring and testing genetic differentiation with ordered versus unordered alleles. Genetics 144:1237–1245
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Provan J, Bennett KD (2008) Phylogeographic insights into cryptic glacial refugia. Trends Ecol Evol 23:564–571
Provan J, Beatty GE, Hunter AM, McDonald RA, McLaughlin E, Preston SJ, Wilson S (2008) Restricted gene flow in fragmented populations of a wind-pollinated tree. Conserv Genet 9:1521–1532
Quézel P (1985) Definition of the Mediterranean region and the origin of its flora. In: Gómez-Campo C (ed) Plant conservation in the Mediterranean area, Geobotany 7. W. Junk, Dordrecht, pp 9–24
Rodríguez-Echeverría S, Freitas H, Van der Putten WH (2008) Genetic diversity and differentiation of Ammophila arenaria (L.) Link as revealed by ISSR markers. J Coastal Res 24:122–126
Rodríguez-Sánchez F, Pérez-Barrales R, Ojeda F, Vargas P, Arroyo J (2008) The Strait of Gibraltar as a melting pot for plant biodiversity. Quaternary Sci Rev 27:2100–2117
Rosselló JA, Cebrián MC, Mayol M (2002) Testing taxonomic and biogeographical relationships in a narrow Mediterranean endemic complex (Hippocrepis balearica) using RAPD markers. Ann Bot 89:321–327
Rubio de Casas R, Besnard G, Schönswetter P, Balaguer L, Vargas P (2006) Extensive gene flow blurs phylogeographic but not phylogenetic signal in Olea europaea L. Theor Appl Genet 113:575–583
Sáez Ll, Rosselló JA (2001) Llibre vermell de la flora vascular de les Illes Balears. Conselleria de Medi Ambient. Govern de les Illes Balears, Palma de Mallorca
Saltonstall K (2001) A set of primers for amplification of noncoding regions of chloroplast DNA in the grasses. Mol Ecol Notes 1:76–78
Soriguer RC (1986) The rabbit as a plant seed disperser. Mammal Rev 16:197–198
Swofford DL (2002) PAUP: phylogenetic analysis using parsimony, Ver. 4.0b10. Sinauer Ass, Sunderland
Taberlet P, Gielly L, Pautou G, Bouvet J (1991) Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol Biol 17:1105–1109
Taberlet P, Fumagalli L, Wust-Saucy A-G, Cosson J-F (1998) Comparative phylogeography and postglacial colonization routes in Europe. Mol Ecol 7:453–464
Terrab A, Schönswetter P, Talavera S, Vela E, Stuessy TF (2008) Range-wide phylogeography of Juniperus thurifera L., a presumptive keystone species of western Mediterranean vegetation during cold stages of the Pleistocene. Mol Phyl Evol 48:94–102
Thompson JD (2005) Plant evolution in the Mediterranean. Oxford University Press, USA
Van der Merwe M, Winfield MO, Arnold GM, Parker JS (2000) Spatial and temporal aspects of the genetic structure of Juniperus communis populations. Mol Ecol 9:379–386
Vekemans X (2002) AFLP-SURV, version 1.0. Laboratoire de Génétique et Ecologie Végétale, Université Libre de Bruxelles, Brussels, Belgium
Wang WP, Hwang CY, Lin TP, Hwang SY (2003) Historical biogeography and phylogenetic relationships of the genus Chamaecyparis (Cupressaceae) inferred from chloroplast DNA polymorphism. Plant Syst Evol 241:13–28
Ward LK (1981) The demography, fauna and conservation of Juniperus communis in Britain. In: Synge H (ed) The Biological aspects of rare plant conservation. Wiley, Chichester, pp 312–329
Ward LK (1982) The conservation of juniper: longevity and old age. J Appl Ecol 19:917–928
Yeh FC, Yang R, Boyle T (1999) POPGENE, version 1.31. Microsoft Window-Based Freeware for Population Genetic Analysis. University of Alberta, Edmonton. Available at: http://www.ualberta.ca/~fyeh/index.htm
Zahreddine H, Clubbe C, Baalbaki R, Ghalayini A, Talhouk SN (2004) Status of native species in threatened Mediterranean habitats: the case of Pancratium maritimum L. (sea daffodil) in Lebanon. Biol Conserv 120:11–18
Zhivotovsky LA (1999) Estimating population structure in diploids with multilocus dominant DNA markers. Mol Ecol 8:907–913
Acknowledgments
We thank Magdalena Vicens (Jardín Botánico de Sóller, Balearic Islands), Antonio Sánchez (Valencia) and the staff from the Plant Microreserves Section (Conselleria de Vivienda y Territorio, Generalitat Valenciana) for collecting some of the samples. We also thank M.A. Alonso, A. Guilló, J.C. Agulló and J.L. Villar (University of Alicante) for their valuable help in the lab and in collecting plant samples. We are very grateful to R. Cowan and R. Smith (Royal Botanic Gardens, Kew) for their kind laboratory assistance. This study was partly carried out in the frame of the research project ‘Estudio de la variabilidad genética de Juniperus oxycedrus subsp. macrocarpa en el litoral de Andalucía’ from the Junta de Andalucía (OG-073/06). A. Juan was supported by a grant from Conselleria de Ciencia y Tecnología (Generalitat Valenciana) to work at Jodrell Laboratory (Royal Botanic Gardens, Kew).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by R. Matyssek.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Juan, A., Fay, M.F., Pastor, J. et al. Genetic structure and phylogeography in Juniperus oxycedrus subsp. macrocarpa around the Mediterranean and Atlantic coasts of the Iberian Peninsula, based on AFLP and plastid markers. Eur J Forest Res 131, 845–856 (2012). https://doi.org/10.1007/s10342-011-0558-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10342-011-0558-5